Abstract
Seventy-eight blood cultures with a Gram stain result of Gram-positive cocci in pairs and/or chains were evaluated with the Nanosphere Verigene Gram-positive blood culture (BC-GP) assay. The overall concordance of the assay with culture was 89.7% (70/78 cultures), allowing for the development of a targeted treatment algorithm.
TEXT
Bloodstream infections (BSI) are a leading cause of morbidity and mortality. In 2009, BSI were the cause of nearly 36,000 deaths in the United States (1). Health care-associated BSI are a major contributor to these statistics, being associated with nearly 75,000 infections a year with a mortality rate of approximately 25% (2). Enterococci are the third leading cause of health care-associated BSI (3), and inappropriate antimicrobial therapy has been shown to be an independent risk factor for the increased mortality of BSI (4). The Nanosphere Verigene Gram-positive blood culture (BC-GP) assay (Nanosphere, Northbrook, IL) is an on-demand FDA-cleared test for the identification of 15 targets from positive blood culture bottles. Previous studies have shown this assay to be an effective method for determining the identity of Gram-positive organisms, showing 92 to 95% overall concordance for organism identification (5, 6); however, the implementation of an algorithm for treatment based on BC-GP test results has not been proposed. The current work focuses on the performance of the assay specifically in cultures where streptococci and enterococci are suspected based on the primary Gram stain and on the algorithm we have developed for treatment based on the BC-GP results.
We first performed an evaluation of the BC-GP assay on the Nanosphere Verigene system. Routine blood cultures were collected using FAN aerobic (FA), pediatric FAN (PF), and standard anaerobic (SA) bottles and subsequently incubated on the BacT/Alert (bioMérieux, Durham, NC) system. Positive blood cultures were Gram stained, plated onto solid medium, and stored upright at 35°C for up to 5 days. Subcultures were evaluated according to standard laboratory protocols, and organisms were identified using a combination of morphological and phenotypic methods as well as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek MS; bioMérieux). Notably, the Vitek MS accurately distinguishes between Streptococcus pneumoniae and Streptococcus mitis/Streptococcus oralis (7). Positive blood cultures with a Gram stain of Gram-positive cocci in pairs and/or chains were further evaluated with the BC-GP assay according to the manufacturer's recommendations. The BC-GP targets analyzed include Enterococcus faecalis, Enterococcus faecium, Streptococcus spp., Streptococcus agalactiae, Streptococcus anginosus group, S. pneumoniae, Streptococcus pyogenes, and the resistance determinants vanA and vanB.
Seventy-eight consecutive positive blood cultures from 74 patients with an initial Gram stain result containing Gram-positive cocci in pairs and/or chains were evaluated with the BC-GP assay, of which 72 had at least one target detected. Of the six cultures for which no target was detected, three grew organisms (Acinetobacter sp., Atopobium sp., B. cereus group) for which the assay does not target, and two grew Streptococcus vestibularis, which has not been evaluated by the manufacturer according to the package insert. The remaining negative BC-GP cultures grew five different organisms (Enterococcus faecalis, Enterococcus gallinarum, Enterobacter cloacae, Candida tropicalis, and Klebsiella pneumoniae) on subculture. See Table 1 for a complete list of organisms that were not detected by the BC-GP assay.
Table 1.
List of organisms recovered but not detected by the BC-GP assay
| Organism | No. of cultures positive | No. detected on original Gram stain |
|---|---|---|
| Klebsiella pneumoniaea | 3 | 3 |
| Stenotrophomonas maltophiliaa | 2 | 1 |
| Bacillus cereus groupa | 2 | 1 |
| Streptococcus vestibularis | 2 | 2 |
| Rothia spp.a | 2 | 0 |
| Atopobium spp. | 1 | 1 |
| Enterococcus faecalis | 1 | 1 |
| Enterobacter cloacaea | 1 | 1 |
| Candida tropicalisa | 1 | 1 |
| Enterococcus gallinaruma | 1 | 1 |
| Acinetobacter spp. | 1 | 1 |
| Serratia marcescensa | 1 | 1 |
| Actinomyces spp.a | 1 | 1 |
Component of polymicrobial culture.
The BC-GP assay performed comparably to routine isolate identification; no target had more than three errors (Table 2). S. agalactiae and S. pyogenes demonstrated 100% agreement between BC-GP and culture identification. The Streptococcus spp. target had 96.2% agreement with isolate identification. One discordant result was a culture that grew a pyrrolidonyl arylamidase (PYR)-negative alpha-hemolytic organism presumptively identified as viridans group Streptococcus by our laboratory; confirmatory testing was not performed, and the blood culture bottle gave a result of Enterococcus faecium by the BC-GP assay. The other two discordant results were two cultures that grew an organism identified as Streptococcus vestibularis by MALDI-TOF MS. Although the manufacturer lists S. vestibularis as an organism for which the performance of the BC-GP has not been evaluated, two additional cultures that grew S. vestibularis were positive for the Streptococcus species target. All S. vestibularis isolates were from distinct patients and were detected using both FA and SN media, indicating potential target variability in different strains of S. vestibularis. S. anginosus group also had one discordant result (98.7% agreement), which was a culture that grew an organism identified by MALDI-TOF MS as Streptococcus mitis/S. oralis, but the BC-GP assay gave a dual result of S. anginosus group and S. pneumoniae. This was one of two S. mitis/S. oralis cultures that gave an identification of S. pneumoniae by the BC-GP assay, equating to 97.4% agreement for the S. pneumoniae target. In total, eight cultures grew S. mitis/S. oralis; the other six were correctly identified by the BC-GP assay as Streptococcus species only. The two cultures that grew S. pneumoniae were correctly identified as such by the BC-GP assay. The only culture that grew an organism that was claimed to be included by the manufacturer yet did not generate a positive result by the BC-GP assay was a culture with an initial polymicrobial Gram stain which grew E. faecalis, E. gallinarum, Enterobacter cloacae, Candida tropicalis, and Klebsiella pneumoniae. This was also the only discordant result for the E. faecalis target and the only discordant polymicrobial culture. Fourteen cultures grew more than one organism, 13 (92.3%) of which were concordant with the BC-GP result. Nine of the 14 polymicrobial cultures grew organisms that are not detected by the BC-GP assay (Table 1). In our study, 10 cultures grew organisms that were phenotypically identified as vancomycin-resistant enterococci (VRE), and all had vanA detected by the BC-GP assay. Additionally, there was a culture that grew vancomycin-susceptible E. faecium, for which the vanA target was positive. The patient who was the source of this culture had a history of VRE-positive rectal screens as well as a concurrent urine sample from which VRE was isolated. Based on the data above, the overall concordance of the BC-GP assay with isolate identification was 89.7%, which notably includes two false-negative results observed with S. vestibularis identified as the Streptococcus species target, which is not claimed to be included by the manufacturer.
Table 2.
Performance of the BC-GP assay by target
| Target | No. positive by culture | % Agreement |
|---|---|---|
| Streptococcus spp. | 39 | 96.2 (75/78) |
| Streptococcus agalactiae | 7 | 100 (78/78) |
| Streptococcus anginosus | 5 | 98.7 (77/78) |
| Streptococcus pneumoniae | 2 | 97.4 (76/78) |
| Streptococcus pyogenes | 1 | 100 (78/78) |
| Enterococcus faecalis | 18 | 98.7 (77/78) |
| Enterococcus faecium | 16 | 98.7 (77/78) |
| Vancomycin resistance (vanA) | 10 | 97.0 (32/33) |
Based on the performance of the assay during our evaluation period, we were able to develop a treatment algorithm in consultation with our antimicrobial stewardship program. For improved algorithm utilization, the results of the BC-GP assay are communicated directly to an on-call pharmacist and entered into the patient's electronic medical record. The algorithm illustrated in Fig. 1 is used as a guide for the pharmacist when recommending therapy. Patients whose cultures are positive by the BC-GP assay for S. pyogenes, S. agalactiae, and S. anginosus group are recommended to receive penicillin or ceftriaxone with or without clindamycin (clindamycin is recommended for patients with signs of septic shock). Ceftriaxone was not our empirical therapy of choice for S. pneumoniae, due to a 73% susceptibility rate at our institution. Reduced susceptibility, along with the false-positive results for the S. pneumoniae target due to S. mitis/S. oralis, led us to recommend vancomycin for empirical therapy for infection with S. pneumoniae along with other Streptococcus spp. not identified to the species level. We note that while we chose to specifically recommend daptomycin for vancomycin-intolerant patients, a recommendation or requirement for infectious disease consultation would be suitable as well. Importantly, early detection of VRE is critical to aid in the switch from a vancomycin-based treatment regimen. Lastly, based on our data, we do not recommend that cultures with mixed Gram stain results (e.g., Gram-negative rods and Gram-positive cocci in pairs/chains, or Gram-positive cocci in clusters and Gram-positive cocci in pairs/chains) be evaluated by the BC-GP assay in conjunction with the recommended treatment algorithm.
Fig 1.
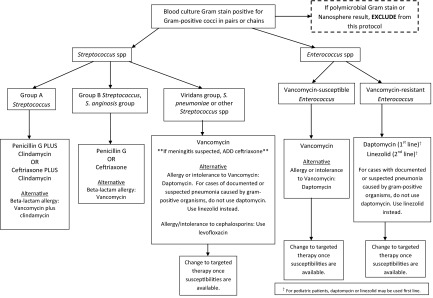
Algorithm for treatment recommendations based on BC-GP assay result.
Our data support previous findings that the Nanosphere BC-GP assay accurately identifies targets from positive blood cultures, with the exception of the differentiation of S. mitis/S. oralis and S. pneumoniae (5, 6). False-positive S. pneumoniae results do not limit the utility of the assay, as we were still able to develop a treatment algorithm to promote the use of targeted regimens based on the results generated by the BC-GP assay. Although we have previously shown that a pharmacy-based intervention strategy is effective at our institution (8), the effectiveness of our BC-GP algorithm for streptococci and enterococci will need to be measured.
ACKNOWLEDGMENT
We acknowledge the kind generosity of Nanosphere for providing the reagents used in this study.
Footnotes
Published ahead of print 28 August 2913
REFERENCES
- 1.Kochanek KD, Xu J, Murphy SL, Minino AM, Kung H-C. 2011. Deaths: final data for 2009. Natl. Vital Stat. Rep. 60:1–116 [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention 2011. Vital signs: central line-associated blood stream infections—United States, 2001, 2008, and 2009. MMWR Morbid. Mortal. Wkly. Rep. 60:243–248 [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 4.Suppli M, Aabenhus R, Harboe ZB, Andersen LP, Tvede M, Jensen J-U. 2011. Mortality in enterococcal bloodstream infections increases with inappropriate antimicrobial therapy. Clin. Microbiol. Infect. 17:1078–1083 [DOI] [PubMed] [Google Scholar]
- 5.Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA. 2013. Evaluation of a microarray based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J. Clin. Microbiol. 51:1188–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. 17 April 2013. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for the rapid detection of bacteria and resistance determinants. J. Clin. Microbiol. 10.1128/JCM.00831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubois D, Segonds C, Prere M-F, Marty N, Oswald E. 2013. Identification of clinical Streptococcus pneumoniae isolates among other alpha and nonhemolytic streptococci by use of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J. Clin. Microbiol. 51:1861–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heil EL, Daniels LM, Long DM, Rodino KG, Weber DJ, Miller MB. 2012. Impact of a rapid peptide nucleic acid fluorescence in situ hybridization assay on treatment of Candida infections. Am. J. Health Syst. Pharm. 69:1910–1914 [DOI] [PubMed] [Google Scholar]


