Highlights
-
•
Host cell proteases activate the spike protein of the SARS-coronavirus.
-
•
Activation is essential for viral infectivity and a potential target for intervention.
-
•
Cathepsin L activates the spike protein in host cell endosomes.
-
•
TMPRSS2 activates the spike protein at the plasma membrane.
-
•
SARS- and MERS-coronavirus exploit the same proteases for activation.
Keywords: SARS, Cathepsin L, TMPRSS2, Spike protein, Protease, MERS
Abstract
The severe acute respiratory syndrome (SARS) pandemic revealed that zoonotic transmission of animal coronaviruses (CoV) to humans poses a significant threat to public health and warrants surveillance and the development of countermeasures. The activity of host cell proteases, which cleave and activate the SARS-CoV spike (S) protein, is essential for viral infectivity and constitutes a target for intervention. However, the identities of the proteases involved have been unclear. Pioneer studies identified cathepsins and type II transmembrane serine proteases as cellular activators of SARS-CoV and demonstrated that several emerging viruses might exploit these enzymes to promote their spread. Here, we will review the proteolytic systems hijacked by SARS-CoV for S protein activation, we will discuss their contribution to viral spread in the host and we will outline antiviral strategies targeting these enzymes. This paper forms part of a series of invited articles in Antiviral Research on “From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses.’’
1. Introduction
Coronaviruses are enveloped, single stranded RNA viruses that cause gastrointestinal, respiratory and neurological symptoms in several mammalian species and birds (Holmes, 2001). Animal coronaviruses can pose a severe threat to their hosts; for instance, transmissible gastroenteritis virus infection of newborn seronegative piglets almost invariably results in a fatal outcome (Laude et al., 1993). In contrast, the human coronaviruses (hCoV) identified so far, NL63, 229E, OC43 and HKU-1, usually induce common cold-like symptoms (Holmes, 2001, Wevers and van der Hoek, 2009), although a more severe disease can develop in young children, the elderly and immunocompromised patients (Chiu et al., 2005, Gorse et al., 2009, Jean et al., 2013, Jevsnik et al., 2012, van der Hoek et al., 2004, Woo et al., 2005). These viruses circulate globally, frequently display seasonality and were mainly studied because of their elaborate regulation of gene expression.
The perception of coronavirus infection as largely harmless for humans, which prevailed since the discovery of the first human coronaviruses, 229E and OC43, in the late 1960s, changed dramatically with the outbreak of the severe respiratory syndrome coronavirus (SARS-CoV) in Southern China in the winter of 2002. Infection with the novel coronavirus caused a severe respiratory disease, SARS, which took a fatal course in roughly 10% of the infected patients, with the risk for a fatal outcome increasing with age (Peiris et al., 2004, Stadler and Rappuoli, 2005). The virus rapidly spread throughout Asia and was introduced by international travel to more than 30 other countries. When the outbreak was finally contained on July 5, 2003, more than 8000 patients were afflicted and almost 800 had lost their lives (Cheng et al., 2007).
The outbreak of SARS was traced back to bats (Lau et al., 2005, Li et al., 2005b), which are believed to be the natural reservoir of SARS-CoV, and civet cats and raccoon dogs (Guan et al., 2003), which may have served as intermediate hosts. These findings revealed for the first time that zoonotic transmission of animal coronaviruses to humans can pose a serious threat to public health (Wang and Eaton, 2007). After the SARS pandemic, only sporadic, zoonotic infections were detected, with the exception of three laboratory infections, suggesting that the threat might have subsided. However, the recent emergence of the novel, highly pathogenic Middle East Respiratory Syndrome (MERS) CoV (Zaki et al., 2012) and the identification of closely related viruses in bats (Annan et al., 2013) demonstrate that the spillover of coronaviruses from animals to humans can occur at any time and with unforeseeable consequences for public health. Therefore, it is essential to understand how these viruses invade their hosts and cause disease and how these processes can be prevented.
Host cell entry is the first step in the viral life cycle and constitutes a target for treatment and prevention. Coronaviruses encode three surface proteins, spike (S), membrane (M) and envelope (E). The M and E proteins play a role in particle assembly and release (Masters, 2006), while the S protein binds to host cell receptors and fuses the viral membrane with a cellular membrane – processes essential for infectious entry (Heald-Sargent and Gallagher, 2012, Hofmann and Pöhlmann, 2004). In addition, the SARS-CoV spike protein (SARS-S) is the major target of the neutralizing antibody response (Hofmann et al., 2004, Simmons et al., 2004, Yang et al., 2004). As a consequence, the viral S protein has been a focus of SARS research. Only months after the discovery of the virus it was shown that SARS-S uses angiotensin-converting enzyme 2 (ACE2) as receptor for entry into host cells (Li et al., 2003, Wang et al., 2004), mainly type II pneumocytes and enterocytes (Hamming et al., 2004, To et al., 2004). More recent research demonstrated that SARS–S interactions with ACE2 are not sufficient for host cell entry. In addition, the S protein needs to be proteolytically activated by host cell proteases to fuse the viral and a target cell membrane and the cellular localization of the responsible enzymes determines in which cellular compartment membrane fusion occurs. The activity of host cell proteases is essential for SARS-CoV infectivity (Simmons et al., 2004), making these enzymes potential targets for intervention. In the present review, we will discuss the proteolytic pathways priming SARS–S for membrane fusion, we will introduce the responsible proteases and we will outline antiviral strategies targeting these proteases.
2. The SARS-coronavirus spike protein: Functional organization and membrane fusion reaction
Coronavirus S proteins, including SARS–S, share several features: Peplomers of coronavirus S proteins protrude from the virion surface and are responsible for the corona-like shape of virions observed upon electron microscopy. The S proteins contain N-terminal signal sequences and are synthesized in the constitutive secretory pathway of infected cells (Fig. 1 ). While they traverse the secretory pathway towards the site of assembly of progeny virions, the endoplasmatic reticulum – Golgi apparatus intermediate compartment (ERGIC), the S proteins trimerize and are extensively modified by N-glycans (Belouzard et al., 2012, Heald-Sargent and Gallagher, 2012). The functional organization of the S proteins resembles that of several other viral glycoproteins, including the influenza hemagglutinin and the human immunodeficiency virus envelope protein, collectively referred to as class I membrane fusion proteins (Harrison, 2008). A hallmark of these proteins is the presence of an N-terminal surface unit (termed S1 in the context of CoV S proteins), which harbors the receptor binding domain and a C-terminal transmembrane unit (termed S2 in the context of CoV S proteins), which contains the functional elements required for membrane fusion: a fusion peptide, heptad repeats and a transmembrane domain (Fig. 1). Notably, the separation of the surface and transmembrane units upon proteolytic cleavage by host cell proteases is a prerequisite to membrane fusion driven by class I membrane fusion proteins. A second characteristic shared by class I membrane fusion proteins is the transition of the transmembrane unit into a thermostable, protease-insensitive conformation termed six-helix-bundle upon successful completion of the membrane fusion reaction (Harrison, 2008).
Fig. 1.

Domain organization of the SARS-coronavirus spike protein. The surface unit, S1, contains an N-terminal signal peptide, which targets the protein for import into the constitutive secretory pathway, and a receptor binding domain (RBD), which mediates binding to ACE2. The transmembrane unit, S2, harbors the functional elements required for membrane fusion, a fusion peptide and two heptad repeats. It also contains a transmembrane domain, which anchors the protein within cellular membranes, and a cytoplasmic tail, which is required for appropriate intracellular trafficking of the spike protein. Arrows indicate amino acid positions cleaved by cellular proteases.
The first indispensable step of the entry cascade is the binding of the S proteins to a receptor. The receptor binding domain (RBD) of SARS–S is located at the C-terminus of the S1 subunit and binds to ACE2 with high affinity (Wong et al., 2004). The structure of the RBD in complex with the catalytic domain of ACE2 has been determined on the atomic level (Li, 2013, Li et al., 2005a). The RBD consists of two subdomains, a core and an extended loop. The core structure is conserved between SARS–S and other CoV S proteins but does not contact ACE2. In contrast, the structure of the extended loop differs among CoV S proteins and this subdomain is responsible for all SARS–S contacts with ACE2 (Li et al., 2005a). Sequence variations in the extended loop determine the affinity of SARS–S binding to ACE2 and have been associated with the potential of SARS-CoV for zoonotic transmission (Li, 2013, Li et al., 2006).
Binding of SARS–S to ACE2 triggers subtle conformational changes within the S protein, which render the S protein susceptible to activation for membrane fusion, as discussed below. Upon activation, a fusion peptide located at the N-terminus of the S2 subunit inserts into a target cell membrane, either the plasma membrane or an endosomal membrane, as described below. At this stage, the S2 subunit is connected with the viral membrane via its transmembrane domain and the cellular membrane via the fusion peptide. Subsequently, the N- and C-terminal heptad repeats within the S2 subunit fold back onto each other, forming the six-helix-bundle structure. As a consequence, the S2 subunit collapses and the viral and cellular membranes are pulled into close proximity and ultimately fuse. A similar mechanism is operative for all class I membrane fusion proteins. However, different stimuli can activate the viral fusion proteins and the one responsible for activation of SARS–S is in the focus of the remainder of this review.
3. Activation of the SARS-coronavirus spike protein by host cell proteases
3.1. The search for the cellular activator of the SARS-coronavirus spike protein
In order to mediate membrane fusion pH-independent viruses, such as HIV, utilize binding to cell surface (co)receptors in order to trigger the glycoprotein reorganization described above, while for pH-dependent viruses the low pH of acidified endosomal organelles is sufficient (Plemper, 2011). SARS-CoV entry is sensitive to lysosomotropic agents (Hofmann et al., 2004, Simmons et al., 2004, Yang et al., 2004), initially suggesting a requirement for acidic pH mediated triggering of SARS–S (Yang et al., 2004), similar to influenza. However, a number of factors suggested that entry might not be so straightforward for SARS-CoV (Simmons et al., 2004). Many other coronaviruses are pH-independent, with entry being insensitive to lysosomotropic agents (Nash and Buchmeier, 1997), and cell-to-cell fusion occurring at neutral pH (Kooi et al., 1991). Likewise, cell–cell fusion occurred in the absence of acidification, requiring addition of trypsin for efficient fusion in conditions of low ACE2 expression on target cells (Simmons et al., 2004). Similarly, the block to infection mediated by lysosomotropic agents could be bypassed by binding virus to the cell surface and then treating with trypsin, elastase or thermolysin (Matsuyama et al., 2005, Simmons et al., 2005). Finally, many pH-dependent viruses, including influenza and dengue, are inactivated by premature exposure to low pH – irreversibly driving the glycoproteins through their conformational rearrangements, and into the post-fusion state prior to contact with target membrane. In contrast, pulsing SARS–S pseudotyped onto lentiviral particles did not inactivate infectivity (Simmons et al., 2004). The pH-dependent rhabdoviruses are also insensitive to premature triggering (Steffen et al., 2013), due to the reversible nature of their glycoprotein (Backovic and Jardetzky, 2011). However, SARS–S has more structural features in common with class I viral glycoproteins such as HIV gp160 and influenza hemagglutinin (HA) than with the rhabdovirus glycoproteins, which belong to the family of class III membrane fusion proteins.
3.2. Cathepsin L activates SARS-CoV spike protein in host cell endosomes
One explanation for these discordant results was that pH-dependent endosomal cellular factors were required for SARS-CoV membrane fusion, rather than the virus requiring an acidic trigger itself. Influenza HA can be primed for infectivity within the endosomes of certain target cells by endosomal proteases (Boycott et al., 1994), suggesting similar circumstances may occur for SARS-CoV. Indeed, using broad-spectrum, followed by more specific inhibitors, it was demonstrated that SARS-CoV requires the activity of an endosomal cysteine protease, most likely cathepsin L (Huang et al., 2006, Simmons et al., 2005) (Fig. 2 ). Cathepsin L is ubiquitously expressed, explaining why once human ACE2 is added to a cell line, no further restrictions appear to exist for efficient entry. Few inhibitors are entirely specific, however, using a partially cell-free model of membrane fusion it was demonstrated that cathepsin L, but not other cathepsins, primes SARS-CoV for membrane fusion, but only after interaction with ACE2 at temperatures above 4 °C (Simmons et al., 2005). Thus, we can theorize a model where engagement of ACE2 by SARS–S triggers subtle conformational changes within the S protein, which render the S protein susceptible to activation by host cell proteases, likely by exposure of a cryptic cleavage site. Cleavage of the S2 subunit of SARS–S by cellular proteases then liberates a conserved fusion peptide, which is located directly adjacent to the cleavage site (Madu et al., 2009). Upon S protein cleavage, the fusion peptide can insert into a target cell membrane and the membrane merger commences as described above.
Fig. 2.

Routes employed by the SARS-coronavirus for entry into target cells. The SARS-coronavirus can employ two routes for host cell entry, which are determined by the localization of the proteases required for activation of the SARS-coronavirus spike protein. Binding of SARS-coronavirus to the cellular receptor, ACE2, can result in uptake of virions into endosomes, where the spike protein is activated by the pH dependent cysteine protease cathepsin L. Activation of the spike protein by cathepsin L can be blocked by lysosomotropic agents, like bafilomycin A1 and ammonium chloride, which indirectly inhibit cathepins L activity by interfering with endosomal acidification, or by compounds which directly block the proteolytic activity of cathepsin L, like MDL28170. Alternatively, the spike protein can be activated by TMPRSS2 at (or close to) the cell surface, resulting in fusion of the viral membrane with the plasma membrane.
The precise sites of proteolysis by cathepsin L are unclear. Unlike other proteases that prime viral glycoproteins, such as furin, cathepsin L is relatively indiscriminate in terms of recognition sites (Biniossek et al., 2011). Thus, it may be that rather than a defined site for proteolysis, S2 is trimmed at multiple cleavage points. A cathepsin L cleavage site was reported close to the predicted S1/S2 boundary, at position T678 in SARS–S (Bosch et al., 2008), however no functional data was shown, such as mutagenesis, to demonstrate that this site was indeed required to prime the S protein for infectivity. Based on data from other proteases, such as trypsin and elastase, although an initial cleave may occur in that site (Belouzard et al., 2009, Belouzard et al., 2010), and introduction of a complete furin cleavage site enhances cell–cell fusion (Follis et al., 2006) the critical sites of proteolysis are located in S2 (positions 795–797), closer to the predicted fusion peptide (Belouzard et al., 2009, Madu et al., 2009).
Other than academic interest, the fact that several coronaviruses utilized cathepsin L for entry (Bertram et al., 2013, Gierer et al., 2013, Hofmann et al., 2006, Kawase et al., 2009, Qiu et al., 2006, Simmons et al., 2005) offered the opportunity for rapid development of antiviral compounds. The cathepsins have long been a target of pharmaceutical interest (Vasiljeva et al., 2007) for diseases such as osteoporosis, cardiovascular disease and cancer, due to their myriad of functions that impact on aspects of disease pathogenesis such as collagen degradation (Chapman et al., 1997) and antigen processing (Pietschmann et al., 2013). Indeed, several libraries of cysteine protease inhibitors from such sources have been compiled, initially with the intent of using them to screen for inhibitors of closely related cathepsin-like proteases found in many parasitic infections (Leslie, 2011). These libraries have proved excellent for mining for both antiparasitic (Ang et al., 2011) and antiviral (Zhou et al., 2011) drug leads. These compounds are more extensively reviewed elsewhere (Zhou and Simmons, 2012). Briefly, inhibitors are generally peptidic and bind in the active site of the enzyme, either reversibly in the case of MDL28170 and a potent tetrahydroquinoline oxocarbazate compound (Shah et al., 2010, Simmons et al., 2005) or irreversibly, as in the case of the dipeptidyl epoxyketone inhibitors (Zhou et al., 2011). In contrast, a small molecule inhibitor of cathepsin L was recently identified in screening assays using SARS-S bearing pseudotypes, and may have more specificity than peptidomimetic inhibitors as cathepsin B activity was not inhibited (Adedeji et al., 2013). However, it is not entirely clear which strategy, if any, would be adventurous for therapeutic treatment, based on results from inhibitor escape mutants. The filoviruses are similar to SARS-CoV in that they require cysteine proteases for efficient viral entry, although cathepsin B plays a greater role than cathepsin L in Ebolavirus glycoprotein priming (Chandran et al., 2005), while unidentified proteases perform this function for other filoviruses (Gnirss et al., 2012, Misasi et al., 2012). Ebolavirus escape mutants can relatively easily be made resistant to inhibitors of cathepsin B, with the mutants remaining sensitive to broad spectrum cysteine protease inhibitors, suggesting the virus has switched protease usage (Wong et al., 2010). Thus, we hypothesize that more broadly active protease inhibitors may lessen or even prevent escape (Zhou and Simmons, 2012).
3.3. Type II transmembrane serine proteases activate the SARS-coronavirus at the cell surface
A key stimulus to investigate whether SARS–S can hijack proteases other than cathepsin L to ensure its activation came from novel insights into the activation of influenza virus by host cell proteases. Influenza viruses, like SARS-CoV, mainly infect respiratory epithelia and depend on the proteolytic activation of their entry-mediating surface protein, hemagglutinin (HA), for acquisition of infectivity (Bertram et al., 2010b, Böttcher-Friebertshauser et al., 2013). On the basis of cell culture studies it has been posited that HA may be activated by several host cell proteases secreted into the lung lumen (Kido et al., 2007). A milestone study by Zhirnov et al. (2002) indicated that this might not be the case: Activation of HA in cultured human adenoid epithelial cells, a model for human respiratory epithelium, was shown to be a cell-associated process occurring during HA biogenesis in infected cells and during viral entry into target cells. However, the identities of the proteases involved remained unclear. Böttcher et al. and Chaipan et al. demonstrated that members of the type II transmembrane serine protease (TTSP) family are prime candidates: TMPRSS2 (transmembrane protease serine 2) and HAT (human airway tryptase) were shown to activate all influenza virus subtypes previously pandemic in humans (Böttcher et al., 2006) and TMPRSS4 was found to activate the HA protein of the 1918 influenza virus (Chaipan et al., 2009). More recent studies added MSPL, TMPRSS13 and matriptase to the list of HA activating TTSPs (Baron et al., 2013, Beaulieu et al., 2013, Hamilton et al., 2012, Okumura et al., 2010). In addition, evidence was reported that TMPRSS2 activates human metapneumovirus by cleavage of the fusion protein (Shirogane et al., 2008), indicating that several respiratory viruses may hijack TMPRSS2 and related proteases to promote their spread in the host.
In humans the TTSP family comprises 19 members for most of which expression in the respiratory tract has been reported (Antalis et al., 2011). The proteins exhibit a modular organization (Fig. 3 ). The N-terminus is located in the cytoplasm and can interact with components of the cytoskeleton and signaling molecules. It is followed by a transmembrane domain, which inserts the proteins into the plasma membrane. The extracellular portion comprises an N-terminal stem domain, which is built of combinations of up to 11 structural domains, and a C-terminal, trypsin-like protease domain (Antalis et al., 2011). The stem region has a regulatory function, including modulation of the enzymatic activity and substrate specificity, and mediates protein–protein interactions. The C-terminal protease domain contains a catalytic triad consisting of serine, histidine and aspartate, which recognizes and cleaves substrates, including hormones, growth and differentiation factors and viral surface proteins (Antalis et al., 2011). The TTSPs are synthesized as inactive proenzymes, zymogens, which are activated upon proteolytic cleavage between the pro-domain and the catalytic domain. In the mature protein both domains remain covalently associated via a disulfide bond (Fig. 3) but shedding of the enzymatically active ectodomain has been reported (Antalis et al., 2011). Although the physiological roles of for instance TMPRSS2 and TMPRSS4 are largely unknown, it is well established that TTSPs regulate diverse biological processes, including development and homeostasis, and that dysregulated TTSP expression is intimately associated with various cancers (Antalis et al., 2011). Thus, TTSPs play an important role in health and disease and a constantly growing body of evidence suggests that viruses exploit these enzymes to promote their spread in and between hosts.
Fig. 3.
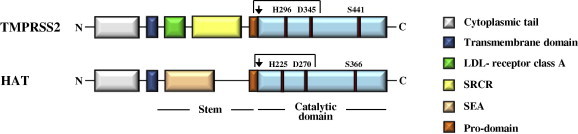
Domain organization of TMPRSS2 and HAT. TMPRSS2 and HAT, both members of the type II transmembrane serine protease family, contain an N-terminal cytoplasmic domain, a transmembrane domain, a stem region and a catalytic domain. The stem region of TMPRSS2 harbors a LDL-receptor class A domain and a scavenger receptor cysteine-rich domain (SRCR), while sperm protein, enterokinase and agrin (SEA) domain is present in the stem region of HAT. The catalytic domains of both proteases contain a catalytic triad, consisting of a serine (S), histidine (H) and aspartate (D) residue, which is essential for enzymatic activity. Both enzymes are synthesized as inactive precursors, zymogens, and transit into their active form upon cleavage between the pro- and catalytic domain (indicated by an arrow). In the mature enzyme, the catalytic domain and the remainder of the protein covalently associated via a disulfide bond.
An important role of TTSPs in SARS-CoV infection was first reported by three studies examining the impact of TMPRSS2 on SARS–S activity. They show that TMPRSS2 expression in ACE2+ target cells results in cleavage and activation of SARS-S for cell–cell and virus–cell fusion (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). Activation by TMPRSS2 renders SARS–S-driven virus–cell fusion independent of cathepsin activity (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011) but increases susceptibility to inhibition by peptides targeting the fusion apparatus in the S2 subunit (Matsuyama et al., 2010). As a consequence, one must assume that SARS–S activation by TMPRSS2 occurs at or close to the cell surface. However, detailed information on the cellular localization of the SARS–S activation reaction is not available at present. The ability of TMPRSS2 to activate SARS–S seems to depend on the spatial orientation of the two proteins relative to each other: If TMPRSS2 is expressed on target cells it can activate SARS–S on adjacent effector cells while coexpression of TMPRSS2 and SARS–S in the same cells does not activate the S protein (Glowacka et al., 2011, Matsuyama et al., 2010). Nevertheless, SARS–S cleavage can be detected upon coexpression of TMPRSS2 in the same cell and results in the shedding of the SARS–S ectodomain into the extracellular space, where it functions as an antibody decoy (Glowacka et al., 2011). Therefore, TMPRSS2 might impact SARS-CoV spread in at least two ways: Activation of SARS–S for membrane fusion could promote viral spread while release of soluble SARS–S could compromise viral control by the humoral immune response. The latter process has so far only been demonstrated in transfected cells and it remains to be investigated whether it is operative in the infected host.
The contribution of TTSPs to SARS-CoV infection is not limited to TMPRSS2, since the related protease HAT can also activate SARS–S (Bertram et al., 2011). However, several important differences between SARS–S activation by HAT and TMPRSS2 were noted: The SARS–S residue R667, which is essential for SARS–S cleavage by trypsin (Belouzard et al., 2009, Kam et al., 2009), is also required for SARS–S processing by HAT but not TMPRSS2 (Bertram et al., 2011). Although formal proof that R667 is indispensable for HAT-mediated SARS–S activation is currently lacking, these results indicate that the S protein is differentially recognized by HAT and TMPRSS2. Whether the differential recognition is due to differences in the protease domains or the remainder of the proteins is currently unknown. Regardless of the determinants controlling cleavage specificity, it is clear that the differential processing of SARS–S by TMPRSS2 and HAT has important consequences for SARS–S activation. For one, HAT but not TMPRSS2 can activate coexpressed S protein for cell–cell fusion (Bertram et al., 2011) and might contribute to syncytia formation in infected tissues. Conversely, TMPRSS2 but not HAT expressed on viral target cells can activate SARS–S for cathepsin-independent virus–cell fusion (Bertram et al., 2011). Differences in the expression levels or cellular localization (Böttcher-Friebertshauser et al., 2010) of these proteases could partially account for these findings. However, it is more likely that differential SARS–S proteolytic processing by TMPRSS2 and HAT results in differential SARS–S activation. Addressing this question requires the identification of the so far elusive TMPRSS2 cleavage sites in SARS–S. Finally, it is noteworthy that TMPRSS4 can activate SARS–S for cell–cell but not virus–cell fusion although SARS–S cleavage by TMPRSS4 was not detected (Glowacka et al., 2011). It is conceivable that inefficient and thus undetectable cleavage of SARS–S is still sufficient to promote cell–cell fusion, which involves the interactions of large cellular surfaces usually harboring high copy numbers of S protein and protease, respectively. Whether inefficient SARS–S processing occurs upon engineered expression of TMPRSS4 and accounts for S protein activation remains to be determined.
Activation of SARS-CoV by TTSPs in the infected host requires expression of these proteins in viral target cells. Analysis of lung tissue from uninfected cynomolgus macaques revealed prominent TMPRSS2 expression in type I pneumocytes while ACE2 expression was detected in type II pneumocytes, although at low levels (Matsuyama et al., 2010). Within severe lesions in the lungs of SARS-CoV infected animals no obvious correlation between the presence of ACE2, TMPRSS2 and SARS-CoV antigen was seen but TMPRSS2 and ACE2 were detected in the cytoplasm of enlarged type II pneumocytes (Matsuyama et al., 2010). Analysis of human tissue is so far limited to material from patients not infected by SARS-CoV. Immunohistochemistry revealed ACE2 and TMPRSS2 coexpression in alveolar macrophages and type II pneumocytes (Glowacka et al., 2011). A more comprehensive study detected extensive coexpression of ACE2, TMPRSS2 and HAT in the epithelia of the aerodigestive tract, although exceptions were noted, including the epithelia of trachea, vocal folds and epiglottis (Bertram et al., 2012). Thus, TMPRSS2 and HAT are present in major viral target cells and could promote viral spread in infected humans.
What is the evidence that TTSP expression in respiratory epithelium is indeed responsible for SARS-CoV activation in this tissue? Studies with cultured primary human respiratory epithelium addressing this question are needed. Interesting information obtained with a lung adenocarcinoma cell line, Calu-3, is already available: SARS-CoV infection of these cells, which express endogenous TMPRSS2 (Böttcher-Friebertshauser et al., 2011), is largely dependent on this protease as determined by inhibitor studies but full inhibition of infection is only achieved upon simultaneous blockade of both TMPRSS2 and cathepsin L (Kawase et al., 2012). Similar results were recently reported for hCoV-229E infection (Bertram et al., 2013) and MERS-CoV infection (Gierer et al., 2013) of a colorectal adenocarcinoma cell line, Caco-2, which endogenously expresses TMPRSS2 (Bertram et al., 2010a). Collectively, these results hint towards an important contribution of TMPRSS2 and, to a lesser degree, cathepsin L to the activation of SARS-CoV, hCoV-229E, MERS-CoV and potentially other human coronaviruses in the infected human host. Further evidence for an important role of TMPRSS2 in the infection by SARS-CoV and other human respiratory viruses has to come from animal models. TMPRSS2 knock-out mice (Kim et al., 2006), which display no obvious phenotype in the absence of infections, are an ideal tool for these endeavors.
The constantly increasing evidence pointing towards activation of SARS-CoV by TMPRSS2 might also have immediate therapeutic implications. The small molecule serine protease inhibitor camostat mesylate efficiently inhibits SARS-CoV activation by TMPRSS2 (Kawase et al., 2012). Camostat mesylate was already shown to inhibit replication of influenza and parainfluenza viruses and to prevent the development of pneumonia and viral myocarditis in infected mice (Zhirnov et al., 2011). In addition, the compound slows down or inhibits chronic pancreatitis in animal models (Jia et al., 2005) and has been used in Japan for treatment of human patients with this condition. Thus, camostat mesylate could also be employed for treatment of patients infected with SARS-CoV or MERS-CoV, as discussed below. In addition, the development of novel synthetic TMPRSS2 inhibitors which suppress influenza virus spread has recently been reported (Meyer et al., 2013). Treatment of SARS-CoV or influenza virus infected patients with these compounds is unlikely to completely abrogate viral spread, since alternative routes of activation are available, as discussed above. However, even a modest reduction of viral amplification, which can be expected based on the cell culture findings detailed above, might well translate into a therapeutic benefit and may tip the balance towards control of viral replication.
3.4. Activation of the SARS-coronavirus spike protein by an unidentified leupeptin-sensitive protease
It is generally accepted that cathepsin L and TTSPs are the major activators of SARS–S. However, studies of SARS–S-driven cell–cell fusion indicate that they might not be the only ones. Fusion of effector 293T cells expressing SARS–S with 293T target cells endogenously expressing low amounts of ACE2 is inefficient but can be rescued either by engineered expression of high levels of ACE2 or by trypsin treatment (Bertram et al., 2011, Simmons et al., 2011). These observations raise the questions whether SARS–S-driven fusion with ACE2-positive target cells in the absence of exogenous trypsin depends on SARS–S activation by a so far uncharacterized cellular protease. Indeed, leupeptin but not other protease inhibitors, including a cathepsin B/L inhibitor, reduced fusion of SARS–S expressing effector 293T cells with unmodified target 293T cells (Simmons et al., 2011). Endogenous expression of SARS–S-activating TTSPs has not been detected in 293T cells. Therefore, the above discussed results suggest that a so far unidentified leupeptin-sensitive protease can active SARS–S for cell–cell fusion. The nature of this protease and its potential contribution to SARS–S activation in other cellular systems and in the infected host remain to be determined.
4. Proteolytic processing of ACE2 promotes SARS-coronavirus entry and lung pathogenesis
The studies discussed above unambiguously demonstrate that proteolytic cleavage of SARS–S is essential for infectious entry of SARS-CoV. An additional layer of complexity has been added to the SARS–S/protease theme by observations indicating that not only SARS–S but also its receptor, ACE2, is proteolytically processed. ACE2 is known to be shed into the extracellular space upon cleavage by the sheddase ADAM17/TACE and shedding can be induced by treatment of cells with PMA and other agents (Jia et al., 2009, Lambert et al., 2005) (Fig. 4 ). Haga et al. (2008) demonstrated that binding of SARS–S also induces ACE2 shedding by ADAM17 and provided evidence that shedding is important for uptake of SARS-CoV into target cells. Furthermore, it was demonstrated that an ADAM17 inhibitor displays modest antiviral activity in SARS-CoV infected mice (Haga et al., 2010). Reduced cellular levels of ACE2 were noted in SARS-CoV infected cell cultures (Glowacka et al., 2010, Haga et al., 2008), animals (Kuba et al., 2005, Rockx et al., 2009) and humans (Oudit et al., 2009) and might be accounted for by SARS–S-induced shedding of ACE2. Notably, ACE2 expression protects mice against experimentally induced lung injury (Imai et al., 2005, Kuba et al., 2005). Therefore, SARS–S-induced ACE2 shedding might not only promote viral uptake but might also contribute to SARS pathogenesis (Fig. 4), although it should be noted that the contribution of ACE2 shedding to viral uptake is controversial (Glowacka et al., 2010).
Fig. 4.
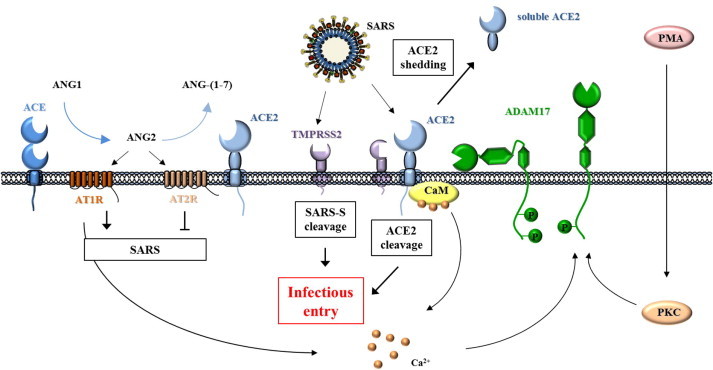
Role of proteases in SARS-coronavirus entry and lung pathogenesis. The dipeptidyl peptidase ACE converts angiotensin 1 (ANG1) into ANG2, which promotes severe lung injury (and thus development of SARS) via the AT1R receptor. Binding of ANG2 to AT2R and conversion of ANG2 into ANG-1(1–7) by ACE2 protect from lung injury. The binding of the SARS-coronavirus spike protein to ACE2 induces ACE2 shedding by ADAM17 and is associated with increased cellular uptake of SARS-coronavirus particles. ADAM17 activity is known to be regulated by AT1R via intracellular calcium levels and by phorbol esters like PMA, which induce phosphorylation of the ADAM17 cytoplasmic tail via PKC activity. In addition, calmodulin is known to associate with the ACE2 cytoplasmic tail and may regulate ACE2 shedding via PKC-dependent activation of ADAM17. TMPRSS2 also cleaves ACE2 and it was proposed that cleavage increases SARS-coronavirus entry. Whether ACE2 cleavage by TMPRSS2 results in ACE2 shedding remains to be investigated. The figure was partially adapted from Imai et al., Cell. Mol. Life Sci., 2007.
Interestingly, ACE2 cleavage is not limited to ADAM17. Shulla and colleagues demonstrated that TMPRSS2, HAT and TMPRSS11a cleave coexpressed ACE2 and cleavage was associated with increased infectivity (Shulla et al., 2011). Augmentation of SARS–S-driven virus–cell entry upon TTSP expression was confirmed by other studies. However, formal proof that ACE2 proteolysis is responsible for increased viral infectivity is missing. In addition, the mechanistic connection between ACE2 cleavage and increased infectivity as well as the potential contribution of these processes to cathepsin-independent SARS–S activation are unclear. Similarly, it is unknown if ACE2 cleavage by TTSPs results in shedding and can be induced by SARS–S. Future studies need to shed light on these interesting questions.
5. The emerging MERS-coronavirus is activated by TMPRSS2 and cathepsins
The novel coronavirus MERS emerged in Jordan in 2012 and up until September 7, 2013, 114 laboratory confirmed infections and 54 deaths have been reported (World-Health-Organization, 2013). Strategies for treatment and prevention of MERS-CoV infection are urgently required, raising the question whether blockade of the proteolytic systems responsible for SARS–S activation could also inhibit MERS-CoV infection. Indeed, an initial study demonstrated that MERS–S-driven infection of certain cell lines depends on the activity of cathepsin B/L and this dependency was rescued by engineered expression of TMPRSS2, which was found to cleave the MERS–S protein (Gierer et al., 2013). In addition, MERS–S-driven infection of Caco-2 cells was largely dependent on the activity of endogenous TMPRSS2, although the additional blockade of cathepsin B/L was required for full inhibition of viral entry (Gierer et al., 2013). These observations suggest that SARS-CoV and MERS-CoV hijack the same proteolytic systems to accomplish their activation and thus share an Achilles heel susceptible to therapeutic inhibition. However, further studies are required to assess whether the MERS-CoV is activated by TMPRSS2 and cathepsin B/L in the respiratory epithelium or if the virus can also hijack other activators, in particular other members of the TTSP family, to promote its spread.
In contrast to SARS-CoV, the MERS-CoV might offer a second target for attack by protease inhibitors targeting viral enzymes. The MERS–S but not SARS–S is efficiently cleaved by an unidentified protease upon ectopic expression in cell lines (Gierer et al., 2013). It will thus be interesting to examine if MERS–S is also cleaved in MERS-CoV infected cells and if cleavage is important for viral infectivity. If so, the protease responsible is a potential target for intervention, although a combination of inhibitors targeting at least two S protein activating proteases might be required to achieve pronounced antiviral effects.
6. Conclusions and future perspectives
The concept that influenza and other respiratory viruses hijack many functionally redundant proteolytic systems for their activation in the host needs to be reconsidered. New studies provide intriguing evidence that influenza viruses, human metapneumovirus and several coronaviruses, including the emerging agents SARS-CoV and MERS-CoV, might hijack TMPRSS2 for activation in the host. Whether these viruses also employ other TTSPs requires further investigation. SARS-CoV and MERS-CoV can also employ cathepsin B/L for activation but the contribution of these proteases to infectious entry into key target cells might be minor. Confirmation of an important role of these proteases in viral spread in the host is required and cathepsin B/L and Tmprss2 knock-out mice can be employed for these studies (Kim et al., 2006, Reinheckel et al., 2001). The recent finding that cathepsin B/L activity is essential for Ebolavirus infection of cell lines (Chandran et al., 2005) but dispensable for viral spread and pathogenesis in mice (Marzi et al., 2012) highlights the need for such studies. Finally, a protease inhibitor, camostat mesylate, which is active against TMPRSS2 and has been used to treat humans, is available and could be employed as a first line defense against the threat imposed by emerging respiratory viruses.
Acknowledgements
GS is supported by grants R01AI074986 and R21AI107165 from the National Institute of Allergy and Infectious Diseases. SP, AH and SG were supported by a grant from BMBF (SARS Verbund, 01KI1005C). PZ was supported by the Leibniz Graduate School Emerging Infectious Diseases.
Contributor Information
Graham Simmons, Email: gsimmons@bloodsystems.org.
Pawel Zmora, Email: PZmora@dpz.eu.
Stefanie Gierer, Email: SGierer@dpz.eu.
Adeline Heurich, Email: AHeurich@dpz.eu.
Stefan Pöhlmann, Email: SPoehlmann@dpz.eu.
References
- Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013;87:8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang K.K., Ratnam J., Gut J., Legac J., Hansell E., Mackey Z.B., Skrzypczynska K.M., Debnath A., Engel J.C., Rosenthal P.J., McKerrow J.H., Arkin M.R., Renslo A.R. Mining a cathepsin inhibitor library for new antiparasitic drug leads. PLoS. Negl. Trop. Dis. 2011;5:e1023. doi: 10.1371/journal.pntd.0001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antalis T.M., Bugge T.H., Wu Q. Membrane-anchored serine proteases in health and disease. Prog. Mol. Biol. Transl. Sci. 2011;99:1–50. doi: 10.1016/B978-0-12-385504-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M., Jardetzky T.S. Class III viral membrane fusion proteins. Adv. Exp. Med. Biol. 2011;714:91–101. doi: 10.1007/978-94-007-0782-5_3. [DOI] [PubMed] [Google Scholar]
- Baron J., Tarnow C., Mayoli-Nussle D., Schilling E., Meyer D., Hammami M., Schwalm F., Steinmetzer T., Guan Y., Garten W., Klenk H.D., Bottcher-Friebertshauser E. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J. Virol. 2013;87:1811–1820. doi: 10.1128/JVI.02320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu A., Gravel E., Cloutier A., Marois I., Colombo E., Desilets A., Verreault C., Leduc R., Marsault E., Richter M.V. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 2013;87:4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010;285:22758–22763. doi: 10.1074/jbc.M110.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., Thiel V., Pöhlmann S. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.Y., Park Y., Schneider H., Schughart K., Pöhlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pöhlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Steffen I., Kuhl A., Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010;20:298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pöhlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniossek M.L., Nagler D.K., Becker-Pauly C., Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 2011;10:5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshauser E., Freuer C., Sielaff F., Schmidt S., Eickmann M., Uhlendorff J., Steinmetzer T., Klenk H.D., Garten W. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 2010;84:5605–5614. doi: 10.1128/JVI.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshauser E., Klenk H.D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. doi. 2013 doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshauser E., Stein D.A., Klenk H.D., Garten W. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J. Virol. 2011;85:1554–1562. doi: 10.1128/JVI.01294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E., Matrosovich T., Beyerle M., Klenk H.D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott R., Klenk H.D., Ohuchi M. Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology. 1994;203:313–319. doi: 10.1006/viro.1994.1489. [DOI] [PubMed] [Google Scholar]
- Chaipan C., Kobasa D., Bertram S., Glowacka I., Steffen I., Tsegaye T.S., Takeda M., Bugge T.H., Kim S., Park Y., Marzi A., Pöhlmann S. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.A., Riese R.J., Shi G.P. Emerging roles for cysteine proteases in human biology. Annu. Rev. Physiol. 1997;59:63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L., Peiris J.S. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pöhlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J., Drosten C., Pöhlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirss K., Kuhl A., Karsten C., Glowacka I., Bertram S., Kaup F., Hofmann H., Pöhlmann S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology. 2012;424:3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., O’Connor T.Z., Hall S.L., Vitale J.N., Nichol K.L. Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J. Infect. Dis. 2009;199:847–857. doi: 10.1086/597122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haga S., Nagata N., Okamura T., Yamamoto N., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antiviral Res. 2010;85:551–555. doi: 10.1016/j.antiviral.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B.S., Gludish D.W., Whittaker G.R. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J. Virol. 2012;86:10579–10586. doi: 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pöhlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Simmons G., Rennekamp A.J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pöhlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80:8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. Coronaviruses. In: Knipe D., editor. Fields Virology. Lippincott, Wiliams & Wilkins; Philadelphia: 2001. pp. 1187–1203. [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean A., Quach C., Yung A., Semret M. Severity and outcome associated with human coronavirus OC43 infections among children. Pediatr. Infect. Dis. J. 2013;32:325–329. doi: 10.1097/INF.0b013e3182812787. [DOI] [PubMed] [Google Scholar]
- Jevsnik M., Ursic T., Zigon N., Lusa L., Krivec U., Petrovec M. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect. Dis. 2012;12:365. doi: 10.1186/1471-2334-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Taguchi M., Otsuki M. Preventive and therapeutic effects of the protease inhibitor camostat on pancreatic fibrosis and atrophy in CCK-1 receptor-deficient rats. Pancreas. 2005;30:54–61. [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B., Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol Lung Cell Mol. Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., Matsuyama S., Taguchi F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009;83:712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H., Okumura Y., Yamada H., Le T.Q., Yano M. Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr. Pharm. Des. 2007;13:405–414. doi: 10.2174/138161207780162971. [DOI] [PubMed] [Google Scholar]
- Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the TMPRSS2-encoded protease. Mol. Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi C., Cervin M., Anderson R. Differentiation of acid-pH-dependent and -nondependent entry pathways for mouse hepatitis virus. Virology. 1991;180:108–119. doi: 10.1016/0042-6822(91)90014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Van Reeth K., Pensaert M. Porcine respiratory coronavirus: molecular features and virus–host interactions. Vet. Res. 1993;24:125–150. [PubMed] [Google Scholar]
- Leslie M. Infectious diseases. Drug developers finally take aim at a neglected disease. Science. 2011;333:933–935. doi: 10.1126/science.333.6045.933. [DOI] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor LI2005. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2–S–protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Reinheckel T., Feldmann H. Cathepsin B & L are not required for ebola virus replication. PLoS Negl. Trop. Dis. 2012;6:e1923. doi: 10.1371/journal.pntd.0001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Sielaff F., Hammami M., Bottcher-Friebertshauser E., Garten W., Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem. J. 2013;452:331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- Misasi J., Chandran K., Yang J.Y., Considine B., Filone C.M., Cote M., Sullivan N., Fabozzi G., Hensley L., Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T.C., Buchmeier M.J. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology. 1997;233:1–8. doi: 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- Okumura Y., Takahashi E., Yano M., Ohuchi M., Daidoji T., Nakaya T., Bottcher E., Garten W., Klenk H.D., Kido H. Novel type II transmembrane serine proteases, MSPL and TMPRSS13, Proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J. Virol. 2010;84:5089–5096. doi: 10.1128/JVI.02605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann P., Foger-Samwald U., Sipos W., Rauner M. The role of cathepsins in osteoimmunology. Crit. Rev. Eukaryot. Gene Expr. 2013;23:11–26. doi: 10.1615/critreveukargeneexpr.2013005929. [DOI] [PubMed] [Google Scholar]
- Plemper R.K. Cell entry of enveloped viruses. Curr. Opin. Virol. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T., Deussing J., Roth W., Peters C. Towards specific functions of lysosomal cysteine peptidases: phenotypes of mice deficient for cathepsin B or cathepsin L. Biol. Chem. 2001;382:735–741. doi: 10.1515/BC.2001.089. [DOI] [PubMed] [Google Scholar]
- Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., Dyer M.D., Teal T.H., Proll S., van den Brand J., Baric R., Katze M.G. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.P., Wang T., Kaletsky R.L., Myers M.C., Purvis J.E., Jing H., Huryn D.M., Greenbaum D.C., Smith A.B., 3rd, Bates P., Diamond S.L. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 2010;78:319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Bertram S., Glowacka I., Steffen I., Chaipan C., Agudelo J., Lu K., Rennekamp A.J., Hofmann H., Bates P., Pöhlmann S. Different host cell proteases activate the SARS-coronavirus spike-protein for cell–cell and virus–cell fusion. Virology. 2011;413:265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Rappuoli R. SARS: understanding the virus and development of rational therapy. Curr. Mol. Med. 2005;5:677–697. doi: 10.2174/156652405774641124. [DOI] [PubMed] [Google Scholar]
- Steffen I., Liss N.M., Schneider B.S., Fair J.N., Chiu C.Y., Simmons G. Characterization of the Bas–Congo virus glycoprotein and its function in pseudotyped viruses. J. Virol. 2013;87:9558–9568. doi: 10.1128/JVI.01183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.F., Tong J.H., Chan P.K., Au F.W., Chim S.S., Chan K.C., Cheung J.L., Liu E.Y., Tse G.M., Lo A.W., Lo Y.M., Ng H.K. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers B.A., van der Hoek L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009;29:715–724. doi: 10.1016/j.cll.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C., Sandesara R.G., Mulherkar N., Whelan S.P., Chandran K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 2010;84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World-Health-Organization, 2013. Novel coronavirus infection – update. http://www.who.int/csr/don/2013_09_07/en/index.html.
- Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. PH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhirnov O.P., Ikizler M.R., Wright P.F. Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J. Virol. 2002;76:8682–8689. doi: 10.1128/JVI.76.17.8682-8689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov O.P., Klenk H.D., Wright P.F. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011;92:27–36. doi: 10.1016/j.antiviral.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Agudelo J., Lu K., Goetz D.H., Hansell E., Chen Y.T., Roush W.R., McKerrow J., Craik C.S., Amberg S.M., Simmons G. Inhibitors of SARS-CoV entry – identification using an internally-controlled dual envelope pseudovirion assay. Antiviral Res. 2011;92:187–194. doi: 10.1016/j.antiviral.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Simmons G. Development of novel entry inhibitors targeting emerging viruses. Expert Rev. Anti Infect. Ther. 2012;10:1129–1138. doi: 10.1586/eri.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]


