Abstract
The recent emergence of influenza A virus (H7N9) emphasizes the need for its rapid detection. While commercial nucleic acid amplification tests (NAATs) are commonly used to detect seasonal influenza virus, this study demonstrated that the analytical sensitivity of commercial assays is highly variable compared to that of CDC-based in-house NAATs for the detection of H7N9.
TEXT
An avian influenza A virus (H7N9) was recently linked to 132 human cases and 32 deaths in China (1). Since rapid and accurate detection of novel influenza viruses is the cornerstone of pandemic preparedness (2, 3), many countries have been validating methods to rapidly identify this pathogen (4, 5). While diagnostic options are available, some methods have significant limitations. Growth and propagation of novel influenza viruses require biosafety level 3 containment, and the poor sensitivity of rapid influenza diagnostic tests (RIDTs) limits their usefulness for patient management (3, 6–9). In contrast, nucleic acid amplification tests (NAATs) such as reverse transcription-PCR (RT-PCR) have become the gold standard and a number of commercial assays are available (2, 10). On the other hand, genetic variations in novel influenza viruses could potentially hamper or prevent detection by molecular methods. In silico analysis can be used to identify sequence mismatches within the target region amplified by NAATs; however, the target sequence is often proprietary in commercial assays. The ability to detect novel influenza viruses using NAATs must be experimentally evaluated.
Recent publications have confirmed the suspicion that antigen-based RIDTs lack sensitivity for the detection of influenza A virus (H7N9) (11, 12). Surprisingly, two commercial NAATs, Resplex II Plus version 2.0 (Qiagen Inc.) and xTAG RVP Fast (Luminex Molecular Diagnostics), showed analytical sensitivities comparable to those of the RIDTs, with sensitivity values approximately 1,000-fold lower than those of an in-house RT-PCR targeting the highly conserved M gene (12). This is concerning, since commercial NAATs have been considered valuable tools for the detection of newly emerged strains of influenza A virus (13). This study compared the performance of several commercially NAATs to the performance of “in-house” real-time RT-PCR assays employed in public health or reference laboratories across Canada that use primer pairs designed by the Centers for Disease Control and Prevention (CDC) or the World Health Organization (WHO) targeting the M, H7, or N9 genes of influenza A virus (Table 1) (14, 15).
Table 1.
Protocol summaries and analytical sensitivity
| Assaya | Protocol summary |
Estimated LoDb |
||||
|---|---|---|---|---|---|---|
| Equipment | RT-PCR reagent | Primer and probe source | Target gene | No. of copies/ml | TCID50/ml | |
| CDC M* (NS) | ABI7500 Fast (Life Technologies) | TaqMan Fast Virus 1-Step Master Mix (Life Technologies) | CDC (modified) | M | 617 | 0.20 |
| CDC H7* (MB) | CFX 96 (Bio-Rad Laboratories) | TaqMan Fast Virus 1-Step Master Mix (Life Technologies) | CDC (modified) | H7 | 1,334 | 0.37 |
| CDC M* (BC) | ABI7500 Fast (Life Technologies) | TaqMan Fast Virus 1-Step Master Mix (Life Technologies) | CDC (modified) | M | 1,340 | 0.37 |
| CDC M* (MB) | CFX 96 (Bio-Rad Laboratories) | TaqMan Fast Virus 1-Step Master Mix (Life technologies) | CDC (modified) | M | 1,429 | 0.40 |
| CDC*M (QC) | IQ5 (Bio-Rad Laboratories) | Quantitect RT-PCR probe (Qiagen Inc.) | CDC | M | 1,496 | 0.42 |
| CDC M (SK) | BioMark HD (Fluidigm Corp.) | Quantitect RT-PCR probe (Qiagen Inc.) | CDC | M | 3,388 | 0.95 |
| CDC M* (SK) | BioMark HD (Fluidigm Corp.) | Quantitect RT-PCR probe (Qiagen Inc.) | CDC (modified) | M | 3,891 | 1.10 |
| CDC M (PE) | LightCycler 2.0 (Roche Diagnostics) | Roche Master mix HybProbe (Roche Diagnostics) | CDC | M | 5,754 | 1.60 |
| WHO N9 (SK) | BioMark HD (Fluidigm Corp.) | Quantitect RT-PCR Probe (Qiagen Inc.) | WHO | N9 | 5,754 | 1.60 |
| CDC*H7 (SK) | BioMark HD (Fluidigm Corp.) | Quantitect RT-PCR probe (Qiagen Inc.) | CDC | H7 | 5,808 | 1.62 |
| CDC M* (QC2) | IQ5 (Bio-Rad Laboratories) | Quantitect RT-PCR probe (Qiagen Inc.) | CDC | H7 | 5,998 | 1.98 |
| CDC M* (AB) | ABI7500 Fast (Life Technologies) | TaqMan Fast Virus 1-Step Master Mix (Life Technologies) | CDC | M | 6,761 | 2.00 |
| WHO H7 (SK) | BioMark HD (Fluidigm Corp.) | Quantitect RT-PCR Probe (Qiagen Inc.) | WHO | H7 | 7,161 | 2.04 |
| CDC M (NS) | ABI7500 Fast (Life Technologies) | Invitrogen SuperScript III Platinum One-Step qRT-PCR system (Life Technologies) | CDC (in duplex with influenza B virus) | M | 7,762 | 2.19 |
| RealStar (ON) | Rotor Gene 6000 (Corbett Research) | RealStar Influenza S&T kit v.3 (Altona Diagnostics) | Undisclosed | M | 11,482 | 3.20 |
| Quidel (ON) | Rotor Gene 6000 (Corbett Research) | Molecular Influenza A+ B (Quidel Corp.) | Undisclosed | M | 13,335 | 3.72 |
| Simplexa (ON) | 3 M cycler (Focus Diagnostics) | Simplexa FluA/B & RSV (Focus Diagnostics) | Undisclosed | M | 118,850 | 33.11 |
| Seegene RVI5 (NS) | DNA Engine (Bio-Rad) | Seeplex RV15 One Step ACE detection kit (Seegene) with agarose gel electrophoresis | Undisclosed | M | 288,403 | 79.43 |
| xTag RVP Fast (QC) | LiquiChip 200 (Qiagen Inc.) | XTag RVP Fast v. 1 | Undisclosed | M | 1,496,236 | 416.87 |
| Seegene RVI5 (MB) | ABI Veriti 96 well (Life Technologies) | Seeplex RV15 One Step ACE detection kit (Seegene) with capillary electrophoresis (QIAxcel, Qiagen Inc.) | Undisclosed | M | 2,985,383 | 1,659.59 |
| xTag RVP Classic | Luminex 200 (Luminex Corp.) | XTag RVP Classic (Luminex Corp.) | Undisclosed | M | ND | ND |
| Flu RT-LAMP (ON) | Genie II (Optigene, U.K.) | Flu RT-LAMP assay (Pro-Lab Diagnostics, Inc.) | Undisclosed | M | ND | ND |
An asterisk (*) in the assay name denotes a modification from the CDC protocols that are summarized in Table S1 in the supplemental material. Abbreviations are as follows: AB, Alberta; BC, British Columbia; CDC, Centers for Disease Control and Prevention; H7, hemagglutinin 7; LoD, limit of detection; M, matrix; MB, Manitoba; N9, neuraminidase 9; ND, not detected; NS, Nova Scotia; ON, Ontario; PE, Prince Edward Island; QC, Québec; SK, Saskatchewan; TCID50, 50% tissue culture infectious dose; U.K., United Kingdom; WHO, World Health Organization.
The estimated LoDs are ranked in descending order of the analytical sensitivity determined at a probability of 95%.
The lower limit of detection (LoD) of each method was determined by testing 10-fold serial dilutions of RNA extracted from influenza A/Anhui/01/2013 virus (H7N9) and tested in five independent runs. To reduce interlaboratory variability, specimen preparation, nucleic acid extraction, and dilutions were carried out at the National Microbiology Laboratory (NML) in Winnipeg (Manitoba, Canada). The virus was grown on embryonated eggs, and the viral stock titer was determined with a 50% tissue culture infective dose (TCID50) assay and the Spearman-Kärber calculation method. The viral stock (140 μl) was extracted using a QIAamp Viral RNA Minikit (Qiagen, Mississauga, Ontario, Canada), as recommended by the manufacturer. Each dilution of RNA was prepared as a single batch with five replicates and shipped to participating sites in sufficient volumes to ensure there would be only a single freeze-thaw. Viral RNA was maintained at −70°C until use. All commercial assays were carried out as recommended by the manufacturer. The in-house real-time RT-PCR assays were performed as described by the CDC or WHO (4, 14, 15), and variations and modifications are summarized in Table 1 and Table S1 in the supplemental material, respectively. The LoD was defined by Probit analysis (16) using a probability of 95%, and values were expressed as TCID50/ml or copies/ml (Table 1).
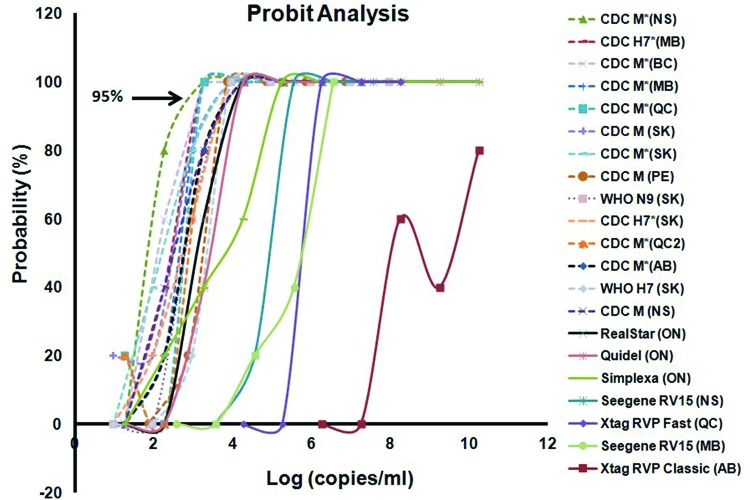
The in-house real-time RT-PCRs were consistently more sensitive than all of the commercial assays, despite some variability in protocols, reagents, and equipment (Table 1; see also Table S1 in the supplemental material). Unlike those of the in-house methods, the LoDs of the different commercial assays were highly variable. While RealStar Influenza S&T RT-PCR assay version 3.0 (Altona Diagnostics) and the Quidel Molecular Influenza A+B assay (Quidel Corp.) were only slightly less sensitive than the in-house RT-PCR assays, the Flu RT-LAMP assay (Pro-Lab Diagnostics, Inc.) was unable to detect H7N9 at any concentration. With a LoD estimated at 6.17 log10 copies/ml, xTagRVP Fast was approximately 200- to 2,000-fold less sensitive than the CDC-based assays (Fig. 1), which is consistent with the findings by Chan et al. (12), who reported a LoD of 7.2 log10 copies/ml for the same strain and 6.8 log10 copies/ml for A/Zhejiang/DTID-ZJU01/2013 virus. Of note, the xTagRVP Classic assay generated only indeterminate results, inconsistently, for the first three most concentrated dilutions of RNA. For this study, indeterminate values were plotted as positive in Fig. 1 but would require confirmation for diagnostic purposes. The poor sensitivity of the xTag RVP Classic assay is concerning, given that this assay is approved by the Food and Drug Administration (FDA) and used in many front-line laboratories in North America. Seeplex RV15 (Seegene) was also 4,800-fold less sensitive than most in-house real-time RT-PCR assays, and this observation was consistent between different laboratories (Fig. 1).
Fig 1.
The analytical sensitivity was estimated using Probit analysis, and values were determined at a probability of 95%. An asterisk (*) in the assay name denotes a modification from the CDC protocols that are summarized in Table S1 in the supplemental material.
Many factors can influence the performance of NAATs, including specimen type, transport, and the nucleic acid extraction method. The panel used in this study consisted of extracted RNA, removing the extraction variable. While freeze-thaw could reduce the overall sensitivity of the molecular assays, each RNA dilution was limited to a single freeze-thaw, and thus, the various methods could be directly compared. Although this study did not compare the clinical performances of specimens collected from patients, our results highlight the analytical variability of the different commercial assays and the importance of verifying the ability to detect newly emerged influenza viruses in reference laboratories. This finding will have major impacts on health care systems, as the methods used in different laboratories may not have equivalent sensitivities. Given the poor sensitivity of some commercial assays, front-line-laboratory personnel, clinicians, and other stakeholders such as public health authorities need to understand the limitations of these assays and that a negative result cannot reliably rule out infection in circumstances in which there is a high index of suspicion for H7N9 infection. In these cases, further testing at a local public health laboratory or other reference laboratories that use the CDC- or WHO-based M, H7, or N9 assays is essential. We believe that specimens from individuals with risks factors for novel influenza virus strains should be tested by laboratories with these in-house reference NAATs or with assays with equivalent performance characteristics. This could be facilitated with specialized screening tools (e.g., education, surveillance, and requisitions) for patients who have animal contacts or a history of travel to regions with emerging influenza A virus strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National Institute for Viral Disease Control and Prevention, China CDC, and CDC Atlanta for providing the H7N9 virus. We are indebted to the many individuals who participated in the laboratory testing, including Amanda Lang (Halifax, Nova Scotia, Canada), Cara MacRae (Halifax, Nova Scotia, Canada), George Moussa (Toronto, Ontario, Canada), and Richard Keenan (Prince Edward Island, Canada). Finally, we acknowledge the various members of Pandemic Influenza Laboratory Preparedness Network (PILPN) within the Canadian Public Health Laboratory Network (CPHLN) who have been instrumental in the planning of this study.
Footnotes
Published ahead of print 21 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01808-13.
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897 [DOI] [PubMed] [Google Scholar]
- 2.LeBlanc JJ, Li Y, Bastien N, Forward KR, Davidson RJ, Hatchette TF. 2009. Switching gears for an influenza pandemic: validation of a duplex reverse transcriptase PCR assay for simultaneous detection and confirmatory identification of pandemic (H1N1) 2009 influenza virus. J. Clin. Microbiol. 47:3805–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatchette TF, Bastien N, Berry J, Booth TF, Chernesky M, Couillard M, Drews S, Ebsworth A, Fearon M, Fonseca K, Fox J, Gagnon JN, Guercio S, Horsman G, Jorowski C, Kuschak T, Li Y, Majury A, Petric M, Ratnam S, Smieja M, Van Caeseele P; Pandemic Influenza Laboratory Preparedness Network 2009. The limitations of point of care testing for pandemic influenza: what clinicians and public health professionals need to know. Can. J. Public Health 100:204–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. 2013. [Accessed 25 July 2013]. http://www.cdc.gov/flu/avianflu/h7n9/detecting-diagnostics.htm.
- 5.Corman VM, Eickmann M, Landt O, Bleicker T, Brünink S, Eschbach-Bludau M, Matrosovich M, Becker S, Drosten C. 2013. Specific detection by real-time reverse-transcription PCR assays of a novel avian influenza A (H7N9) strain associated with human spillover infections in China. Euro Surveill. 18:20461. [PubMed] [Google Scholar]
- 6.Balish A, Garten R, Klimov A, Villanueva J. 2013. Analytical detection of influenza A (H3N2)v and other A variant viruses from the USA by rapid influenza diagnostic tests. Influenza Other Respi. Viruses 7:491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KH, Lam SY, Puthavathana P, Nguyen TD, Long HT, Pang CM, Chan KM, Cheung CY, Seto WH, Peiris JS. 2007. Comparative analytical sensitivities of six rapid influenza A antigen detection test kits for detection of influenza A subtypes H1N1, H3N2 and H5N1. J. Clin. Virol. 38:169–171 [DOI] [PubMed] [Google Scholar]
- 8.Fedorko DP, Nelson NA, McAuliffe JM, Subbarao K. 2006. Performance of rapid tests for detection of avian influenza A virus types H5N1 and H9N2. J. Clin. Microbiol. 44:1596–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurt AC, Baas C, Deng YM, Roberts S, Kelso A, Barr IG. 2009. Performance of influenza rapid point-of-care tests in the detection of swine lineage A (H1N1) influenza viruses. Influenza Other Respi. Viruses 3:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahony JB, Petrich A, Smieja M. 2011. Molecular diagnosis of respiratory virus infections. Crit. Rev. Clin. Lab. Sci. 48:217–249 [DOI] [PubMed] [Google Scholar]
- 11.Baas C, Barr I, Fouchier R, Kelso A, Hurt A. 2013. A comparison of rapid point-of-care tests for the detection of avian influenza A (H7N9) virus. Euro Surveill. 18:pii=20487. [PubMed] [Google Scholar]
- 12.Chan KH, To KK, Chan JF, Li CP, Chen H, Yuen KY. 19 June 2013. Evaluation of analytical sensitivity of seven point-of-care influenza detection kits and two molecular tests for detection of avian-origin H7N9 and swine-origin H3N2 variant influenza A viruses. J. Clin. Microbiol. [Epub ahead of print] 10.1128/JCM.01222-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahony JB, Hatchette T, Ojkic D, Drews SJ, Gubbay J, Low DE, Petric M, Tang P, Chong S, Luinstra K, Petrich A, Smieja M. 2009. Multiplex PCR tests sentinel the appearance of pandemic influenza viruses including H1N1 swine influenza. J. Clin. Virol. 45:200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO 2013. CDC protocol for real-time RT-PCR for swine influenza A H1N1. http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf Accessed 25 July 2013
- 15.WHO 2013. Real-time RT-PCR protocol for the detection of avian influenza A(H7N9) virus. http://www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf Accessed 25 July 2013
- 16.Finney DJ. 1971. Probit analysis, 3rd ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.