Abstract
The oral bacterium, Campylobacter rectus, is an etiological agent of periodontitis. The virulence genes of C. rectus are largely unknown. The aim of this study was to query C. rectus for the presence of an invasion antigen B (ciaB) gene, which is needed for cell invasion by the related species Campylobacter jejuni. PCR and PCR-walking identified a ciaB from C. rectus. In silico analyses of C. rectus 314 ciaB (Cr-ciaB) revealed an ORF of 1,830 base pairs. The Cr-CiaB protein shared significant sequence identity (BLASTx and phylogeny) with CiaB from related campylobacters. Cr-CiaB is predicted to lack membrane helices, signal peptides, and localizes to the cytoplasm; which are consistent with CiaB proteins. Expression of Cr-ciaB was confirmed with RT-PCR; and potential ciaB genes were detected in eight additional strains of C. rectus. Cr-ciaB is the first CiaB identified from the oral campylobacters.
Keywords: Campylobacter rectus, Periodontitis, Invasion antigen B, Virulence factor
Introduction
Periodontitis is an inflammatory disorder of connective tissue and bone that support the teeth [1]. Periodontitis is common, affecting 35 % of adults [1, 2]. Approximately 13 % of patients develop severe forms of periodontitis, which, if untreated, may result in tooth loss and systemic complications including an increased risk of heart attack and an increased risk of pregnancy complications [1, 2].
Campylobacter rectus is a poorly described Gram-negative, oral bacterium that has been implicated as a cause of periodontitis [3–5]. Recent studies indicate that women with periodontitis are seven times more likely to experience preterm labor than their healthy counterparts [6, 7]. Serological data from human studies has implied that C. rectus, as part of the poly-microbial oral community, plays a role in the preterm labors of mothers with periodontitis [8]. Additionally, a pregnant mouse model has shown the association of C. rectus strain 314, isolated from a patient with periodontitis [9], with fetal growth restriction and a decreased survival of pups [10, 11]. Although the aforementioned human and mouse studies have established C. rectus as an agent of periodontitis, the pathogenic mechanisms of C. rectus are not well elucidated.
Although C. rectus strain 33238 has been partially sequenced (NCBI GenBank #ACFU00000000), the genes important to the virulence and pathogenic mechanisms of C. rectus have not been identified. To understand C. rectus as an oral pathogen, querying multiple strains for the presence of virulence factors may be significant as strain-to-strain genomic variability has been seen among campylobacters [12, 13]. Recently, a few studies have identified potential virulence factors in C. rectus, including potential toxin genes (csx A and B; csxC and csxD) and a surface array protein (crsA gene) thought to play a role in allowing the bacteria to avoid the host immune system [14–16]. While these studies demonstrate that C. rectus encodes potential virulence genes; it is unknown whether C. rectus contains genes that have been shown to significantly contribute to the pathogenesis of related campylobacters, including ciaB.
Within the campylobacters, virulence factors have been best characterized from the gastrointestinal pathogen Campylobacter jejuni [17]. Among the best studied C. jejuni virulence factors is the ciaB gene, which has been shown to play a critical role in host cell invasion; and therefore pathogenesis [18–20]. In addition, ciaB genes have been identified from additional campylobacters including Campylobacter lari, Campylobacter coli, and Campylobacter upsaliensis [21, 22]. Given the conservation of ciaB, and a recent study showing C. rectus 314 can invade host cells [23], we hypothesized that the C. rectus 314 contains a ciaB gene. In this study, a combination of PCR, PCR-walking, reverse transcriptase PCR, and in silico tools were used to characterize a ciaB gene (Cr-ciaB) from C. rectus 314. The identification of C. rectus CiaB suggests that CiaB might contribute to the pathogenesis of C. rectus during host cell invasion [23]; and raises the possibility of developing novel periodontal therapeutics that disrupt CiaB functionality.
Materials and Methods
Bacterial Strains and Genomic DNA Isolation
Campylobacter curvus (ATCC 33273) and the C. rectus strains listed in Table 1 were grown under standard anaerobic conditions. C. curvus, like C. rectus, is an oral bacteria associated with periodontitis [36]. Bacteria were grown on tryptic soy blood agar with sodium formate (0.3 %). Genomic DNA was isolated from bacterial pellets using CTAB [24].
Table 1.
Summary of Campylobacter rectus strains used in this study
| Strain | Description |
|---|---|
| 33238 | ATCC type strain (http://www.atcc.org) |
| 314 | Periodontal isolate [9] |
| CCUG 11645 | Periodontal isolatea |
| CCUG 11643 | Periodontal isolatea |
| CCUG 11642 | Periodontal isolatea |
| CCUG 11640 | Periodontal isolatea |
| CCUG 27948 | Periodontal isolatea |
| CCUG 48803 | Periodontal isolatea |
| BCCM 7615 | Periodontal isolateb |
aUniversity of Göteborg, Sweden (http://www.ccug.se)
bBelgian collection of microorganisms (http://bccm.belspo.be/index.php)
Degenerate PCR
Genomic DNA from C. rectus 314 and 33238 were scanned for ciaB genes using oligonucleotides CiaB-DF and CiaB-DR (Table 2). CiaB-DF and CiaB-DR were designed to amplify conserved regions within known ciaB [25]. PCR was then used to screen C. rectus for the presence of ciaB. Each 50 μl PCR reaction contained 1.0 μM of each primer. The cycling conditions used were as follows: 94 °C for 1 min, 40 °C for 1 min, and 72 °C for 1 min (10 cycles); followed by 30 cycles of 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min. Amplicons were purified (QIAquick gel extraction kit, Qiagen, USA) and cloned for sequencing into pCR2.1-TOPO (TOPO-TA kit, Invitrogen, USA).
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ → 3′) | Description |
|---|---|---|
| CiaB-DF | TAYACNCAYGGNGTNGC | Degenerate PCR |
| CiaB-DR | TCRTGNCCKATNGTNGA | |
| CiaB-F | GTGAACGATTTTAAAAGATTAAACG | Strain PCR |
| CiaB-R | CTATCTCTCGCTCGCTTGCC | |
| CiaB-RTF1 | CTAGAACGCAAACCTACATCA | RT-PCR detection |
| CiaB-RTR1 | CCGCTAAGCTTCATAAACGG | |
| M13 F | CGCCAGGGTTTTCCCAGTCACGA | DNA Sequencing |
| M13 R | TCACACAGGAAACAGCTATGAC | |
| CiaB-F5 | CAAACTATAAAAATTTGGCGCTTC | |
| CiaB-R5 | GAAAAGCTTTGCGAAAGTCG | |
| CiaB-Walk-5R | TACATCGCGTTTAGTTCGTCTTTGC | Genome Walker |
| CiaB-Walk-3F | CTATAGGAAACGAGCTTGACGAGAGC | |
| AP1 | GAAAAGCTTTGCGAAAGTCG | |
| AP2 | ACTATAGGGCACGCGTGGT |
N A, T, G, or C; Y C or T; R A or G; K G or T (IUPAC nucleotide ambiguity codes)
RNA Isolation and Reverse Transcription PCR
RNA was isolated from bacterial pellets using a QIAgen miniprep kit (Qiagen, USA). The RNA was then used for RT-PCR using the Access RT-PCR kit (Promega, USA).
PCR-Walking
A Genome Walker kit (Clontech, USA) was used to obtain the ORF of ciaB. Genome Walker is a PCR method for identifying sequences adjacent to known sequences. The kit was used according to the manufacturer’s protocol. In brief, the sequences identified by PCR were used to design gene-specific primers (Table 2). Next, 2.5 μg of 314 genomic DNA was digested with one of four blunt-ended restriction endonucleases: DraI, EcoRV, PvuII or StuI. The digested DNA reactions was then ligated to adapter oligonucleotides. The ligated products were then used as templates for PCR using primers complementary to the adapters (Table 2, AP1) and complementary to ciaB (Table 2, CiaB-Walk-5R or CiaB-Walk-3F). A primer combination of AP1 and CiaB-Walk-5R was used to obtain the 5′-end of ciaB; while AP1 and CiaB-Walk-3R was used to obtain the 3′-end of ciaB. PCR products were purified (QIAquick gel extraction kit) and cloned for sequencing into pCR2.1-TOPO.
Sequence Analysis
DNA chromatographs were edited manually and assembled using Vector NTI software (Invitrogen, USA). Obtained sequences were then used as templates for sequence homology searches using BLAST. Searches were performed using the default parameters for BLASTx [26]. Alignments were generated using CLUSTAL [27]. Membrane-spanning helices were predicted using TMHMM [28]. Potential signal peptide cleavage sites were predicted using SignalP [29]. PSORT-B was used to predict the cellular location of proteins [30].
Phylogenetic Analysis
A phylogenetic tree of C. rectus 314 CiaB was generated using Phylogeny-fr [31]. BLAST-EXPLORER [32] was used to retrieve and align sequences related to CiaB. After alignment, ambiguous regions were removed with Gblocks [33], and the resultant alignment was used to construct a tree using the maximum likelihood method [34, 35].
Conservation of CiaB Among C. rectus Strains
PCR was used to screen C. rectus DNA for the presence of ciaB genes. Primers specific to the 5′ and 3′ ends of ciaB from C. rectus 314 were used to PCR DNA from the C. rectus strains listed in Table 1.
Results
Identification of C. rectus 314 ciaB
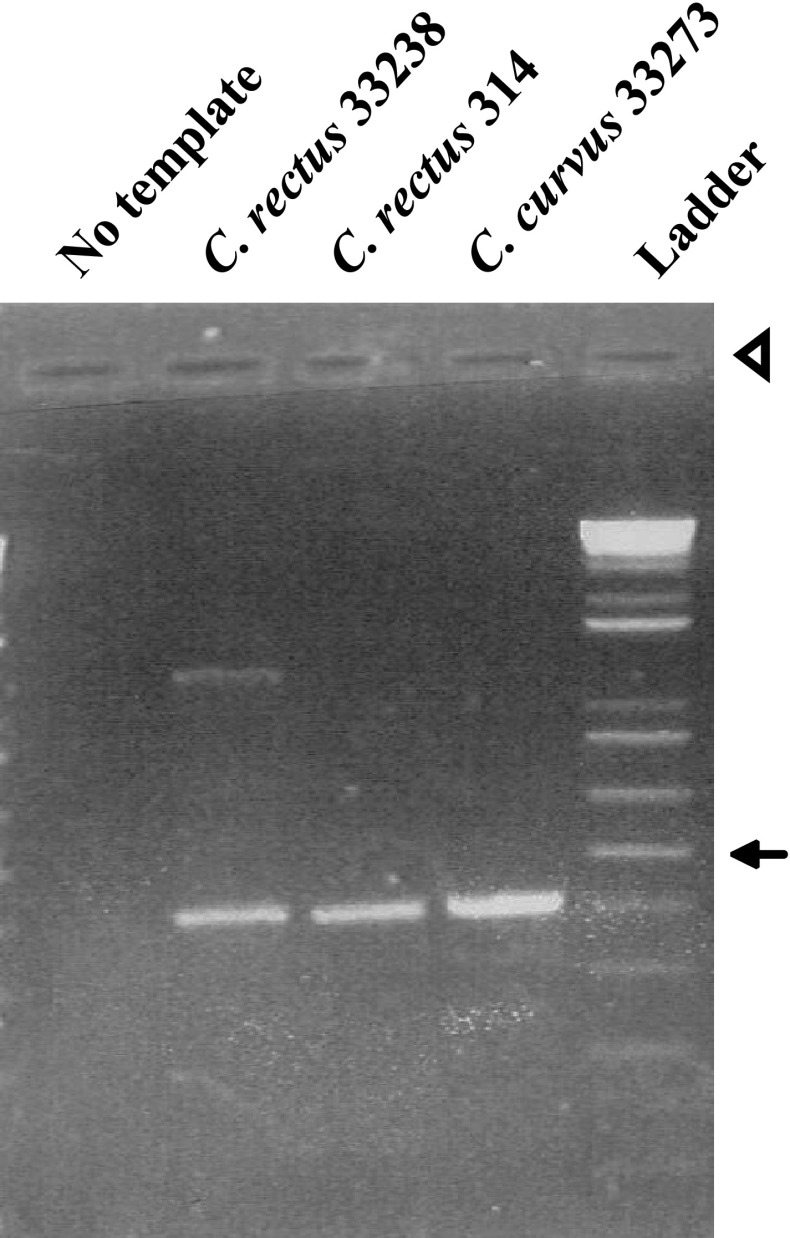
Degenerate PCR was used to amplify a potential ciaB gene from DNA of C. rectus 314. PCR yielded an amplicon of the expected size (448 base pairs, Fig. 1). Amplicons of the correct size were also detected from C. rectus 33238 and C. curvus 33273. C. curvus, like C. rectus, is an oral bacteria associated with periodontitis [36]. BLASTx analysis of the amplicon from strain 314 revealed a gene fragment sharing sequence identity and similarity (53 and 70 %, respectively) to the [21] ciaB gene from C. jejuni (NCBI, AAD38497.1). The sequence of the 448 base pair amplicon was used as a template for PCR-based gene walking. PCR-based gene walking generated sequences upstream and downstream of the 448 base pair amplicon. Amplicons of 1,060 base pairs (downstream) and 892 base pairs (upstream) were cloned into pCR2.1-TOPO for sequencing (data not shown).
Fig. 1.
Agarose gel of C. rectus or Campylobacter curvus DNA amplified by degenerate PCR (Table 1). An arrow marks 500 base pairs. Ladder 1 Kb plus DNA ladder (Invitrogen); and the arrowhead marks loading wells
In Silico Analyses of C. rectus ciaB
Assembly of DNA sequences from PCR and gene walking revealed a ciaB ORF of 1,830 base pairs. The 1,830 base pair sequence is available at NCBI (#JN588592). The ORF included a start codon (GTG), stop codon (TAG), and ribosome-binding site 6 bases pairs upstream of the start codon (Shine-Delgaro sequence, AAAAGG). Translation of the ORF reveals a protein of 609 amino acids, with a molecular weight of 69.8 kDa. BLASTx analysis of the C. rectus 314 ciaB, named Cr-ciaB, revealed sequence identity and similarity to genes annotated as a hypothetical protein, a lipoprotein signal peptidase, and invasion antigen B (ciaB) proteins (Table 3). An alignment (Fig. 2) of Cr-CiaB showed homology with CiaB proteins, including the presence of a zinc-binding domain conserved among CiaB [22].
Table 3.
BLASTx analysis of C. rectus strain ciaB
| Speciesa | Protein length (aa) | E-value | Annotation (GenBank #) | Identity (%) |
|---|---|---|---|---|
| C. rectus (strain 33238) | 609 | 0.0 | Lipoprotein signal peptidase (ZP03610758.1) | 100 |
| C. curvus | 608 | 0.0 | Hypothetical protein (YP001407631.1) | 64 |
| C. concisus | 614 | 0.0 | Invasion antigen B (EAT98433.1)b | 60 |
| C. fetus | 606 | 0.0 | Invasion antigen B (YP892436.1) | 60 |
| C. jejuni | 610 | 0.0 | Invasion antigen B (AAD38497.1)c | 53 |
| C. lari | 611 | 0.0 | Invasion antigen B (BAH23913.1)d | 51 |
Fig. 2.
Alignment of Campylobacter rectus 314 CiaB (GenBank # JN588592) with putative homologues from: Campylobacter concisus (EAT98433.1, Table 3); Campylobacter fetus (YP 892436.1, Table 3); and Campylobacter jejuni (AAD38497.1, Table 3). An asterisk indicates a conserved residue, moderate conservation is marked by a colon, and a period indicates weak conservation. Grey denotes a zinc-binding domain conserved among CiaB [19]
To further characterize Cr-CiaB, the protein was analyzed using tools designed to predict select cellular properties of genes. Specifically, Cr-ciaB was predicted not to contain membrane helices, did not contain a signal peptide, and is localized to the cytoplasm (Table 4). An identical analysis using related sequences (Table 3), including the lipoprotein signal peptidase from C. rectus 33238, a hypothetical protein from C. curvus, and CiaB proteins from related campylobacters; predicted a lack of membrane helices, a lack of signal peptides, and a cytoplasmic localization (Table 4). For predictions using PSORT-B, a score of greater than 7.5 is considered significant.
Table 4.
Predicted properties of C. rectus CiaB and related sequences
| Species/genea | Membrane helicesb | Signal peptide probabilityc | Location (score)d |
|---|---|---|---|
| C. rectus 314 CiaB (JN588592) | No | 0.0 | Cytoplasm (8.96) |
| C. rectus 33238 signal peptidase (ZP03610758.1) | No | 0.0 | Cytoplasm (8.96) |
| C. curvus hypothetical protein (YP 001407631.1) | No | 0.0 | Cytoplasmic (8.96) |
| C. concisus CiaB (EAT98433.1) | No | 0.0 | Cytoplasmic (8.96) |
| C. fetus CiaB (YP 892436.1) | No | 0.0 | Cytoplasmic (8.96) |
| C. jejuni CiaB (AAD38497.1) | No | 0.0 | Cytoplasmic (8.96) |
| C. lari CiaB (BAH23913.1) | No | 0.0 | Cytoplasmic (8.93) |
Analysis of Cr-ciaB showed sequence identity (Table 3, ZP03610758.1) with a gene from C. rectus 33238 annotated as a lipoprotein signal peptidase (Cr-LspA). However, in comparing the predicted properties of Cr-CiaB and Cr-LspA to the characteristics of bacterial LspA from Escherichia coli and C. jejuni [37], significant differences were observed. In contrast to Cr-CiaB and Cr-LspA, the bacterial LspA are predicted to contain membrane helices and are likely to be located within membranes (Table 5). For E. coli LspA, the membrane helices and cellular location have been demonstrated [38]. In addition, bacterial LspA are less than 200 amino acids long. In contrast, CiaB proteins are greater than 600 amino acids (Table 5).
Table 5.
Properties of CiaB versus lipoprotein signal peptidases (LspA)
| Species/genea | Membrane helicesb | Protein length (aa) | Cellular location (score)c | |
|---|---|---|---|---|
| C. rectus 314 CiaB (JN588592) | No | 609 | Cytoplasm (8.96) | |
| C. rectus 33238 signal peptidase (ZP03610758.1)d | No | 609 | Cytoplasm (8.96) | |
| C. concisus CiaBd (EAT98433.1) | No | 614 | Cytoplasm (8.96) | |
| C. jejuni CiaBd (AAD38497.1) | No | 610 | Cytoplasm (8.96) | |
| Escherichia coli LspA (P00804) | Yes, 4 | 164 | Membrane (9.82) | |
| C. jejuni LspA (Q9PIE1) | Yes, 4 | 156 | Membrane (10.0) | |
| C. rectus 33238 signal peptidase II (ZP03609438.1)e | Yes, 4 | 188 | Membrane (9.86) | |
Expression of Cr-ciaB and Conservation Among C. rectus Strains
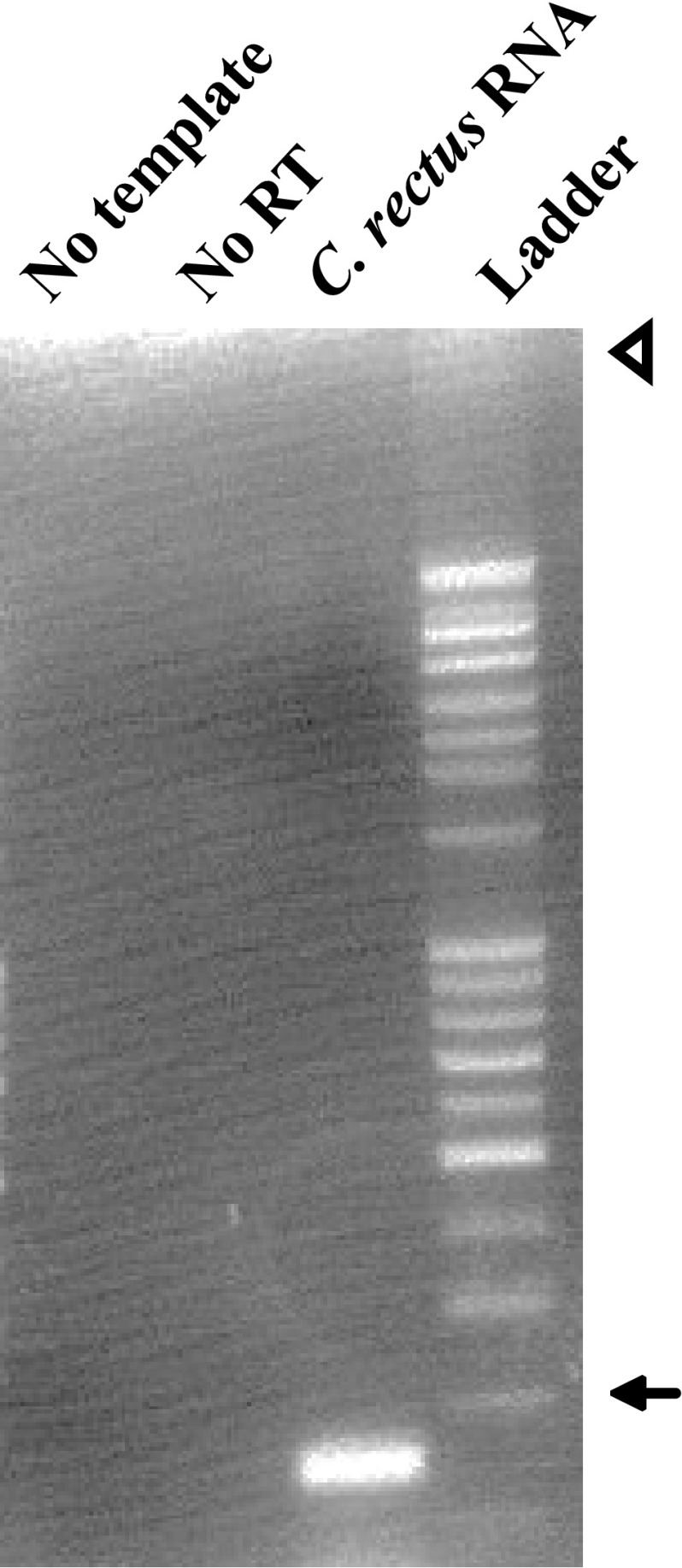
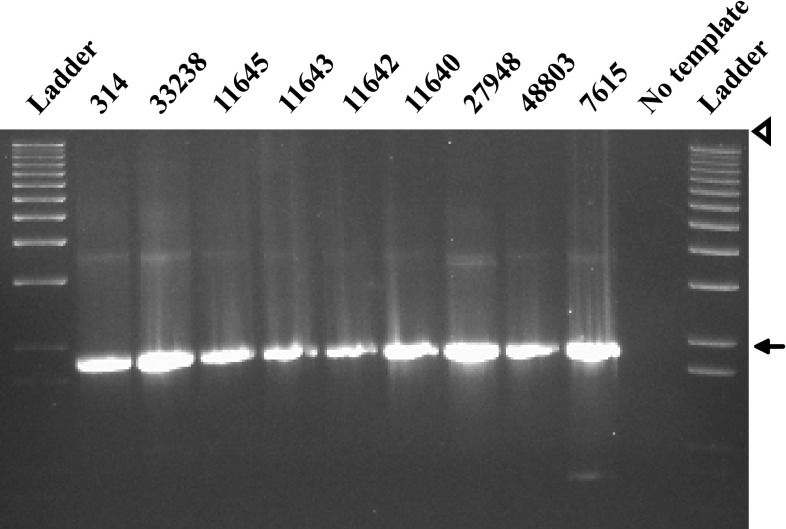
Expression of Cr-ciaB was shown by RT-PCR (Fig. 3). In addition, PCR suggests that CiaB is conserved among C. rectus strains (Fig. 4).
Fig. 3.
Agarose gel of C. rectus strain 314 RNA amplified with RT-PCR. No template no RNA template control, No RT no reverse-transcriptase control, Ladder 1 Kb plus DNA ladder. An arrow marks 100 base pairs; and the arrowhead marks loading wells
Fig. 4.
Agarose gel of C. rectus DNA amplified using primers designed to amplify the complete ciaB gene (Table 2). Each number refers to a different strain of C. rectus (Table 1). An arrow marks 2,000 base pairs. Ladder = 1 Kb Plus DNA ladder; and the arrowhead denotes loading wells
Phylogeny of Cr-CiaB
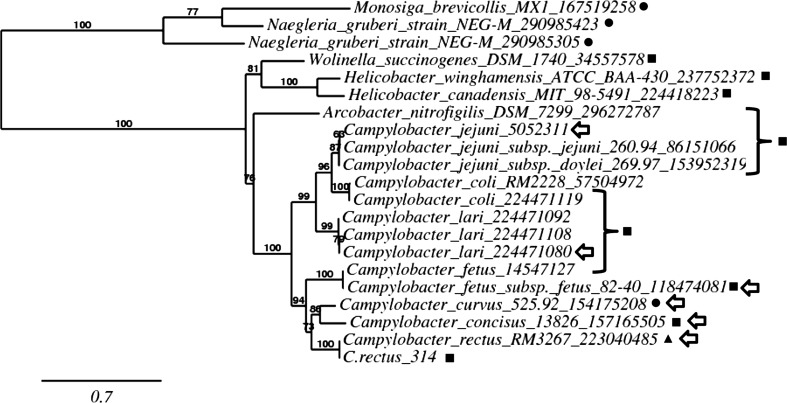
A maximum likelihood tree of Cr-CiaB and related sequences is depicted in Fig. 5. As shown, CiaB proteins from related bacteria form a major branch of the tree that is significantly supported (100 %). Within the main branch, a cluster of related CiaB proteins from campylobacters was noted. The CiaB proteins among the campylobacters are sorted according to species, including a group of CiaB from the oral campylobacters. Significantly, Cr-CiaB and the gene from C. rectus 33238 annotated as a lipoprotein signal peptidase clearly places both genes among CiaB proteins.
Fig. 5.
Phylogram of C. rectus strain 314 CiaB and related genes. Values at nodes represent branch support [35]. The NCBI gene identifier is indicated for each species. Proteins marked by a square are annotated as CiaB. Proteins marked with a circle are annotated as hypothetical proteins. The protein marked by a triangle is annotated as a lipoprotein signal peptidase. Proteins marked with an arrow are those presented in Table 3. The scale bar represents evolutionary distance (0.7)
Discussion
In this study a ciaB gene from C. rectus 314, a poorly understood periodontal pathogen, was identified and characterized. Initial analysis of C. rectus 314 ciaB (Cr-ciaB) was surprising as BLASTx revealed 100 % sequence identity to a C. rectus 33238 gene annotated as a lipoprotein signal peptidase. However, further characterization of the 33238 gene; along with parallel studies of C. curvus YP001407631.1 and Campylobacter concisus EAT98433.1, suggests that all three genes represent CiaB proteins, not lipoprotein signal peptidases. The possibility that the genes from C. rectus 33238, C. curvus, and C. concisus were annotated incorrectly is plausible. Indeed, between 30 and 40 % of annotated microbial genes are assigned incorrect functions using automated software [39]. Of course, future biochemical and genetic studies are needed to fully define the function of C. rectus 314 ciaB.
CiaB genes have been identified from several species of campylobacters that do not typically reside in the oral cavity of humans including C. jejuni, C. lari, C. coli, C. fetus, and C. upsaliensis [21, 22]. However, during the course of characterizing the ciaB from C. rectus 314, potential ciaB genes were identified among campylobacters that do reside within the oral cavity of humans (Table 3) including C. rectus 33238 (ZP03610758.1, annotated at NCBI as a lipoprotein signal peptidase), C. curvus (YP001407631.1, annotated as a hypothetical protein), and C. concisus (EAT98433.1, annotated as both lipoprotein signal peptidase and an invasion antigen B). The conservation of CiaB proteins among oral and non-oral species suggests that CiaB may be important to the lifestyle of campylobacters, independent of niche preference. In addition, this study represents the first effort to characterize a ciaB gene from any of the oral campylobacters.
Among the best studied virulence factors of campylobacters is the invasion antigen gene B (ciaB) of C. jejuni, which has been shown to play a key role in host cell invasion [18–20]. In C. jejuni, CiaB has been shown to be a secreted protein, although it lacks a predictable signal peptide [19]. Interestingly, C. rectus 314 CiaB also appears to lack a signal peptide (Table 3). As previously mentioned, C. rectus 314, like C. jejuni, has been shown to invade eukaryotic cells, including placental trophoblasts [23]. Invasion of placental trophoblasts is important, as previous studies have suggested that C. rectus plays a significant role in the preterm labors of mothers with periodontitis [10, 11] and has the ability to translocate to the placenta, from a distant site of infection, in mouse models of periodontitis.
Conclusions
The suite of genes that play a role in the pathogenesis of C. rectus, including those that contribute to the process of cell invasion, are unknown. However, the discovery of C. rectus 314 CiaB in this study, taken together with the observed cell invasion by C. rectus, suggests that CiaB may play a role in this phenotype [23]. Future studies aim to verify the function of CiaB biochemically. In addition, the identification of C. rectus CiaB raises the possibility of developing novel periodontal therapeutics that disrupt CiaB functionality.
Acknowledgments
Michael J. LaGier was supported in part by Florida Gulf Coast University (Internal Grant #09101). This study was also supported in part by the NSF-POWRE MCB-9973861 to Deborah S. Threadgill.
References
- 1.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 2.Teng YT, et al. Periodontal health and systemic disorders. J Can Dent Assoc. 2002;68:188–192. [PubMed] [Google Scholar]
- 3.Colombo AV, et al. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 4.Dzink JL, et al. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;5:648–659. doi: 10.1111/j.1600-051X.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 5.Lai CH, et al. Wolinella recta in adult gingivitis and periodontitis. J Periodontal Res. 1992;27:8–14. doi: 10.1111/j.1600-0765.1992.tb02079.x. [DOI] [PubMed] [Google Scholar]
- 6.Bobetsis YA, Barros SP, Offenbacher S. Exploring the relationship between periodontal disease and pregnancy complications. J Am Dent Assoc. 2006;137:7S–13S. doi: 10.14219/jada.archive.2006.0403. [DOI] [PubMed] [Google Scholar]
- 7.Offenbacher S, et al. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol. 1996;67:1103–1113. doi: 10.1902/jop.1996.67.10s.1103. [DOI] [PubMed] [Google Scholar]
- 8.Madianos PN, et al. Maternal periodontitis and prematurity Part II: maternal infection and fetal exposure. Ann Periodontol. 2001;6:175–182. doi: 10.1902/annals.2001.6.1.175. [DOI] [PubMed] [Google Scholar]
- 9.Borinski R, Holt SC. Surface characteristics of Wolinella recta ATCC 33238 and human clinical isolates: correlation of structure with function. Infect Immun. 1990;58:2770–2776. doi: 10.1128/iai.58.9.2770-2776.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Offenbacher S, et al. Effects of maternal Campylobacter rectus infection on murine placenta fetal and neonatal survival and brain development. J Periodontol. 2005;76:2133–2143. doi: 10.1902/jop.2005.76.11-S.2133. [DOI] [PubMed] [Google Scholar]
- 11.Yeo A, et al. Campylobacter rectus mediates growth restriction in pregnant mice. J Periodontol. 2005;76:551–557. doi: 10.1902/jop.2005.76.4.551. [DOI] [PubMed] [Google Scholar]
- 12.Hofreuter D, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson BM, et al. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828) J Bacteriol. 2007;189:8402–8403. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun M, et al. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J Bacteriol. 1999;181:2501–2506. doi: 10.1128/jb.181.8.2501-2506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaGier MJ, Threadgill DS. Identification of novel genes in the oral pathogen Campylobacter rectus. Oral Microbiol Immunol. 2008;2008(23):406–412. doi: 10.1111/j.1399-302X.2008.00443.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Kraig E, Kolodrubetz D. Use of defined mutants to assess the role of the Campylobacter rectus S-layer in bacterium-epithelial cell interactions. Infect Immun. 2000;68:1465–1473. doi: 10.1128/IAI.68.3.1465-1473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 18.Konkel ME, et al. Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 19.Konkel ME, et al. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouts DE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onozato J, et al. Cloning sequencing and expression of full-length Campylobacter invasion antigen B gene operon from Campylobacter lari. J Basic Microbiol. 2009;49:342–349. doi: 10.1002/jobm.200800214. [DOI] [PubMed] [Google Scholar]
- 23.Arce RM, et al. Characterization of the invasive and inflammatory traits of oral Campylobacter rectus in a murine model of fetoplacental growth restriction and in trophoblast cultures. J Reprod Immunol. 2010;84:145–153. doi: 10.1016/j.jri.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F, et al. Current protocols in molecular biology. New York: Wiley; 1990. [Google Scholar]
- 25.Wren BW, et al. Degenerate PCR primers for the amplification of fragments from genes encoding response regulators from a range of pathogenic bacteria. FEMS Microbiol Lett. 1992;78:287–291. doi: 10.1111/j.1574-6968.1992.tb05583.x. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh A, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 29.Emanuelsson O, et al. Locating proteins in the cell using TargetP SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 30.Rey S, et al. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–D168. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dereeper A, et al. Phylogenyfr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dereeper A, et al. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2008;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;2007(56):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 34.Guindon S, et al. PHYML online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chevenet F, et al. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mucach P, Tanner AC. Campylobacter species in health gingivitis and periodontitis. J Dent Res. 2000;79:785–792. doi: 10.1177/00220345000790021301. [DOI] [PubMed] [Google Scholar]
- 37.Paetzel M, et al. Signal peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 38.Yu F, et al. Nucleotide sequence of the lspA gene the structural gene for lipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1984;173:264–268. doi: 10.1016/0014-5793(84)81060-1. [DOI] [PubMed] [Google Scholar]
- 39.Poptsova MS, Gogarten JP. Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology. 2010;156:1909–1917. doi: 10.1099/mic.0.033811-0. [DOI] [PubMed] [Google Scholar]