Abstract
Age-related changes in DNA methylation have been demonstrated in mammals, but it remains unclear as to the generality of this phenomenon in vertebrates, which is a criterion for the fundamental cause of senescence. Here we showed that the zebrafish genome gradually and clearly lost methylcytosine in somatic cells, but not in male germ cells during aging, and that age-dependent hypomethylation preferentially occurred at a particular domain called the CpG island shore, which is associated with vertebrates’ genes and has been shown to be hypomethylated in humans with age. We also found that two CpG island shores hypomethylated in zebrafish oocytes were de novo methylated in fertilized eggs, which suggests that the zebrafish epigenome is reset upon fertilization, enabling new generations to restart with a heavily methylated genome. Furthermore, we observed an increase in cleavage of the zebrafish genome to an oligonucleosome length in somatic cells from the age of 12 months, which is suggestive of an elevated rate of apoptosis in the senescent stage.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-013-9548-5) contains supplementary material, which is available to authorized users.
Keywords: Aging, Zebrafish, DNA methylation, CpG island shore
Introduction
The majority of cytosines in CpG dinucleotides in vertebrate genomes are methylated at the 5 position by cytosine methyltransferases, except for CpGs in the CpG-rich region called the “CpG island,” which often encompasses the promoters and transcription start sites of many vertebrate genes (Illingworth and Bird 2009). Most promoter region CpG islands are in an unmethylated state irrespective of transcriptional activity, while a small fraction of these is densely methylated and associated with gene silencing (Shen et al. 2007). The artificial methylation of CpG island promoters was shown to repress transcription in living cells (Stein et al. 1982), and the demethylation of endogenous methylated CpG islands using DNA methyltransferase inhibitors restored the expression of the gene (Hansen and Gartler 1990). Thus, the aberrant methylation or demethylation of CpG islands can be a cause of gene misregulation and has been attributed to human pathogenic conditions represented by cancers (Jones and Baylin 2007). Similarly, changes in DNA methylation have been considered to be a cause of senescence since the Vanyushin group first found an age-related decrease in methylcytosine levels in the somatic tissues of the salmon (Berdishev et al. 1967). This phenomenon was later shown to be true for cellular senescence and mammalian aging (Wilson and Jones 1983). Subsequently, with the advent of bisulfite sequencing, age-related methylation changes have been located at distinctive positions in mammalian genomes: certain CpG islands were more likely to be methylated with age (Christensen et al. 2009; Hernandez et al. 2011), whereas age-related hypomethylation was observed at the CpG island “shore” region of a comparatively low CpG density that is present near traditional CpG islands, typically within 2 kb (Alisch et al. 2012; Pirazzini et al. 2012; Heyn et al. 2012). CpG island shores were first recognized as regions showing human tissue- and cancer-specific methylation (Irizarry et al. 2009). Besides CpG island shores, several reports have described age-related hypomethylation at repetitive sequences in human blood samples (Bollati et al. 2009; Jintaridth and Mutirangura 2010; Gentilini et al. 2012). Furthermore, a comprehensive study that performed whole genome bisulfite sequencing of a centenarian and newborn showed that cytosines unmethylated in the centenarian, but not in the newborn, covered all genomic compartments, such as exonic, intronic, and intergenic regions (Heyn et al. 2012). Thus, loci showing age-related methylation changes appear to be scattered in mammalian genomes, and it is still elusive if any specific hyper- or hypomethylated domains exist relevant to senescence. In addition, it remains unclear if age-related methylation changes are widely seen in vertebrates, which will be a requirement for the fundamental cause of senescence. To address these issues, we examined the zebrafish, Danio rerio, in this study based on the indication that if a certain type(s) of methylation change exists that is primarily involved in senescence, it should be seen in the lower vertebrate, and such a change(s) may be more easily found by comparing distantly related species rather than closely related ones. Other merits of utilizing fish included the availability of basic information regarding zebrafish senescence (Kishi et al. 2003; Tsai et al. 2007) and possibilities for future genetic and pharmacological manipulations. We did not detect global hypomethylation in the zebrafish genome during development, which had been previously shown (Macleod et al. 1999); however, we detected a clear loss of methylation at the CpG island shores of some genes after the developmental stage in the tissues of aging zebrafish. Thus, methylation changes in this domain may be directly involved in vertebrate senescence. Furthermore, we observed genome DNA fragmentation in zebrafish of an advanced age, a likely result of apoptosis, which is another phenomenon also observed in aged mammals (Taglialatela et al. 1996; Itzhaki et al. 2003).
Experimental procedures
Zebrafish maintenance
Zebrafish of the wild-type AB/Tü were used throughout this study. Fish at the same stage were derived from a single clutch of eggs. Fish were bred naturally and early stage fry were fed with paramecia, and then with living brine shrimp three times per day. Water temperature was maintained at 28.5 °C with a 14-h light/10-h dark cycle in a central fish facility. At around 10 weeks of age, male and female fish were separately housed at a density of ~20 fish per 3-l glass tank. They were crossed twice per month on average from 3 to 4 months old until reproductive activity declined (12 ~ 15 months old).
Preparation of genomic DNA
Zebrafish embryos and adult tissues were digested with Proteinase K and SDS to extract genomic DNA. Zebrafish oocytes were obtained by gently pushing the female’s belly with our fingers. Follicle cells were carefully removed using forceps under a dissecting microscope. Chorions were removed by Pronase E before the isolation of genomic DNA from fertilized eggs and cleavage stage (128-cell) embryos (Westerfield 2000).
Methylation analysis
After digestion with methylation-sensitive enzymes, 100 ng of DNA fragments were separated on 1.2 % agarose gels with one or two DNA size markers (New England Biolabs, Ipswich, MA, USA), stained with ethidium bromide (0.5 μg/ml), and then destained in water. All enzymes used were purchased from New England Biolabs. For bisulfite sequencing, 200 ng of genomic DNA was processed using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) to convert unmethylated cytosines into uracils. We used the MethPrimer program to choose sequences of primer pairs for bisulfite sequencing (Li and Dahiya 2002). Primer sequences and PCR conditions are shown in the Supplemental Table. PCR fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and were used for the transformation of Escherichia coli. Plasmids isolated from 24 transformants were sequenced with the DYEnamic ET Terminator Cycle Sequencing kit (GE Healthcare, Piscataway, NJ, USA) on an ABI 310 genetic analyzer or ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA). QUMA was used for the analysis of sequence data and drawing figures (Kumaki et al. 2008).
Statistical analysis
All analyses were performed using Prism 5 software (GraphPad Software, San Diego, CA, USA). The methylation level of each amplicon was the ratio of methylated cytosines to the total number of CpG dinucleotides present in 20 or more clones. The Kruskal–Wallis test was used to make nonparametric comparisons of the median methylation levels of a region to be investigated between more than two groups. A two-tailed t test was employed for comparisons between two groups.
Southwestern hybridization for the detection of DNA fragmentation
One hundred nanograms of genomic DNA was labeled with 50 pmol of digoxigenin (DIG)-dUTP (Roche Diagnostics, Mannheim, Germany) by 0.5 U of Klenow fragment (3′→5′ exo−) (New England Biolabs) in a volume of 4 μl (10 mM Tris–HCl, 5 mM MgCl2, 7.5 mM DTT, pH 7.5 at 25 °C) at 37 °C for 15 min. After the addition of 1 μl of 5× sample loading buffer (25 % glycerol, 5 mM EDTA, pH 8.0, 0.5 % SDS), the reaction mixture was directly loaded onto a 2 % agarose gel and run at 50 V for 60 min. The gel was stained with EtBr for 10 min, and then rinsed in water. DNA was transferred and fixed onto Biodyne B (Pall, Port Washington, NY, USA) by Stratalinker UV Crosslinker (Agilent Technologies, Santa Clara, CA, USA). The membrane was rinsed with 2× SSC, and DIG-labeled DNA fragments were detected by an anti-DIG antibody conjugated with alkaline phosphatase (Roche Diagnostics) following the manufacturer’s instructions.
Results
Changes in global methylation levels in the zebrafish genome with age
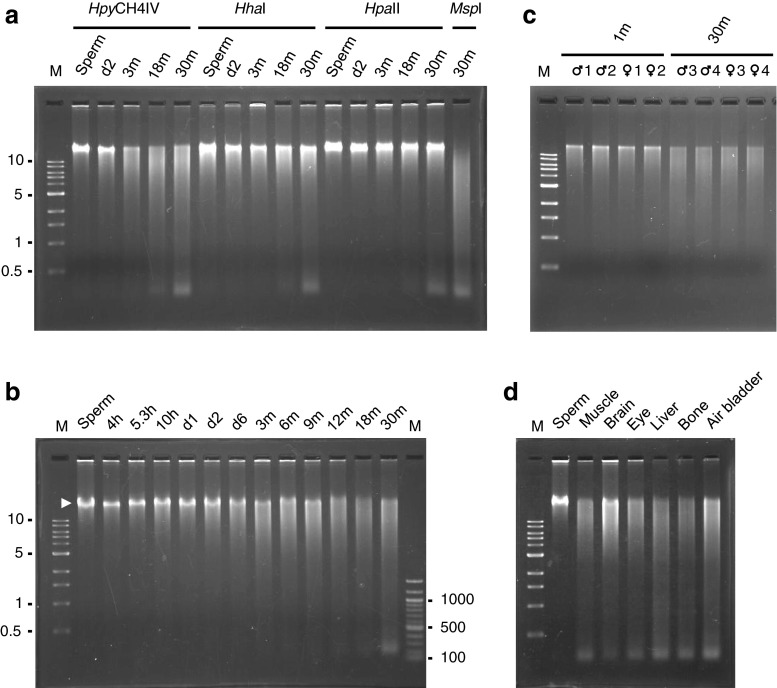
To examine if global methylation levels in the zebrafish genome changed with aging, we extracted genomic DNA from the sperm, 2-day post fertilization (d2) embryos, and muscle of 3-, 18-, and 30-month-old fish. We regarded 30-month-old zebrafish as aged fish since they live for approximately 3 years on average in conventional aquarium systems including ours (Tsai et al. 2007). Muscle was chosen as a representative of somatic cells since it is a predominant structure of the fish species. Sperm was extracted from the same 30-month-old fish from which muscle was isolated. DNA was digested with each of three methylation-sensitive restriction enzymes, HpyCH4IV, HhaI, and HpaII, neither of which was able to cut at each recognition site if there was a methylated cytosine inside. Digested DNA was then run on an agarose gel and stained with ethidium bromide (Fig. 1a). One of the enzymes, HpyCH4IV, whose recognition site is ACGT, digested the genomes of adult fish (3 months or older) more frequently than those of sperm and d2 embryos, as judged by DNA smearing patterns on gels. Furthermore, the enzyme cleaved the genomes of 18- and 30-month-old fish slightly more frequently than the genomes of 3-month-old fish. These results indicated that age-dependent genomic hypomethylation occurred in zebrafish somatic cells, but not in male germ cells.
Fig. 1.
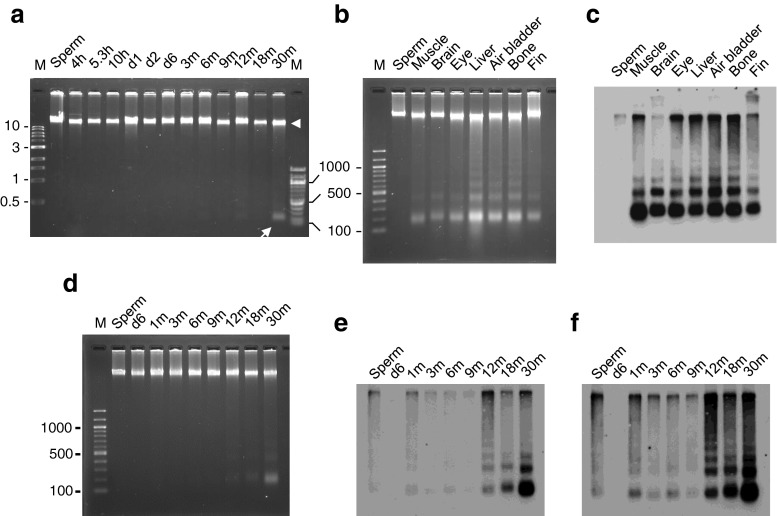
Age-dependent DNA hypomethylation in the zebrafish genome. a The genomic DNA of zebrafish sperm, 2-day post fertilization embryos (d2), and muscle of 3-, 12-, and 30-month-old fish (3 m, 12 m, and 30 m, respectively) were digested with each of the three methylation-sensitive restriction enzymes indicated and were run on an agarose gel. HpyCH4IV digestion clearly showed age-dependent hypomethylation. MspI, an isoschizomer of HpaII, is a methylation-insensitive enzyme. b Genomic DNAs of sperm, embryos (h; hours post fertilization), and adult muscle of the indicated ages were digested with HpyCH4IV. Hypomethylation was recognizable as early as d2 and became evident at d6 (larval stage). A high molecular weight DNA band was seen until 9 months (lanes 2 ~ 11, white arrowhead). No discernible difference in the smearing pattern was observed after 12 months (lanes 12 ~ 14). c Liver DNA from four individuals of 1- and 30-month-old zebrafish were digested and electrophoresed as in b. d Genomic DNA isolated from various tissues of an aged zebrafish (30-month-old) was digested with HpyCH4IV and run on an agarose gel. The molecular marker of DNA loaded on the first lane of each gel was a 1-kb ladder. The 100-bp DNA ladder was also loaded on the last lane in b
HhaI, whose recognition site is GCGC, also cleaved the genomic DNA of 18- and 30-month-old fish slightly more than that of 3-month-old fish. However, this enzyme did not reveal an appreciable difference in digestion patterns between embryos and 3-month-old fish. The other enzyme, HpaII, whose recognition site is CCGG, detected little increase in DNA cleavage with age. This was consistent with the result of a previous study in which HpaII was used to investigate differences in global DNA methylation between zebrafish embryos and adults (Macleod et al. 1999). These results showed that age-related demethylation events were governed by the immediate nucleotide composition, and it should be noted that age-demethylated CpG dinucleotides in humans were preferably preceded and followed by A and T, respectively (Alisch et al. 2012), which constitutes the tetranucleotide digested by HpyCH4IV.
To trace age-dependent hypomethylation in detail, we prepared DNA from the embryos of several developmental stages and muscle of aging adult fish at various stages and digested them with HpyCH4IV (Fig. 1b). We found that the genome was heavily methylated in early zebrafish embryos, from the sphere stage (4 hpf—hours post fertilization) to d1. Hypomethylation of the genome could be recognized as early as d2, the end of the developmental stage, and became evident by d6, the early larval stage. Hypomethylation of the genome then appeared to proceed gradually until 12 months, the stage when high molecular weight fragments became obscure, after which no further digestion was discernible by smearing patterns on agarose gels. As shown in Fig. 1c, the liver DNAs of 30-month-old fish were cleaved more than those of 1-month-old fish, indicating that age-related hypomethylation was not restricted to muscle. We next compared the methylation levels of different tissues in a single 30-month-old zebrafish by digestion with HpyCH4IV (Fig. 1d). DNA from somatic tissues was cleaved as much as muscle DNA, except for brain DNA, which was cleaved less than the others. In contrast to somatic DNA, the sperm genome appeared to be resistant to digestion.
Changes in regional methylation levels of the zebrafish genome with age
We first suspected that the regions of genomic hypomethylation that we detected may be located at repetitive sequences since age-dependent hypomethylation at repeats have been found in human blood samples (Bollati et al. 2009; Jintaridth and Mutirangura 2010; Gentilini et al. 2012). To examine this possibility, we performed bisulfite sequence analysis against four short interspersed nuclear elements (SINEs) in zebrafish (Supplementary Fig. S1): three individual DANA elements (Izsvák et al. 1996; Shimoda et al. 1996) and a conserved portion of SINE3-1a elements (Kapitonov and Jurka 2003). To compare interindividual differences in methylation levels and to test the significance of methylation changes between groups of different stages, we purified genomic DNA from four individual embryos (sex was unknown) and four adult fish (two males and two females) and analyzed at least 20 clones per region from each of four embryos or adult fish, which came to at least 80 clones per region in total. However, all the repeats examined were heavily methylated both in embryos and in aged fish (Supplementary Fig. S1).
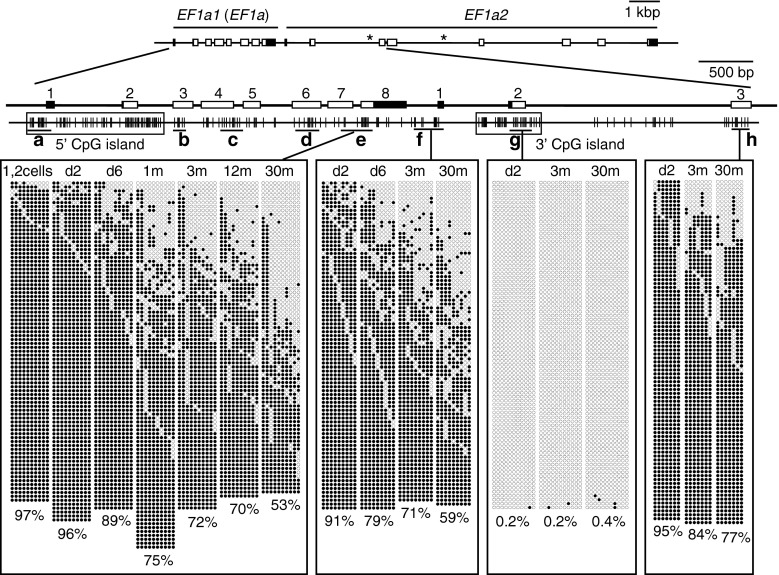
We then turned our attention to unique regions of the genome and noticed that one of our previous studies, which was undertaken for another purpose, had recorded lower methylation levels at a region spanning exons 7 and 8 (exon 7 ~ 8) in the eukaryotic translation elongation factor 1 α1, like 1 (commonly referred to as EF1α) gene in adult zebrafish (67 %) than those at the same region in embryos (97 %) (Yamakoshi and Shimoda 2003). This prompted us to analyze the methylation levels of the EF1α locus in embryos and adult fish of different ages by bisulfite sequencing (Fig. 2 and Supplementary Fig. S2a, b). Results showed that apparent age-related hypomethylation occurred in a region that flanks a CpG island located ~1 kb downstream of the EF1α gene (Fig. 2). Such a region was defined as the “CpG island shore” (Irizarry et al. 2009). No clear decline in methylation with age was observed in regions outside the CpG island shore region (Supplementary Fig. S2a, b). A slight decrease in methylation was already observed in the region of the d6 larvae relative to d2 embryos (Fig. 2). This was in accordance with the HpyCH4IV digestion results of the genome that showed decreases in global methylation levels between d2 and d6 (Fig. 1b). In contrast to the 3′ CpG island shore, the shore region of the promoter-associated 5′ CpG island showed no age-related methylation changes (Supplementary Fig. S2a).
Fig. 2.
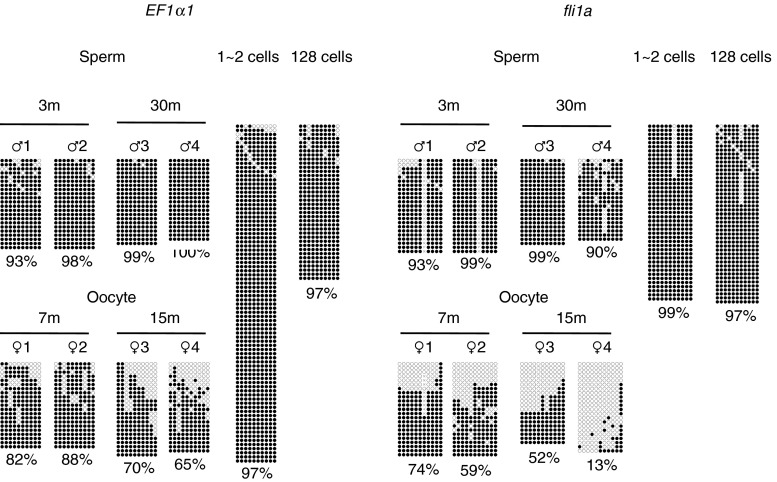
Age-dependent hypomethylation at a CpG island shore region in the EF1α locus. The methylation levels of various regions in the zebrafish EF1α locus in fertilized eggs (1, 2 cells), embryos (d2), young larvae (d6), and muscle of immature (1-month-old) and adult fish (3-, 12-, and 30-month-old) were analyzed by bisulfite sequencing. Schematics of EF1α1 (EF1α) and EF1α2 are shown at the top. Asterisks represent positions where unread sequences in the Sanger database were determined in this study. Filled and open rectangles indicate untranslated and translated regions, respectively, and numbers on exons are also shown. Vertical lines indicate the positions of CpG dinucleotides. Black horizontal bars with small letters indicate the regions analyzed. Filled and open circles show methylated and unmethylated cytosines, respectively. Crosses indicate the positions where CpG was absent due to DNA polymorphism. A total of four single embryos, four larvae, and four adult fish were analyzed to obtain methylation profiles for each region, and at least 20 clones were sequenced per fish for each region. In addition to a conventional CpG island that covered exons 1 and 2 of the EF1α1 gene (5′ CpG island, Fig. S2A), there was another CpG island downstream of the gene (3′ CpG island). Age-related hypomethylation was observed at a limited region juxtaposed to the 3′ CpG island, a region called the “CpG island shore.” The percentages of mean methylation levels were shown at the bottom
Presence of a tandemly duplicated EF1α homologue
It is possible that the 3′ CpG island was a CpG island of another gene localized closely to EF1α. Indeed, the GenBank database annotated an EF1α homologue immediately downstream of the EF1α gene, eukaryotic translation elongation factor 1 α1, like 2, which we referred to as EF1α2, and it was clear that the 3′ CpG island of the locus was an authentic CpG island of EF1α2 (Fig. 2 and Supplementary Fig. S2a, b). Hereafter, we referred to upstream EF1α as EF1α1.
Two isoforms of EF1α have been identified in mammals and are referred to as eEF1A-1 (eukaryotic elongation factor) and eEF1A-2. eEF1A-1 is expressed ubiquitously as is zebrafish EF1α1, whereas eEF1A-2 is specifically expressed in the muscle and heart and weakly in the brain (Khalyfa et al. 2001). Since zebrafish EF1α2 is highly expressed in muscle but not in the liver (data not shown), EF1α2 appears to be the orthologue of mammalian eEF1A-2.
Age-dependent hypomethylation incident to CpG island shores
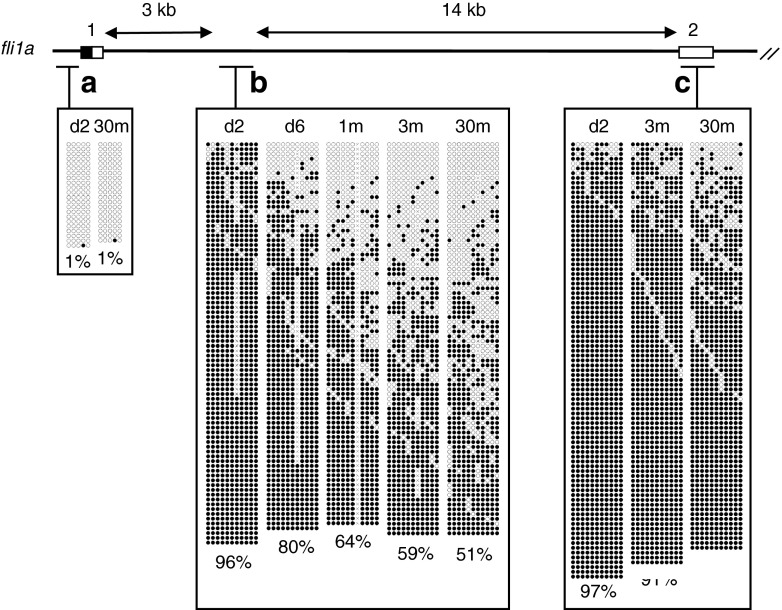
We then examined if age-related hypomethylation occurred at other CpG island shores in muscle. For this analysis, four single-copy zebrafish genes were randomly chosen: fli1a (friend leukemia integration), fgf3 (fibroblast growth factor), β-actin2, and rpl13a (ribosomal protein L13a). fli1a and fgf3 are tissue-specific genes that are exclusively expressed in the vasculature and subsets of neurons in the brain in adult zebrafish, respectively (Lawson and Weinstein 2002; Topp et al. 2008). A CpG island overlapped with exon 1 of fli1a, free of methylation at both embryonic and adult stages (Fig. 3). In contrast, a region in intron 1 of fli1a, which corresponds to the CpG island shore region and was heavily methylated at the embryonic stage, showed a marked reduction in methylation in d6 larvae, which gradually became pronounced with aging (Fig. 3). Differences in methylation levels in this region were significant (p < 0.05) between groups consisting of four 1-month-old and four 30-month-old fish (Fig. 4). Exon 2 of fli1a was heavily methylated irrespective of age.
Fig. 3.
Age-dependent hypomethylation at a CpG island shore region in the fli1a locus. The methylation levels of three regions in the zebrafish fli1a locus in embryos (d2), young larvae (d6), and muscle of immature (1-month-old) and adult fish (3- and 30-month-old) were analyzed and presented as in Fig. 2. Regions a, b, and c correspond to the CpG island, CpG island shore, and non-CpG island, respectively. Only a single embryo and an adult fish were analyzed for region a. A total of four single embryos and four fish were analyzed to obtain methylation profiles for regions b and c. At least 20 clones were sequenced per sample for each region
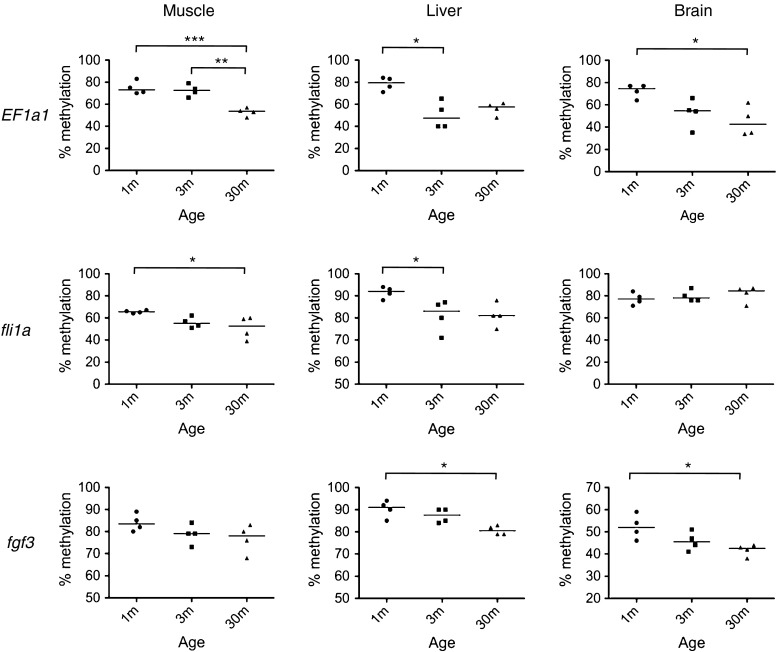
Fig. 4.
CpG island shore regions showed age-related hypomethylation not only in muscle, but also in the liver and brain. The methylation levels of CpG island shore regions in EF1α1 (region e in Fig. 2), fli1a (region b in Fig. S3), and fgf3 (region b in Fig. S4) in the zebrafish muscle, liver, and brain were examined by bisulfite sequencing. *p < 0.05, **p < 0.01, and ***p < 0.001. Four fish per stage were analyzed. Horizontal bars show medians
The second gene, fgf3, had a typical CpG island at the 5′ region that covers exons 1 and 2 (Supplementary Fig. S3; Yamakoshi and Shimoda 2003). Although exon 3 of fgf3 was located within 500 bp of exon 2, it was heavily methylated in embryos. We found that an upstream part of exon 3, which corresponded to the CpG island shore, showed approximately a 20 % reduction in methylation in muscle cells by 3 months. This reduction was less than that observed in the CpG island shores of EF1α2 and fli1a, and no further hypomethylation was observed in the muscle of 30-month-old fish. No significant hypomethylation was detected in the middle part of exon 3.
The third gene, β-actin2, which encodes a cytoskeletal structural protein, had a cluster of CpG dinucleotides at the 5′ region that constituted a CpG island (Supplementary Fig. S4). We found that exon 3, where CpG dinucleotides were sparsely distributed, was heavily methylated in d2 embryos, but showed approximately a 50 % reduction in methylation in muscle cells by 3 months. At this CpG island shore region, the methylation level was further reduced by 30 months. Exon 5 also showed age-dependent hypomethylation, albeit to a lesser extent.
The fourth gene, rpl13a, which codes a subunit of ribosomal 60S protein, had a CpG island at the 5′ region covering exons 1 and 2 (Supplementary Fig. S5). In rpl13a, CpG dinucleotides were sparsely distributed outside the CpG island, and we did not detect age-related methylation changes at exon 6 of the rpl13a gene, which corresponded to the CpG island shore region. All together, age-associated hypomethylation was observed in muscle at four out of six shores.
Hypomethylation at CpG island shores in nonmuscle cells
We next investigated methylation levels at the CpG island shores of EF1α2 and fli1a in other tissues of a 30-month-old male zebrafish (Supplementary Fig. S6) and found that CpG island shores were more or less hypomethylated in any type of somatic cells, but not in germ cells. This result appeared to reflect the global methylation patterns shown in Fig. 1d. To see if hypomethylation in these tissues was age related, we quantified methylation levels at the CpG island shores of EF1α2, fli1a, and fgf3 in the livers and brains of 1-, 3-, and 30-month-old fish using the bisulfite sequencing method (Fig. 4). As in muscle, methylation levels at the CpG island shores of these tissues decreased with age, except for the CpG island shore of fli1a in the brain, and differences in methylation levels between these groups at different ages were significant at most regions examined.
De novo methylation of CpG island shores upon fertilization
We then compared the methylation levels of sperm and oocytes (Fig. 5). Both EF1α2 and fli1a shores were densely methylated in the germ cells of young and aged male fish, which indicated that male germ cells were exempt from age-related hypomethylation in adult zebrafish. In contrast, the same regions were moderately methylated in oocytes. Since repetitive sequences, SINE3-1a and four long interspersed nuclear elements (Sugano et al. 2006; Rai et al. 2008), in oocytes were heavily methylated as in sperm (Supplementary Fig. S7), it is possible that the oocyte genome manifested age-related methylation changes at CpG island shores similar to somatic cell genomes. It should be noted that EF1α2 and fli1a CpG island shores were also fully methylated in fertilized eggs and cleavage stage embryos (Fig. 5). These results imply that the maternal genome is de novo methylated in fertilized eggs and that new generations restart with a heavily methylated genome.
Fig. 5.
CpG island shores moderately methylated in oocytes were heavily methylated in fertilized eggs. The methylation levels of CpG island shores in EF1α1 (region e in Fig. 2) and fli1a (region b in Fig. 3) were determined in germ cells extracted from adult fish of the indicated ages and in early embryos
Fragmentation of the zebrafish genome with age
Although we demonstrated age-related increases in DNA digestion by methylation-sensitive enzymes (Fig. 1), there was a possibility that DNA samples from adult muscle were degraded. To confirm the intactness of DNA samples, we ran the same genomic DNA as the ones used in Fig. 1, but in an undigested state, on a 1 % agarose gel. As shown in Fig. 6a, a high molecular DNA band was observed in each lane with a slight smearing pattern, indicating the relative intactness of the samples. However, we noticed that a small distinct band of approximately 200 bp existed in the lane to which DNA from a 30-month-old fish was applied (Fig. 6a). This indicated that this small DNA was already present in the genomic DNA before digestion. To characterize the small DNA band in detail, we ran undigested genomic DNA from various tissues of the same zebrafish on a 2 % agarose gel (Fig. 6b). The results showed that DNA from all somatic cells, but not from sperm, contained approximately 200 bp DNA, and larger DNA fragments spacing at ~200 bp were also detected in some tissues. This periodic DNA banding was reminiscent of the characteristic DNA ladder observed in apoptosis (Wyllie 1980), in which some specific DNase digested genomic DNA at nucleosome intervals of 180 bp (Enari et al. 1998).
Fig. 6.
Age-related DNA fragmentation occurred in somatic cells, but not in sperm. The undigested genomic DNAs of sperm, embryos (h; hours post fertilization), and adult muscle of the indicated ages were run on a 1 % agarose gel and stained with ethidium bromide (a). In addition to a high molecular weight DNA band (arrowhead), a small DNA fragment was seen in an aged fish (arrow). Small DNA fragments in the tissues of an aged zebrafish were visualized by ethidium staining (b) and by the end-labeling method we developed (c). The same procedure was employed to reveal the stage at which small DNA fragments appeared (d–f). The times for exposure to X-ray films were 20 and 60 s for e and f, respectively. Similar to DNA hypomethylation, this age-related phenomenon was observed in all of the somatic cells examined, but not in male germ cells. The fragments showed a 180 ~ 200-bp periodicity, resembling the DNA cleavage observed in apoptosis. Lane M, 1 kb or 100 bp DNA ladder
To increase the sensitivity for detecting small DNA fragments, we developed a method that utilized DIG to end-label the free 3′ ends of DNA fragments. This method revealed some DNA bands that were hardly detectable by EtBr staining (Fig. 6c). To investigate the stage at which the fragments first appeared in muscle DNA in the lifetime of zebrafish, we used the end-labeling method for the genomic DNA of different stages. Although small bands were hardly detectable even in 12- and 18-month-old zebrafish DNA by EtBr staining (Fig. 6d), after exposure to X-ray films, we found that small DNA fragments already existed in 1-month-old zebrafish muscle and they increased after 12 months old (Fig. 6e, f). No small DNA fragments were detected in sperm or in d6 larvae with this method.
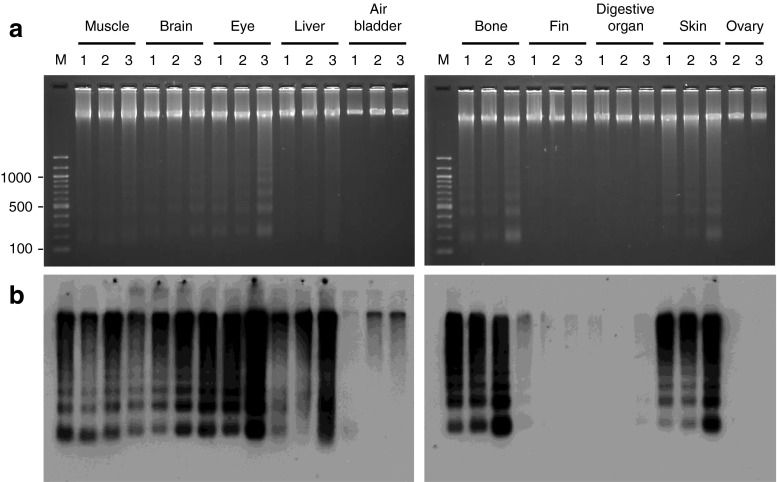
To examine if the generation of small DNA fragments varied among individual fish, we performed the same analysis on various tissues of three 15-month-old fish (Fig. 7a, b). We found that not only the presence or absence of ladders, but also ladder patterns appeared to be very similar among the same tissues, indicating that there was little interindividual variance in DNA fragmentation among the same tissues. The liver was the only organ in which the degradation of genomic DNA occurred without periodicity, and even this pattern was common among the three fish.
Fig. 7.
Interindividual variations in DNA fragmentation in zebrafish tissues. Ethidium bromide staining (a) and the end-labeling method (b) were employed to compare DNA fragmentation patterns in the tissues of three 15-month-old zebrafish: one male (lane 1) and two female fish (lanes 2 and 3). These fish were raised from the same batch of embryos. Lane M, 100 bp DNA ladder
There were two clear differences in DNA fragmentation between the tissues of 15-month-old fish and those of 30-month-old fish. First, the DNA ladder was hardly visible from the air bladder and fin of a 15-month-old fish (Fig. 7b), but appeared in these tissues in a 30-month-old fish (Fig. 6c). This result suggested that each tissue has its own stage at which the level of DNA fragmentation increases. Second, a monomer-sized DNA band (200 bp) was predominant in the DNA ladders of all the tissues from a 30-month-old fish, except for the sperm (Fig. 6c); however, this was not the case for the 15-month-old fish (Fig. 7b). The activity of the enzyme(s) involved in DNA fragmentation may possibly be higher in aged cells than in younger cells; therefore, the genome may be more thoroughly cleaved in aged cells. Alternatively, the genome in aged cells may be more susceptible to such enzyme(s).
Discussion
In this study, we detected the loss of methylation in the zebrafish genome with aging at borders between CpG island and non-CpG island regions, known as CpG island shores (Irizarry et al. 2009). In contrast, no significant methylation changes were observed in the repetitive sequences surveyed. In mammals, regions corresponding to CpG island shores have also been shown to be vulnerable to age-related hypomethylation (Ono et al. 1989; Alisch et al. 2012; Heyn et al. 2012; Pirazzini et al. 2012). Thus, the CpG island shore is the region where age-related hypomethylation changes are more likely to occur in common beyond species and, hence, whose methylation changes may be directly involved in the senescence of vertebrates as we suspected. Since the number of loci we investigated in this study was limited, it is unknown if global hypomethylation detected by enzymatic digestions can be solely explained by hypomethylation at CpG island shores. However, judging from the high probability of age-dependent hypomethylation at CpG island shores, decreases in methylation at CpG island shores should markedly influence global methylation levels during zebrafish aging.
It is currently unclear how hypomethylation preferentially occurs at CpG island shores in the somatic cells of aging zebrafish. It is not likely to be caused by the reduced activity of the maintenance methyltransferase Dnmt1 as it results in global, not focal hypomethylation (Shimoda et al. 2005). Since global methylation levels began to decrease well before maturation and were prominent at the juvenile stage when cellular proliferation was active, the fidelity of methylation maintenance at replication forks may somehow be reduced at CpG island shores. Alternatively, age-related hypomethylation at CpG island shores may be executed by the extended action of the Tet1 protein or a protein complex of MBD4, AID, and Gadd45, both of which eliminate unwanted methylation in CpG islands and maintain the unmethylated status of CpG islands (Rai et al. 2008; Wu and Zhang 2011).
How sperm DNA is exempt from age-dependent hypomethylation has also not been elucidated yet. This is not explained by the exceptional expression of any de novo methyltransferases in the testes because all zebrafish paralogues of the mouse de novo methyltransferase genes dnmt3a and dnmt3b are expressed not only in the testes, but also in somatic tissues such as the brain and liver (Campos et al. 2012). It was previously demonstrated that the zebrafish sperm genome, relative to the fibroblast genome, was packaged in chromatin characterized by abundant linker histones and the absence of H4K16ac (Wu et al. 2011). The distinctive configuration of the sperm genome may allow for a de novo methyltransferase(s) accessible to CpG island shores or prevent the demethylation activities described above.
DNA fragmentation is another phenomenon that we observed in the somatic cells of aging zebrafish. Similar to methylation changes, it has also been observed in aged mammals (Taglialatela et al. 1996; Itzhaki et al. 2003). An increase in DNA fragmentation started in zebrafish muscle at the age of 12 months. In our fish facility, this is around the age at which fecundity begins to drop. Since the decline in reproductive activity is a hallmark of zebrafish senescence (Tsai et al. 2007), the increase in DNA fragmentation observed in the present study may be related to the onset of senescence in zebrafish.
The periodic appearance and length of the periodicity of DNA fragmentation strongly suggested that it was generated by apoptosis because a prominent biochemical event in apoptosis is the internucleosomal cleavage of genomic DNA, initially producing 50 to 200 kb segments and then fragments in multiples of approximately 185 bp (Wyllie 1980; Oberhammer et al. 1993). Apoptosis provides an efficient mechanism for excluding cells that are potentially harmful to the animal. Since global hypomethylation affects genome integrity and gene regulation, cells with hypomethylated genomes in the zebrafish, mouse, and Xenopus were shown to be eliminated by apoptosis (Jackson-Grusby et al. 2001; Stancheva et al. 2001; Chen et al. 2007; Koji et al. 2008; Anderson et al. 2009). This, together with the observations of age-related increases in apoptotic cells in mammals (Jara et al. 2004; Leeuwenburgh et al. 2005; Whitman et al. 2005; Rice and Blough 2006), makes it tempting to hypothesize that, once a continued methylation change in the somatic cells of vertebrates exceeds a critical threshold, cells die by apoptosis due to inadequate expression profiles, which in turn causes a functional decline in tissues, the prominent feature of senescence. If this is the case, somatic cells need to begin with excess DNA methylation so that they can lose a certain amount before they are phenotypically affected (Cooney 1993). This is in line with the observation that the de novo methylation of some genes occurred in fertilized eggs (Fig. 5). The identification of chemicals or genetic mutations that accelerate or delay age-related hypomethylation at CpG island shores in the somatic cells of zebrafish should help to examine this hypothesis experimentally.
Electronic supplementary material
(DOC 6802 kb)
Contributor Information
Nobuyoshi Shimoda, Phone: +81-562-468629, FAX: +81-562-468629, Email: shimoda@ncgg.go.jp.
Naohiro Hashimoto, Phone: +81-562-468629, FAX: +81-562-468629, Email: nao@ncgg.go.jp.
References
- Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22:623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, Stainer DYR. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdishev GD, Korotaev GK, Bojarskikh GV, Vanyushin BF. Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning. Biokhimiia. 1967;32:988–993. [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos C, Valente LMP, Fernandes JMO. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. 2012;500:93–100. doi: 10.1016/j.gene.2012.03.041. [DOI] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney CA. Are somatic cells inherently deficient in methylation metabolism? A proposed mechanism for DNA methylation loss, senescence and aging. Growth Dev & Aging. 1993;57:261–273. [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Gentilini D, Mari D, Castaldi D, Remondini D, Ogliari G, Ostan R, Bucci L, Sirchia SM, Tabano S, Cavagnini F, Monti D, Franceschi C, Di Blasio AM, Vitale G. Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age. 2012 doi: 10.1007/s11357-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Gartler SM. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5′ CpG island. Proc Natl Acad Sci USA. 1990;87:4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki O, Skutelsky E, Kaptzan T, Sinai J, Michowitz M, Huszar M, Leibovici J. Ageing–apoptosis relation in murine spleen. Mech Ageing Dev. 2003;124:999–1012. doi: 10.1016/S0047-6374(03)00171-4. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Ivics Z, Garcia-Estefania D, Fahrenkrug SC, Hackett PB. DANA elements: a family of composite, tRNA-derived short interspersed DNA elements associated with mutational activities in zebrafish (Danio rerio) Proc Natl Acad Sci USA. 1996;93:1077–1081. doi: 10.1073/pnas.93.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jara M, Carballada R, Esponda P. Age-induced apoptosis in the male genital tract of the mouse. Reproduction. 2004;127:359–366. doi: 10.1530/rep.1.00092. [DOI] [PubMed] [Google Scholar]
- Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics. 2010;41:194–200. doi: 10.1152/physiolgenomics.00146.2009. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Bourbeau D, Chen E, Petroulakis E, Pan J, Xu S, Wang E. Characterization of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wasted mice. J Biol Chem. 2001;276:22915–22922. doi: 10.1074/jbc.M101011200. [DOI] [PubMed] [Google Scholar]
- Kishi S, Uchiyama J, Baughman AM, Goto T, Lin MC, Tsai SB. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38:777–786. doi: 10.1016/S0531-5565(03)00108-6. [DOI] [PubMed] [Google Scholar]
- Koji T, Kondo S, Hishikawa Y, An S, Sato Y. In situ detection of methylated DNA by histo endonuclease-linked detection of methylated DNA sites: a new principle of analysis of DNA methylation. Histochem Cell Biol. 2008;130:917–925. doi: 10.1007/s00418-008-0487-7. [DOI] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucl Acid Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Leeuwenburgh C, Gurley CM, Strotman BA, Dupont-Versteegden EE. Age-related differences in apoptosis with disuse atrophy in soleus muscle. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1288–R1296. doi: 10.1152/ajpregu.00576.2004. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet. 1999;23:139–140. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- Oberhammer F, Wilson JW, Dive C, Morris ID, Hickman JA, Wakeling AE, Walker PR, Sikorska M. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;12:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Takahashi N, Okada S. Age-associated changes in DNA methylation and mRNA level of the c-myc gene in spleen and liver of mice. Mutat Res. 1989;219:39–50. doi: 10.1016/0921-8734(89)90039-8. [DOI] [PubMed] [Google Scholar]
- Pirazzini C, Giuliani C, Bacalini MG, Boattini A, Capri M, Fontanesi E, Marasco E, Mantovani V, Pierini M, Pini E, Luiselli D, Franceschi C, Garagnani P. Space/population and time/age in DNA methylation variability in humans: a study on IGF2/H19 locus in different Italian populations and in mono- and di-zygotic twins of different age. Aging. 2012;4:509–520. doi: 10.18632/aging.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KM, Blough ER. Sarcopenia-related apoptosis is regulated differently in fast- and slow-twitch muscles of the aging F344/N × BN rat model. Mech Ageing Dev. 2006;127:670–679. doi: 10.1016/j.mad.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:e181. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda N, Chevrette M, Ekker M, Kikuchi Y, Hotta Y, Okamoto H. Mermaid: a family of short interspersed repetitive elements widespread in vertebrates. Biochem Biophys Res Commun. 1996;220:226–232. doi: 10.1006/bbrc.1996.0385. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Yamakoshi K, Miyake A, Takeda H. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev Dyn. 2005;233:1509–1516. doi: 10.1002/dvdy.20455. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci USA. 1982;79:3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano T, Kajikawa M, Okada N. Isolation and characterization of retrotransposon-competent LINEs from zebrafish. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Taglialatela G, Gegg M, Perez-Polo JR, Williams LR, Rose GM. Evidence for DNA fragmentation in the CNS of aged Fischer-344 rats. NeuroReport. 1996;7:977–980. doi: 10.1097/00001756-199604100-00004. [DOI] [PubMed] [Google Scholar]
- Topp S, Stigloher C, Komisarczuk AZ, Adolf B, Becker TS, Bally-Cuif L. Fgf signaling in the zebrafish adult brain: association of Fgf activity with ventricular zones but not cell proliferation. J Comp Neurol. 2008;510:422–439. doi: 10.1002/cne.21802. [DOI] [PubMed] [Google Scholar]
- Tsai SB, Tucci V, Uchiyama J, Fabian NJ, Lin MC, Bayliss PE, Neuberg DS, Zhdanova IV, Kishi S. Differential effects of genotoxic stress on both concurrent body growth and gradual senescence in the adult zebrafish. Aging Cell. 2007;6:209–224. doi: 10.1111/j.1474-9726.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. 4. Eugene: University of Oregon Press; 2000. [Google Scholar]
- Whitman SA, Wacker MJ, Richmond SR, Godard MP. Contributions of the ubiquitin–proteasome pathway and apoptosis to human skeletal muscle wasting with age. Pflugers Arch. 2005;450:437–446. doi: 10.1007/s00424-005-1473-8. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Yamakoshi K, Shimoda N. De novo DNA methylation at the CpG island of the zebrafish no tail gene. Genesis. 2003;37:195–202. doi: 10.1002/gene.10245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 6802 kb)