Abstract
The performance of a visual slide-based DNA microarray for the identification of non-albicans Candida spp. was evaluated. Among 167 isolates that had previously been identified by Vitek 2, the agreement between DNA microarray and sequencing results was 97.6%. This DNA microarray platform showed excellent performance.
TEXT
Candida species are the predominant cause of systemic fungal infections in hospitalized patients and represent the fourth most common microorganisms found in blood cultures (1, 2). In a large, laboratory-based study conducted in Brazilian hospitals, the incidence of candidemia was found to be 2.49 cases per 1,000 patient admissions (3), which is higher than that in the United States (0.28/1,000), Europe (0.2/1,000), or France (0.17/1,000) (4–7). In our setting, the reported candidemia incidence was 0.54 cases per 1,000 patient days, with 56% non-albicans Candida spp. (8). An increase in the proportion of non-albicans Candida spp. has been observed worldwide with distinctive patterns of antifungal susceptibility (9, 10).
The development of automated microbiology systems has represented a significant improvement in the identification of Candida spp. However, their diagnostic accuracy has been reported to range from 43 to 95% (11–20). Considering the frequency of occurrence, the high mortality rate, and the susceptibility patterns of non-albicans Candida spp., novel point-of-care molecular technologies are urgently needed in clinical practice. Our study evaluated the ability of a novel plastic slide-based DNA microarray to identify non-albicans Candida spp. isolated from blood cultures.
(Partial results of this study were presented at the 18th Congress of the International Society for Human and Animal Mycology, Berlin, Germany, 2012.)
A retrospective cohort surveillance study was performed that included 167 non-albicans Candida bloodstream infection isolates obtained by the Mycology Section of the Hospital and Clinics of the State University of Campinas from 2006 to 2010. Bact/ALERT (bioMérieux, Marcy l'Etoile, France) and the Vitek 2 system (YST REF 21343 154 card; Lab Equipment bioMérieux, Inc., Durham, NC) were used for primary detection and identification, respectively. Four American Type Culture Collection (ATCC) type strains (Candida parapsilosis ATCC 22019, C. glabrata ATCC MYA 2950, C. krusei ATCC 6258, and C. albicans ATCC 76615) and C. dubliniensis CBS 7987 were included as control strains.
The novel DNA microarray platform was developed by the Medical Mycology Research Center at Chiba University (MMRC-Chiba) to identify the following 12 genera and 32 fungal species, including 9 non-albicans Candida spp.: C. guilliermondii, C. lusitaniae, C. krusei, C. glabrata, C. parapsilosis, C. tropicalis, C. dubliniensis, C. famata, C. kefyr, C. albicans, Histoplasma capsulatum, Coccidioides posadasii, Paracoccidioides brasiliensis, Blastomyces dermatitidis, Cryptococcus spp., Trichosporon cutaneum, T. asteroides, T. inkin, T. asahii, T. faecale, T. mucoides, Malassezia furfur, Aspergillus nidulans, A. terreus, A. niger, A. flavus, A. fumigatus, Trichophyton rubrum, T. mentagrophytes, T. tonsurans, and Penicillium marneffei.
For DNA microarray platform fabrication, the oligonucleotide probes used, consisting of 14 to 20 species-specific nucleotide sequences with biotin-labeled poly(T) anchors at the end of each nucleotide (Invitrogen, Showajima, Japan), were designed based on the internal transcribed spacer (ITS) sequences (ITS1 and ITS2) of the type strains (GenBank database, ATCC, Centraalbureau voor Schimmelcultures [CBS], and MMRC-Chiba). Multiple-sequence alignments were performed with the BioEdit software (version 7.1.3; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Conserved regions were also used as targets for genus-specific probes or as controls. Four to 11 oligonucleotide probe sequences were included for each Candida sp.; six probes were designed for common sequences, and two were designed for biotin labeling. The probe sequences were spotted onto a plastic slide (NGK Insulators Ltd., Aichi, Japan) with a KCS minimicroarray printer (Kubota Comps Corporation, Amagasaki, Japan).
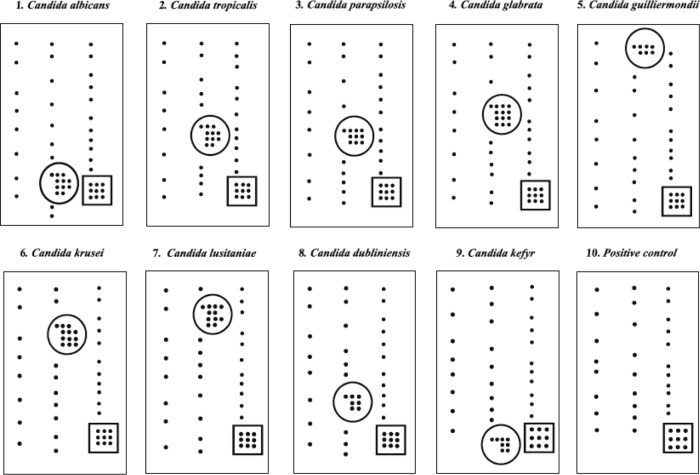
For fungal identification, PCR assays with universal fungal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Sigma-Aldrich, St. Louis, MO) were used to amplify the ITS1 and ITS2 regions and the 5.8S rRNA gene (21), followed by hybridization, conjugation, staining, and direct visualization of specifically positioned spots on the slide (Fig. 1). For DNA sequencing, the PCRs were performed with the universal fungus-specific pairs ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3) (Sigma-Aldrich, St. Louis, MO), followed by purification with ExoSAP-IT (Affymetrix USB, Cleveland, OH), and the products were then sequenced with the BigDye Terminator reagent kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocols on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The sequence data were assembled with ATSQ version 6.0.1 software (Genetix Corporation, Tokyo, Japan). The sequences obtained were then submitted to GenBank.
Fig 1.
Representative hybridization patterns of C. albicans, non-albicans Candida spp., and a positive control. Each number represents a species-specific slide. A group of specific hybridization spots is shown inside the circle, the remaining spots are representative of biotin hybridization (negative for others species), and the positive control is shown inside the square. The positive control represents a sequence common to all fungal species.
Of the 167 non-albicans Candida isolates tested, the new MMRC-Chiba DNA microarray identified 163 (97.6%), Vitek 2 identified 161 (96.4%), and DNA sequencing identified all of them (Table 1). The agreement between the DNA microarray and sequencing results was 97.6%. The DNA microarray did not identify the following yeast strains: five C. orthopsilosis, one C. metapsilosis, one C. lipolytica, one C. rugosa, one C. haemulonii, and one Dipodascus capitatus. Of note, the MMRC-Chiba DNA microarray identified one C. glabrata, one C. guilliermondii, one C. albicans, and one C. kefyr isolate that were previously identified as Candida spp. or non-albicans Candida spp. by Vitek 2. All 39 of the Candida strains of the C. parapsilosis complex were identified as C. parapsilosis. The probe sequences for C. haemulonii, C. rugosa, C. lipolytica, and D. capitatus were absent from the platform's composition.
Table 1.
Molecular identification by DNA microarray compared to that by DNA sequencing of non-albicans Candida yeasts previously identified by Vitek 2
| Yeast | No. (%) identified by: |
% Agreement | ||
|---|---|---|---|---|
| Vitek 2 | DNA microarraya | DNA sequencing | ||
| C. tropicalis | 66 (39.5) | 66 (39.5) | 66 (39.5) | 100 |
| C. parapsilosis complex | 39 (23.3) | 39 (23.3) | 39 (23.3) | 100 |
| C. parapsilosis | 39 (23.3) | 33 (19.7) | 85 | |
| C. orthopsilosis | 5 (3.0) | |||
| C. metapsilosis | 1 (0.6) | |||
| C. glabrata | 34 (20.3) | 35 (20.9) | 35 (20.9) | 100 |
| C. krusei | 11 (6.6) | 11 (6.5) | 11 (6.5) | 100 |
| C. guilliermondii | 5 (2.9) | 6 (3.5) | 6 (3.5) | 100 |
| C. kefyr | 3 (1.8) | 4 (2.4) | 4 (2.4) | 100 |
| C. dubliniensis | 1 (0.6) | 1 (0.6) | 1 (0.6) | 100 |
| C. lipolytica | 1 (0.6) | 1 (0.6) | ||
| C. rugosa | 1 (0.6) | 1 (0.6) | ||
| C. haemulonii | 1 (0.6) | |||
| D. capitatus | 1 (0.6) | |||
| Not identified | 6 (3.5) | 4 (2.4) | ||
C. rugosa, C. lipolytica, and C. haemulonii were absent from the tested DNA microarray slide.
Previous studies have demonstrated that conventional and commercial systems frequently fail to identify the less common species and to discriminate between closely related species (22, 23). Borman et al. (23) showed 18% misidentification in a large surveillance study. Conversely, Leaw et al. (24) showed high sensitivity and specificity of a DNA microarray platform of 100 and 97%, respectively, in a study including 452 isolates. More recently, studies with blood culture isolates (18, 25, 26) identified optimal assay parameters (Table 2). Fungal identification by the DNA microarray approach can reduce the time required for a diagnosis (25) and allows the identification of mixed infections and rare species with direct visual interpretation (24). Another aspect is fungal identification to the species level, as well as susceptibility pattern determination, directly from clinical blood samples (20). Therefore, a multipathogen (fungi, viruses, and bacteria) DNA microarray platform has been proposed for syndromic investigation in clinical settings (18, 27).
Table 2.
A summary of studies of fungal identification using DNA microarrays
| Reference | DNA microarray composition (no. of fungal species) | No. of isolates included | Specimen(s) included | No. of non-albicans Candida species | % Sensitivity | % Specificity |
|---|---|---|---|---|---|---|
| 18 | 7 | 112 | Blood | 4 | 93 | NRc |
| 19 | 20 | 122 | Blood, CSF,a skin, secretions | 4 | 100 | NR |
| 20 | 12 | 21 | Reference strains, clinical isolatesb | 7 | NR | NR |
| 24 | 77 | 452 | Reference strains, clinical isolatesb | 44 | 100 | 97 |
| 25 | 17 | 88 | Blood | 13 | 100 | 99.8 |
| 26 | 76 | 116 | Blood | 40 | 100 | 100 |
| 31 | 14 | 91 | Blood, tissue, BALd fluid | 4 | NR | NR |
| 32 | 24 | 20 | Blood, urine, other | 12 | NR | NR |
| 33 | 26 | 40 | Skin | 10 | 92 | NR |
CSF, cerebrospinal fluid.
Isolation site unspecified.
NR, not reported.
BAL, bronchoalveolar lavage.
A DNA microarray for fungal identification should be validated in terms of cost effectiveness at the point of care compared to others methodologies like Vitek 2 and the novel matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) method. Vitek 2 is accurate, useful, and user friendly in routine laboratories (28). Although MALDI-TOF MS requires expensive equipment, it presents low operational costs (29). Studies addressing these issues should be performed in the near future (30). The MMRC-Chiba DNA microarray platform demonstrated excellent accuracy in the identification of non-albicans Candida spp. The good agreement observed allows the pursuit of the next step, which consists of fungal identification directly in clinical samples. This approach can reduce the turnaround time for cultures and should be evaluated in terms of costs and benefits in the management of patients with a high risk of candidemia.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the NCBI Probe database and assigned accession numbers Pr031796813 to Pr031796896.
ACKNOWLEDGMENTS
Project 02P-29548-05 was supported by JST (Japan Science and Technology Agency)/JICA (Japan International Cooperation Agency); SATREPS (Science and Technology Research Partnership for Sustainable Development); the Faculty of Medical Sciences, State University of Campinas-UNICAMP; and the São Paulo Research Foundation (FAPESP). Michela De Luca Ferrari received a Ph.D. scholarship from the Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation—FAPESP 2010/50958-8).
Footnotes
Published ahead of print 19 June 2013
REFERENCES
- 1.Perlroth J, Choi B, Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infection in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 3.Colombo AL, Nucci M, Park BJ, Nouér SA, Arthington-Skaggs B, da Matta DA, Warnock D, Morgan J. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittet D, Wenzel RP. 1995. Nosocomial bloodstream infections. Secular trends in rates, mortality, and contribution to total hospital deaths. Arch. Intern. Med. 155:1177–1184 [DOI] [PubMed] [Google Scholar]
- 5.Jarvis WR. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526–1530 [DOI] [PubMed] [Google Scholar]
- 6.Tortorano AM, Peman J, Bernhardt H, Klingspor L, Kibbler CC, Faure O, Biraghi E, Canton E, Zimmermann K, Seaton S, Grillot R, ECMM Working Group on Candidaemia 2004. Epidemiology of candidemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23:317–322 [DOI] [PubMed] [Google Scholar]
- 7.Richet H, Roux P, Des Champs C, Esnault Y, Andremont A; French Candidemia Study Group 2002. Candidemia in French hospitals: incidence rates and characteristics. Clin. Microbiol. Infect. 8:405–412 [DOI] [PubMed] [Google Scholar]
- 8.Moretti ML, Trabasso P, Lyra L, Fagnani R, Resende MR, Cardoso LGO, Schreiber AZ. 2013. Is the incidence of candidemia caused by Candida glabrata increasing in Brazil? Five-year surveillance of Candida bloodstream infection in a university reference hospital in southeast Brazil. Med. Mycol. 51:225–230 [DOI] [PubMed] [Google Scholar]
- 9.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montagna MT, Caggiano G, Lovero G, De Giglio O, Coretti C, Cuna T, Iatta R, Giglio M, Dalfino L, Bruno F, Puntillo F. 2013. Epidemiology of invasive fungal infections in the intensive care unit: results of a multicenter Italian survey (AURORA Project). Infection 41:645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meletiadis J, Arabatzis M, Bompola M, Tsiveriotis K, Hini S, Petinaki E, Velegraki A, Zerva L. 2011. Comparative evaluation of three commercial identification systems using common and rare bloodstream yeast isolates. J. Clin. Microbiol. 49:2722–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borst A, Maurine A, Leverstein V, Jan V, Fluit C. 2001. Detection of Candida spp. in blood culture using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39:155–160 [DOI] [PubMed] [Google Scholar]
- 13.Khan ZU, Mustafa AS. 2001. Detection of Candida species by polymerase chain reaction (PCR) in blood samples of experimentally infected mice and patients with suspected candidemia. Microbiol. Res. 156:95–102 [DOI] [PubMed] [Google Scholar]
- 14.Ahmad S, Khan Z, Mustafa AB, Khan ZU. 2002. Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection and biochemical methods for species identification. J. Clin. Microbiol. 40:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreira-Oliveira MS, Mikami Y, Miyaji M, Imai T, Schreiber AZ, Moretti ML. 2005. Diagnosis of candidemia by polymerase chain reaction and blood culture: prospective study in a high-risk population and identification of variables associated with development of candidemia. Eur. J. Clin. Microbiol. Infect. Dis. 24:721–726 [DOI] [PubMed] [Google Scholar]
- 16.Wahyuningsih R, Freisleben HJ, Sonntag HG, Schnitzler P. 2000. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J. Clin. Microbiol. 38:3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flahaut M, Sanglard D, Monod M, Bille J, Rossier M. 1998. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions form C. albicans secreted aspartic proteinase genes. J. Clin. Microbiol. 36:395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo SM, Choi JY, Yun JK, Choi JK, Shin SY, Lee K, Kim JM, Lee SY. 2010. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell. Probes 24:44–52 [DOI] [PubMed] [Google Scholar]
- 19.Huang A, Li JW, Shen ZQ, Wang XW, Jin M. 2006. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J. Clin. Microbiol. 44:3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leinberger DM, Schumacher U, Autenrieth IB, Bachmann TT. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J. Clin. Microbiol. 43:4943–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 22.Linton CJ, Borman AM, Cheung G, Holmes AD, Szekely A, Palmer MD, Bridge PD, Campbell CK, Johnson EM. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 45:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borman AM, Szekely A, Palmer MD, Johnson EM. 2012. Assessment of accuracy of identification of pathogenic yeasts in microbiology laboratories in the United Kingdom. J. Clin. Microbiol. 50:2639–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leaw SN, Chang HC, Barton R, Bouchara J-P, Chang TC. 2007. Identification of medically important Candida and non-Candida yeast species by an oligonucleotide array. J. Clin. Microbiol. 45:2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farina C, Russello G, Andreoni S, Bonetti C, Conte M, Fazi P, Lombardi G, Luzzaro F, Manso E, Marone P, Passera M, Rocchetti A, Sanna S, Viganò EF. 2012. Microarray technology for yeast identification directly from positive blood cultures. A multicenter Italian experience. Med. Mycol. 50:549–555 [DOI] [PubMed] [Google Scholar]
- 26.Hsiue HC, Huang YT, Kuo YL, Liao CH, Chang TC, Hsueh PR. 2010. Rapid identification of fungal pathogens in positive blood cultures using oligonucleotide array hybridization. Clin. Microbiol. Infect. 16:493–500 [DOI] [PubMed] [Google Scholar]
- 27.Lin B, Blaney KM, Malanoski AP, Ligler AG, Schnur JM, Metzgar D, Russell KL, Stenger DA. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45:443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata DJ, Hall L, Fothergill AW, Larone DH, Wengenack NL. 2007. Multicenter evaluation of the new VITEK 2 advanced colorimetric yeast identification card. J. Clin. Microbiol. 45:1087–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev. Proteomics 10:151–164 [DOI] [PubMed] [Google Scholar]
- 30.Miller MB, Tang YW. 2009. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 22:611–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiess B, Seifarth W, Hummel M, Frank O, Fabarius A, Zheng C, Mörz H, Hehlmann R, Buchheidt D. 2007. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J. Clin. Microbiol. 45:3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campa D, Tavanti A, Gemignani F, Mogavero CS, Bellini I, Bottari F, Barale R, Landi S, Senesi S. 2008. DNA microarray based on arrayed-primer extension technique for identification of pathogenic fungi responsible for invasive and superficial mycoses. J. Clin. Microbiol. 46:909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Takayanagi A, Nagao K, Tomatsu N, Fukui T, Kawaguchi M, Kudoh J, Amagai M, Yamamoto N, Shimizu N. 2010. Simple PCR-based DNA microarray system to identify human pathogenic fungi in skin. J. Clin. Microbiol. 48:2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]