Abstract
During 2012, a novel pandemic GII.4 norovirus variant, Sydney 2012, emerged worldwide. A signature of the variant was a GII.Pe ORF1, in association with GII.4 Apeldoorn 2008-like ORF2-ORF3 genes. We report the detection of recombinant GII.4 Sydney 2012 strains, possessing the ORF1 gene of the former pandemic variant New Orleans 2009.
TEXT
Noroviruses (NoVs) are a major cause of acute gastroenteritis in both children and adults. NoV can be classified genetically into at least six genogroups, GI to GVI (1, 2). Although more than 30 genotypes within genogroups GI, GII, and GIV may infect humans (3), a single genotype, GII.4, has been associated with the vast majority of NoV-related outbreaks and sporadic cases of gastroenteritis worldwide (4).
GII.4 NoV strains continuously undergo a process of genetic/antigenic diversification and periodically generate novel strains via accumulation of punctate mutations or recombination, with novel GII.4 variants emerging every 2 to 3 years (5, 6). Since 1996, distinct GII.4 variants have been associated with pandemics or major epidemics of NoV gastroenteritis, including US95/96 1996, Farmington Hills 2002, Asia 2003, Hunter 2004, Yerseke 2006a, Den Haag 2006b, and New Orleans 2009 (3). In late 2012, an increased incidence of NoV-related outbreaks and/or illness in various countries was related to the emergence of a novel GII.4 variant, Sydney 2012. This variant was first identified in March 2012 in Australia (7), and it was found to have originated via recombination by acquiring a GII.Pe polymerase (pol) gene (ORF1) from a GII.4 variant Osaka 2007 strain and ORF2 and ORF3 from a GII.4 Apeldoorn 2008-like strain (8).
The Italian Study Group for Enteric Viruses (ISGEV; http://isgev.net) monitors the epidemiology of enteric viruses in children through hospital-based surveillance (9–12). Monitoring and characterization of NoVs are achieved by a multitarget analysis in the diagnostic regions A, B, C, and D of the NoV genome (13) and interrogation of the Norovirus Typing Tool database (http://www.rivm.nl/mpf/norovirus/typingtool).
During late 2012 (November-December) and January 2013, ISGEV monitored a 28.9% prevalence (90/311) of NoV infection in children hospitalized or presenting for gastroenteritis, versus a prevalence of 25.2% (77/305) in the same period (November-January) of the 2011-2012 winter season. The apparent increased prevalence rate was not significant statistically (P value of >0.1 by the chi-square test). A subset of about half of the NoV-positive samples representative of the two winter periods was randomly selected for sequence analysis and characterized in both region A (ORF1, polymerase) and region C (ORF2, capsid). In the 2012-2013 winter season, about 74.3% of the fully typed strains were characterized as GII.4 Sydney 2012, confirming that, in Italy as in other European and extra-European countries, this new NoV variant was becoming predominant (12). However, the novel variant had already started circulating in Italy at the end of 2011 (12).
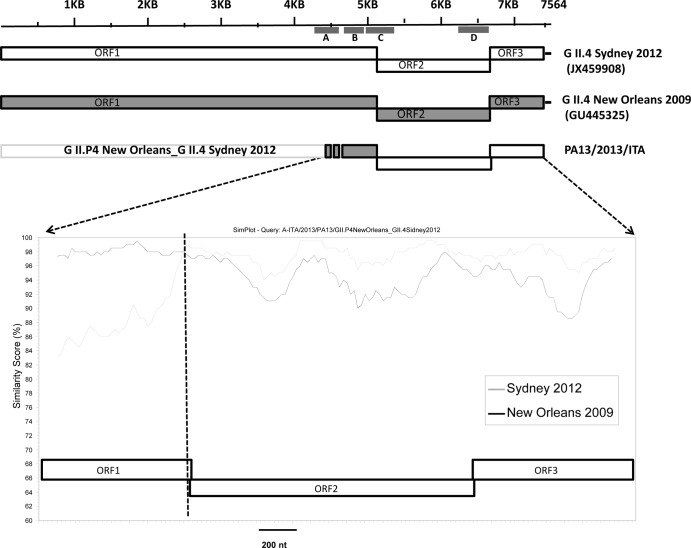
For four samples (PA13/2013/ITA, PA83/2012/ITA, PR4200/2012/ITA, and PR343/2013/ITA), inconsistencies were observed between region A- and region C-based characterizations, suggesting either mixed infections or a recombinant origin. These strains were found to have a GII.4 New Orleans 2009 ORF1 and a GII.4 Sydney 2012 ORF2. A 3′ RACE-PCR (rapid amplification of cDNA ends-PCR) protocol (14) was used to generate a 3.2-kb amplicon encompassing the 3′ end of ORF1, the full-length ORF2 and ORF3, and the 3′ untranslated region (UTR) through the poly(A) tail. Briefly, cDNA was synthesized by the SuperScript III First-Strand cDNA synthesis kit (Invitrogen Ltd., Paisley, United Kingdom) with primer VN3T20 (5′-GAGTGACCGCGGCCGCT20-3′). PCR was then performed with TaKaRa La Taq polymerase (TaKaRa Bio Europe SAS, Saint-Germain en-Laye, France) with forward primer JV12Y (15) and the reverse primer VN3T20 (14). The amplicons were purified and cloned using the TOPO XL cloning kit (Invitrogen Ltd., Paisley, United Kingdom). Additional primers were designed to determine the complete 3.2-kb sequence by an overlapping strategy. Sequence editing and multiple codon-based (translation) alignments were performed with Geneious software v6.2 (Biomatters, Auckland, New Zealand). With the 3′ RACE protocol, it was possible to amplify and sequence a 3.2-kb portion of the genome of strain PA13/2013/ITA, while strains PA83/2012/ITA, PR4200/2012/ITA, and PR343/2013/ITA could not be amplified. For these strains, contiguous fragments encompassing regions A and C (about 1.1 kb in length) were generated by reverse transcription-PCR (RT-PCR) using primers JV12Y (15) and Cog2R (16) and sequenced directly. Nucleotide sequence identity among the four Italian strains was 96.3 to 99.6% in region A and 98.7 to 99.6% in region C. The sequences of the four recombinant strains are available in GenBank under accession numbers KF378731 (PA13/2013/ITA), KF378732 (PR4200/2012/ITA), KF378733 (PA83/2012/ITA), and KF386146 (PR343/2013/ITA). By SimPlot analysis (17), the recombination event was mapped to the ORF1/ORF2 junction region (Fig. 1). This region is highly prone to recombination in NoVs (18), although other recombination hot spots have been identified in the ORF2/ORF3 overlap and in the junction of the shell and protruding capsid domains (8). In the four recombinant strains, the GII.Pe ORF1 (pol) was replaced by a GII.P4 pol derived from the former pandemic NoV GII.4 variant New Orleans 2009. This variant was still dominant in Italy in the winter season 2011-2012 (41.7% of the detected NoV strains), and its circulation was documented until September 2012. The NoV variant Sydney 2012 was already circulating in Italy in November 2011 and accounted for 10.4% of NoV isolates in the winter season 2011-2012 (12). Therefore, the prolonged cocirculation of the two pandemic NoV strains created the opportunities for the emergence of interpandemic recombinant NoV strains. Notably, one recombinant strain was detected as early as January 2012, almost contemporaneously with the emergence of the pandemic variant Sydney 2012.
Fig 1.
SimPlot analysis of partial ORF1 (RdRp) and complete ORF2 (VP1) and ORF3 (VP2) sequences of the GII.4 recombinant strain PA13/2013/ITA. Window size, 200 bp; step, 20 bp. The vertical axis indicates the nucleotide identities between the query strain, PA13/2013/ITA, and the reference strains, expressed as percentages. The horizontal axis indicates the nucleotide (nt) positions. The vertical dashed line indicates the recombination site. Dark gray line, GII.4 variant New Orleans 2009 reference strain Hu/GII.4/New Orleans1805/2009/USA (GU445325); light gray line, GII.4 variant Sydney 2012 reference strain Hu/GII.4/Sydney/NSW0514/2012/AU (JX459908). The positions of the diagnostic regions A to D in the NoV genome are mapped on the schematic representation of the NoV genome.
Also, we observed several distinctive punctate mutations in the GII.P4 New Orleans 2009 pol gene between the recombinant strains detected in northern and southern Italy, suggesting that independent recombination events occurred. Our results notate analogous findings reported recently in Denmark (19).
What we observed in this study is relevant for diagnosis, as these novel interpandemic recombinants can be identified only by multitarget analysis, i.e., by combined analysis of the ORF1 and ORF2. The GII.Pe pol, considered a signature of the pandemic NoV variant Sydney 2012, in some strains can be replaced. Even more importantly, these findings are relevant for the understanding of the evolutionary pathways followed by NoV during its evolution. Continued surveillance for NoV infections and additional data on clinical and epidemiologic features will enable precise assessment of the public health implications of the new variant GII.4 Sydney 2012 and of its recombinant relative strains.
Nucleotide sequence accession numbers.
The sequences of the four recombinant strains are available in GenBank under accession numbers KF378731 (PA13/2013/ITA), KF378732 (PR4200/2012/ITA), KF378733 (PA83/2012/ITA), and KF386146 (PR343/2013/ITA).
ACKNOWLEDGMENTS
This study was supported by the grants “Ricerca Scientifica FIL 2012,” University of Parma, Italy; “Studio dei meccanismi evolutivi dei calicivirus umani” (Italian Scientific Research Fund PRIN 2008); “Caratterizzazione molecolare di norovirus circolanti nella popolazione pediatrica” (University of Palermo, Italy, Research Fund 2007); “Norovirus: caratterizzazione molecolare ed epidemiologia” (University of Palermo, Italy, Research Fund 2012/2013); and MicroMap (PON01_02589).
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Green KY. 2007. Caliciviridae, p 949–979 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Martella V, Decaro N, Lorusso E, Radogna A, Moschidou P, Amorisco F, Lucente MS, Desario C, Mari V, Elia G, Banyai K, Carmichael LE, Buonavoglia C. 2009. Genetic heterogeneity and recombination in canine noroviruses. J. Virol. 83:11391–11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroneman A, Vega E, Vennema H, Vinjé J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. 25 April 2013. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. [Epub ahead of print.] 10.1007/s00705-013-1708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok K, Abente EJ, Realpe-Quintero M, Mitra T, Sosnovtsev SV, Kapikian AZ, Green KY. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull RA, Eden JS, Rawlinson WD, White PA. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. 10.1371/journal.ppat.1000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Beek J, Ambert-Balay K, Botteldoorn N, Eden J, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinje J, White P, Koopmans M. 2013. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 18:8–9 [PubMed] [Google Scholar]
- 8.Eden JS, Tanaka MM, Boni MF, Rawlinson WD, White PA. 2013. Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 87:6270–6282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medici MC, Tummolo F, Albonetti V, Abelli LA, Chezzi C, Calderaro A. 2012. Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008–2009. J. Med. Virol. 84:643–650 [DOI] [PubMed] [Google Scholar]
- 10.Giammanco GM, Rotolo V, Medici MC, Tummolo F, Bonura F, Chezzi C, Martella V, De Grazia S. 2012. Recombinant norovirus GII.g/GII.12 gastroenteritis in children. Infect. Genet. Evol. 12:169–174 [DOI] [PubMed] [Google Scholar]
- 11.De Grazia S, Martella V, Chironna M, Bonura F, Tummolo F, Calderaro A, Moschidou P, Giammanco GM, Medici MC. 2013. Nationwide surveillance study of human astrovirus infections in an Italian paediatric population. Epidemiol. Infect. 141:524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giammanco GM, De Grazia S, Tummolo F, Bonura F, Calderaro A, Buonavoglia A, Martella V, Medici MC. 2013. Norovirus GII.4/Sydney/2012 in Italy, winter 2012–2013. Emerg. Infect. Dis. 19:1348–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroneman A, Vennema H, Deforche K, Avoort HVD, Penaranda S, Oberste MS, Vinje J, Koopmans M. 2011. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51:121–125 [DOI] [PubMed] [Google Scholar]
- 14.Wang QH, Han MG, Cheetham S, Souza M, Funk JA, Saif LJ. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vennema H, de Bruin E, Koopmans M. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233–235 [DOI] [PubMed] [Google Scholar]
- 16.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bull RA, Tanaka MM, White PA. 2007. Norovirus recombination. J. Gen. Virol. 88:3347–3359 [DOI] [PubMed] [Google Scholar]
- 19.Fonager J, Hindbaek L, Fischer T. 2013. Rapid emergence and antigenic diversification of the norovirus 2012 Sydney variant in Denmark, October to December, 2012. Euro Surveill. 18:pii.20413 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20413 [PubMed] [Google Scholar]