Abstract
We describe the first case of bacteremia due to Gallibacterium anatis. The patient, a 26-year-old woman, developed bacteremia and diarrhea. The origin of infection was possibly due to a diet contaminated by G. anatis in this highly immunocompromised patient.
CASE REPORT
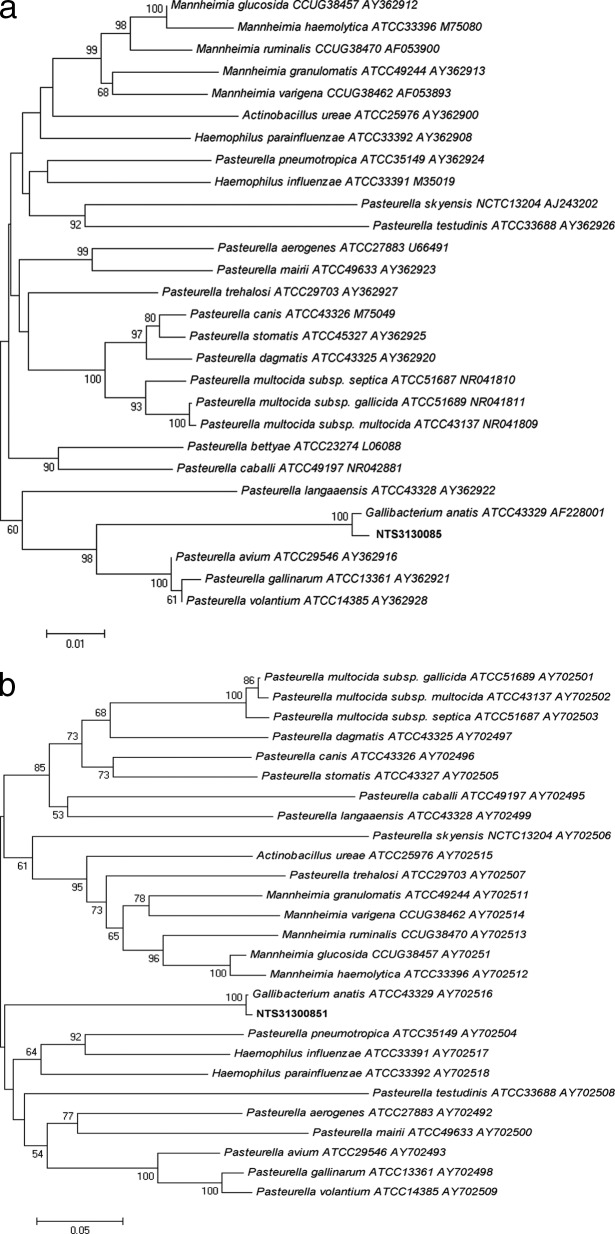
On 24 January 2013, a 26-year-old woman was admitted to the nephrology ward at Nantes University Hospital, in order to have monitored her chronic kidney insufficiency (renal biopsy) due to cystic fibrosis disease. In May 2011, she received a bipulmonary transplant. The immunosuppressive treatment included mycophenolate mofetil (2 g per day), tacrolimus (10 mg per day), and corticoids (9 mg per day). During hospitalization, she suffered abdominal pain, fever, and recent diarrhea. A complete blood cell count showed bicytopenia with neutropenia (0.93 × 109 cells/liter) and anemia (hemoglobin, 80 g/liter). Her C-reactive protein level was 46.4 mg/liter (normal range, <5 mg/liter). An empirical antimicrobial treatment was started with imipenem-cilastatin (1.5 g per day) and tobramycin (650 mg every 72 h, adapted to chronic kidney insufficiency). Four aerobic and anaerobic blood cultures (Bactec FX; Becton, Dickinson, Sparks, MD) were performed on the peripheral site over the course of 48 h. Concomitantly, a primary cytomegalovirus (CMV) infection with gastrointestinal, liver, and lung involvement was diagnosed. The viral investigation revealed a high cytomegalovirus viremia by real-time PCR (viral load, 501,971 copies/ml). Despite the broad-spectrum antimicrobial therapy, the first aerobic blood culture yielded Gram-negative bacilli (laboratory reference no. NTS31300851) after 17 h of incubation. Other blood cultures remained negative after 5 days of incubation. This bacterium was not hemolytic on blood agar (bioMérieux, Marcy l'Etoile, France), with negative oxidase and indole tests. The semiautomated api20E gallery (bioMérieux, Marcy l'Etoile, France) provided low discrimination identification with different taxons (Escherichia coli 2, Shigella species, Pantoea sp. 1, Pasteurella pneumotropica/Mannheimia haemolytica, which are species in the bioMérieux database associated with the semiautomated api20E), whereas the ID-GN card of the Vitek2 system (bioMérieux) failed to identify this bacterium (E. coli, Shigella species, Pantoea species). The 16S rRNA gene amplification and sequencing were performed with universal primers 27f and 1378r as previously described (1). The 1,360-bp fragment obtained matched that of G. anatis, with 99% similarity according to BIBI database (http://umr5558-sud-str1.univ-lyon1.fr/lebibi/lebibi.cgi) or BLAST (http://www.ncbi.nlm.nih.gov/BLAST) analysis. To confirm the identification of this unusual bacterium in human clinical specimens, sodA amplification and sequencing were also performed as previously described (2). The 451-bp fragment obtained matched that of G. anatis, with 99% similarity according to BIBI database (http://umr5558-sud-str1.univ-lyon1.fr/lebibi/lebibi.cgi) or BLAST (http://www.ncbi.nlm.nih.gov/BLAST) analysis. Phylogenetic analysis with either the neighbor-joining or maximum-parsimony algorithm matched the NTS31300851 strain to the genus Gallibacterium and the species G. anatis by using MEGA5 software with tree constructions (Fig. 1a and b). The strain reduced nitrate to nitrite, produced α-glucosidase, and presented positive glucose, mannitol, and saccharose assimilation with api20E and ID-GN biochemical galleries. Unlike E. coli, the strain was negative for lysine decarboxylase, ornithine decarboxylase, and indole. Also, negative reactions for sorbitol, rhamnose, amygdaline, and arabinose assimilation differentiated the strain from Pantoea sp. 2.
Fig 1.
(a and b) Neighbor-joining (NJ) tree showing the phylogenetic placement of strain NTS31300851 (in boldface) among members of the Gallibacterium anatis species. Twenty-seven 16S rRNA gene and sodA sequences selected from the GenBank database were aligned with that of strain NTS31300851 by using MEGA5 (www.megasoftware.net). Accession numbers are indicated after the species name. The evolutionary history was inferred using the NJ method. The figure shows the optimal tree; the sums of the branch lengths for 16S rRNA genes and sodA genes were 0.61902303 and 2.62153347, respectively. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The final data set contains 1,360 and 451 positions, for 16S rRNA genes and sodA genes, respectively. Phylogenetic analyses were conducted in MEGA5. NJ and parsimony trees were globally congruent with the distance tree and confirmed the placement of the NTS31300851 strain in the Gallibacterium anatis species. Scale bar indicates substitutions per nucleotide position. (a) 16S rRNA gene NJ tree showing the phylogenetic placement of strain NTS31300851 (in boldface); (b) sodA NJ tree showing the phylogenetic placement of strain NTS31300851 (in boldface).
Concerning Gallibacterium species, no European Committee on Antimicrobial Susceptibility Testing (EUCAST) specific recommendation for susceptibility testing exists. Therefore, in vitro susceptibility testing was performed using the Vitek2 system, and results were interpreted according to EUCAST guidelines. The bacterium was fully susceptible, except to fluoroquinolones (ofloxacin MIC ≥ 8 mg · liter−1; ciprofloxacin MIC ≥ 4 mg · liter−1), and cotrimoxazole (MIC ≥ 320 mg · liter−1). Therefore, antimicrobial drug treatment was changed, on January 27, to ceftriaxone (1 g per day) and tobramycin (650 mg every 72 h).
On January 30, the patient was admitted to the pneumology ward but developed hypoxemia. She was transferred on February 4 to the intensive care unit due to bacteremia, hypoxemia, CMV sepsis, and metabolic acidosis. The patient died on March 3 due to pulmonary fibrosis, CMV sepsis, and anuric renal failure.
The genus Gallibacterium, first described by Christensen et al. in 2003, belongs to the family Pasteurellaceae, which includes diverse bacteria, including Haemophilus, Aggregatibacter, Actinobacillus, Pasteurella, Mannheimia, Lonepinella, and Phocoenobacter, classified into different genera based on metabolic properties (3, 4). Gallibacterium contains six different species that demonstrate similarities in 16S rRNA gene sequences and in phenotypic and polyamine contents: Gallibacterium melopsittaci, Gallibacterium trehalosifermentans, Gallibacterium genospecies III, Gallibacterium salpingitidis, Gallibacterium group V, and Gallibacterium anatis (5–7). Gallibacterium anatis has been isolated from various birds or other animals, such as chickens, ducks, geese, guinea fowl, turkeys, psittacine birds, partridges, cattle, budgerigars, and pigs (8). G. anatis was found to be prevalent in the upper respiratory tracts and the lower genital tracts of healthy chickens (5). However, isolates have also been recovered from a range of pathological lesions in chickens with bacteremia, oophoritis, follicle degeneration, salpingitis, peritonitis, enteritis, and respiratory tract diseases (5). The ATCC 43329 type strain was recovered from the intestinal track of a duck (3). The genus consists of Gram-negative, nonmotile, rod-shaped, or pleomorphic bacteria. The bacteria are catalase, oxidase, and phosphatase positive. Nitrate is reduced and acid is produced without gas formation from glycerol, (−)d-ribose, (+)d-xylose, (−)d-mannitol, (−)d-fructose, (+)d-galactose, (+)d-glucose, (+)d-mannose, sucrose, and raffinose. The Gallibacterium genus can be separated from other genera of Pasteurellaceae by differences in catalase, in symbiotic growth, in hemolysis, in urease, in indole, in acid production from (+)d-xylose, (−)d-mannitol, (−)d-sorbitol, and (+)d-mannose, in maltose, in raffinose, in dextrin, and in o-nitrophenyl-β-d-galactopyranoside (ONPG) and p-nitrophenyl-β-d-glucoside (PNPG) tests (3).
We describe in this report a case of human bacteremia due to G. anatis. This veterinary pathogen behaving as an opportunistic pathogen was able to cause an infection in a severely immunosuppressed woman. The source of bacteremia was not clearly established, but the most likely hypothesis is the contamination of meal. Moreover, there was no contact with farm animals.
The digestive tract could have been weakened by the cytomegalovirus primary infection, facilitating G. anatis translocation (CMV-seronegative patient before the graft). Virulence factors contributing to the pathogenicity of G. anatis have not yet been well defined, except for an atypical RTX (repeat in toxin) toxin, GtxA, responsible for the hemolytic activity and likely to be a major virulence factor (6, 9). Recently, a fimbrial GalF-A was also characterized as a key structure during colonization and invasion events of mucosal surfaces (10). This last structure could have been involved in digestive translocation leading to bacteremia. Indeed, GalF-A showed sequence similarity to the F17-like fimbrial protein precursor identified in the human extraintestinal pathogenic Escherichia coli (ExPEC) (11). Moreover, the infection has been likely favored by the immunosuppressive therapy overdose, the leucopenia, and the cytomegalovirus primary infection.
Like most environmental organisms, this bacterium was susceptible, except to fluoroquinolones and cotrimoxazole. The intrinsic or acquired resistance of G. anatis to antimicrobial drugs has not yet been elucidated. There has been much debate about the use of fluoroquinolones in veterinary medicine, but the proximity of G. anatis with poultry may be a hypothesis of this resistance. At last, the lack of commercially available biochemical gallery databases makes correct identification of this environmental organism difficult. It also underlines the usefulness of sequencing specific gene targets (16S rRNA genes, sodA, hsp65) for identification of unusual Gram-negative bacilli isolated from immunocompromised hosts (1). In the future, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry should allow more powerful and faster discrimination between species.
Nucleotide sequence accession numbers.
The 16S and sodA nucleotide sequences of the isolate have been deposited in the GenBank database under accession numbers KF032910 and KF032911, respectively.
ACKNOWLEDGMENTS
There are no conflicts of interests for any authors.
We received no financial support for the study.
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Crémet L, Bemer P, Zambon O, Reynaud A, Caroff N, Corvec S. 2009. Chitinophaga terrae bacteremia in human. Emerg. Infect. Dis. 15:1134–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen H, Bisgaard M, Bojesen AM, Mutters R, Olsen JE. 2003. Genetic relationships among avian isolates classified as Pasteurella haemolytica, “Actinobacillus salpingitidis” or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 53:275–287 [DOI] [PubMed] [Google Scholar]
- 4.Bisgaard M, Korczak BM, Busse H-J, Kuhnert P, Bojesen AM, Christensen H. 2009. Classification of the taxon 2 and taxon 3 complex of Bisgaard within Gallibacterium and description of Gallibacterium melopsittaci sp. nov., Gallibacterium trehalosifermentans sp. nov. and Gallibacterium salpingitidis sp. nov. Int. J. Syst. Evol. Microbiol. 59:735–744 [DOI] [PubMed] [Google Scholar]
- 5.Bojesen AM, Torpdahl M, Christensen H, Olsen JE, Bisgaard M. 2003. Genetic diversity of Gallibacterium anatis isolates from different chicken flocks. J. Clin. Microbiol. 41:2737–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen BM, Frees D, Bojesen AM. 2011. Expression and secretion of the RTX-toxin GtxA among members of the genus Gallibacterium. Vet. Microbiol. 153:116–123 [DOI] [PubMed] [Google Scholar]
- 7.Tang YW, Ellis NM, Hopkins MK, Smith DH, Dodge DE, Persing DH. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic Gram-negative bacilli. J. Clin. Microbiol. 36:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisgaard M. 1993. Ecology and significance of Pasteurellaceae in animals. Zentralbl. Bakteriol. 279:7–26 [DOI] [PubMed] [Google Scholar]
- 9.Johnson TJ, Danzeisen JL, Trampel D, Nolan LK, Seemann T, Bager RJ, Bojesen AM. 2013. Genome analysis and phylogenetic relatedness of Gallibacterium anatis strains from poultry. PLoS One 8:e54844. 10.1371/journal.pone.0054844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bager RJ, Nesta B, Pors SE, Soriani M, Serino L, Boyce JD, Adler B, Bojesen AM. 2013. The fimbrial protein FlfA from Gallibacterium anatis is a virulence factor and vaccine candidate. Infect. Immun. 81:1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]