Abstract
Human sirtuins are seven proteins with deacetylase activity that are emerging as key modulators of basic physiological functions. Some evidence links SIRT3 to longevity in mammals. This study aimed to investigate whether variants within SIRT3 gene were associated to human longevity. We analyzed 549 genomic DNA collected during the prospective study “Treviso Longeva,” including elderly over 70 years of age from the municipality of Treviso, a small city in the northeast of Italy. We genotyped SIRT3 rs3825075, rs4980329, and rs11555236 single nucleotide polymorphisms (SNPs) by real-time polymerase chain reaction allelic discrimination assay. A cross-sectional analysis performed by comparing people over and under 85 years of age did not evidence association among the SIRT3 SNPs and longevity. However, when we performed a longitudinal analysis considering mortality as a dependent variable, we observed an association of SIRT3 rs11555236 and rs4980329 with longevity in the whole population (p values corrected for potential confounders = 0.04 and 0.03, respectively). After stratification according to gender, the same SNPs were associated to female longevity only (p values corrected for potential confounders = 0.03 and 0.02, respectively). Finally, as rs11555236 was reported to be in linkage disequilibrium with a putative functional enhancer within the SIRT3 gene, we assessed whether rs11555236 genotypes correlated with a different level of SIRT3 protein in peripheral blood mononuclear cells. We found an increased level of SIRT3 in subjects homozygous for the (T) allele. We suggest that SIRT3 genetic variability might be relevant for the modulation of human longevity in the Italian population.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-013-9559-2) contains supplementary material, which is available to authorized users.
Keywords: SIRT3, Aging, Longevity, Longitudinal study, TRELONG, Genetics
Background
Human sirtuins (SIRTs) are a family comprising seven proteins sharing NAD+-dependent deacetylase activity that seems involved in several basic physiologic mechanisms and relevant for age-linked diseases (Albani et al. 2010; Polito et al. 2010). Among SIRTs, SIRT3 is a mitochondrial protein involved in defensive mechanisms against oxidative stress (Yu et al. 2012; Bause and Haigis 2012; Iwahara et al. 2012). Considering that oxidative stress increase is a basic feature of aging, SIRT3 may have a role in modulating longevity, a hypothesis supported by evidence in an animal model, where the disruption of the gene coding for angiotensin II receptor type 1A resulted in marked prolongation of life span in mice, with an increased number of mitochondria in the kidney and upregulation of prosurvival genes (including SIRT3) (Benigni et al. 2009). Moreover, genetic association studies pointed out a role for SIRT3 in human longevity. The SIRT3 single nucleotide polymorphism (SNP) rs11555236 minor allele (T) increased male survival in an Italian cohort from Calabria and was found in linkage disequilibrium with a putative enhancer located within SIRT3 intron 5 (Bellizzi et al. 2005; Rose et al. 2003). In a different multicentre study that recruited centenarians from Italy and Germany (for a total of 1,321 centenarians and 1,140 younger subjects), some SNPs in SIRT3 showed significant association with longevity in the Italian female and in the German male subgroups, although this result was not supported by a replication enrolling centenarians of French ancestry (Lescai et al. 2009). Moreover, in a study analyzing genetic variants of the oldest old, a list of SIRT3 SNPs was detailed in long-living individuals of Caucasian ancestry (Halaschek-Wiener et al. 2009). Starting from the evidence supporting an involvement of SIRT3 in longevity, we genotyped three SNPs of SIRT3 in an ongoing prospective study enrolling an elderly population (Treviso Longeva (TRELONG) study; Gallucci et al. 2007).
Materials and methods
Population
A thorough description of the TRELONG study has already been reported (Gallucci et al. 2007). In short, after the selection of the 13,861 Treviso inhabitants over 70 years of age (17 % of total inhabitants) from the residents listed in the Registry Office of Treviso, participants were systematically sampled, planning to include 100 participants according to gender and 10-year-age group, 125 women and 125 men aged 70–79 years, and all people over 100 years. A total of 668 people were selected: 311 men and 357 women (mean age 84.0 ± 8.0 years, range 70.0–105.5 years). A 7-year follow-up was then performed. Included subjects underwent examination from the biologic, clinical, and socioeconomic point of view. A blood sample was collected and each participant was administered with a structured interview. The study protocol, blood collection procedure, and the questionnaire to be administered at home were submitted to and approved by the ethical committee of the National Institute on Research and Care of the Elderly (INRCA, Italy). The protocol included a written informed consent for clinical and genetic studies.
Biological sample preparation. SIRT3 rs3825075, rs4980329, and rs11555236 genotyping
Fasting peripheral blood samples (30 mL) were collected by venipuncture; one aliquot was used to separate peripheral blood mononuclear cells (PBMCs) by a standard Ficoll centrifugation procedure. PBMC pellets were washed with ice-cold phosphate-buffered saline (PBS), divided into aliquots and stored at −80 °C for further analysis. From the enrolled population of 668 subjects, 590 gave their consent for blood collection and PBMC preparation.
Genomic DNA was extracted from PBMC pellet using a vacuum-based semiautomated nucleic acid extractor (AB6100, Applied Biosystems, Foster City, CA, USA), checked for concentration by a UV spectrophotometer (Eppendorf, Hamburg, Germany), and stored at 4 °C.
We decided to assess SIRT3 rs3825075, rs4980329, and rs11555236 as the first two ones (which we genotyped in a previous study (Polito et al. 2013)) captured the variation of rs511744 and rs11246020, respectively, that were reported to be overrepresented in long-living elderly of Caucasian ancestry (Halaschek-Wiener et al. 2009). Rs11555236 was already linked to longevity in the Italian population (Bellizzi et al. 2005; Rose et al. 2003). To assess rs3825075, rs4980329, and rs11555236 genotypes, a gDNA aliquot (about 20 ng) was used in an allelic discrimination assay using a real-time PCR apparatus and TaqMan technology according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). The successful genotyping rate was around 92 %. Subjects' genotypes were independently confirmed in a random sample representing 10 % of the population, with 100 % replication rate. Subjects who were not genotyped were equally distributed in the TRELONG population (subjects belonged to almost all the age brackets) and had no peculiar demographic or survival features in comparison to the genotyped population.
Protein extraction and Western blotting
One aliquot of PBMC pellet was lysed in 1× Laemmli sample buffer, subjected to SDS-PAGE electrophoresis, and transferred to a nitrocellulose membrane (BioRad Laboratories, Hercules, CA, USA). The membrane was incubated overnight with a primary antibody diluted in 5 % nonfat dried milk in PBS + 0.1 % Tween-20 (PBS-T). After washing, the membrane was incubated for 1 h with a secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnologies, Santa Cruz, CA) diluted 1:5,000 in 5 % nonfat dried milk in PBS-T. The membrane was finally subjected to chemiluminescence detection using an enhanced reagent (electrochemiluminescence (ECL) Millipore, Billerica, CA, USA). To quantitatively assess SIRT3 immunoreactivity, the ECL signal was measured by a densitometric method setup by a free image analyzer program (ImageJ, http://rsbweb.nih.gov/ij/), using an internal normalization control (α-tubulin).
Antibodies
The following primary antibodies were used to run immunoblotting: anti-α-tubulin monoclonal antibody (dilution 1:5,000) (Abcam, Cambridge, UK) and anti-SIRT3 polyclonal antibody (dilution 1:1,000) (Abcam, Cambridge, UK).
Statistical analysis
Genotypic and allelic frequency distributions and departure from Hardy–Weinberg equilibrium were assessed by χ2 test. Calculations were done using GraphPad Prism program version. 5.04.
Survival curves were estimated by the Kaplan–Meier method. Hazard ratios (HRs) were calculated using Cox proportional hazard model. The proportional hazard has been tested using Schoenfeld's residual test, and it has never been rejected, thus confirming the suitability of the model. Multivariate regression analysis was performed considering mortality as dependent variable. These statistic analyses were computed using the package “survival” of the “R” software. After correction for possible confounders (diabetes, cardiovascular diseases, cerebral vasculopathies, cancer, cholesterol level, scholarity, age, and gender), results were considered significant at p < 0.05, using two-tailed tests of significance.
Results
Cross-sectional analysis
We started evaluating the correlation of SIRT3 rs3825075, rs4980329, and rs11555236 with longevity by splitting the TRELONG population around 85 years of age, basing on the previous evidence from the same study (Albani et al. 2009, 2011). Results of this analysis are reported in Table 1. Each SNP was in Hardy–Weinberg equilibrium (HWE) (p = 0.82, 0.71, and 0.93 for χ2 test assessing departure from HWE, respectively). Genotypic and allelic frequencies did not differ between the considered groups, even after gender stratification (data not shown).
Table 1.
Cross-sectional analysis of the genotypic and allelic frequencies of SIRT3 single nucleotide polymorphisms rs3825075, rs4980329, and rs11555236
| SNP (no.) | Genotype no. (%) | Allele no. (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| ≤85y | >85y | χ 2 statistics | ≤85y | >85y | χ 2 statistics | |||
| rs3825075 (n = 549) | GG | 136 (43.9) | 117 (48.9) | χ 2 = 3.1; df = 2; p = 0.21 | G | 418 (67.4) | 329 (68.8) | χ 2 = 0.24; df = 1; p = 0.62 |
| GA | 146 (47.1) | 95 (37.4) | A | 202 (32.6) | 149 (31.2) | |||
| AA | 28 (9.0) | 27 (13.7) | ||||||
| rs4980329 (n = 542) | GG | 167 (54.7) | 138 (58.2) | χ 2 = 2.0; df = 2; p = 0.37 | G | 458 (75.1) | 360 (75.9) | χ 2 = 0.10; df = 1; p = 0.74 |
| GA | 124 (40.6) | 84 (35.4) | A | 152 (24.9) | 114 (24.1) | |||
| AA | 14 (4.7) | 15 (6.4) | ||||||
| rs11555236 (n = 542) | GG | 169 (54.9) | 137 (58.5) | χ 2 = 2.5; df = 2; p = 0.28 | G | 454 (73.7) | 361 (77.1) | χ 2 = 1.7; df = 1; p = 0.19 |
| GT | 116 (37.7) | 87 (37.2) | T | 162 (26.3) | 107 (22.9) | |||
| TT | 23 (7.4) | 10 (4.3) | ||||||
no. number, 85y 85 years of age, df degree of freedom, p p value
Prospective analysis
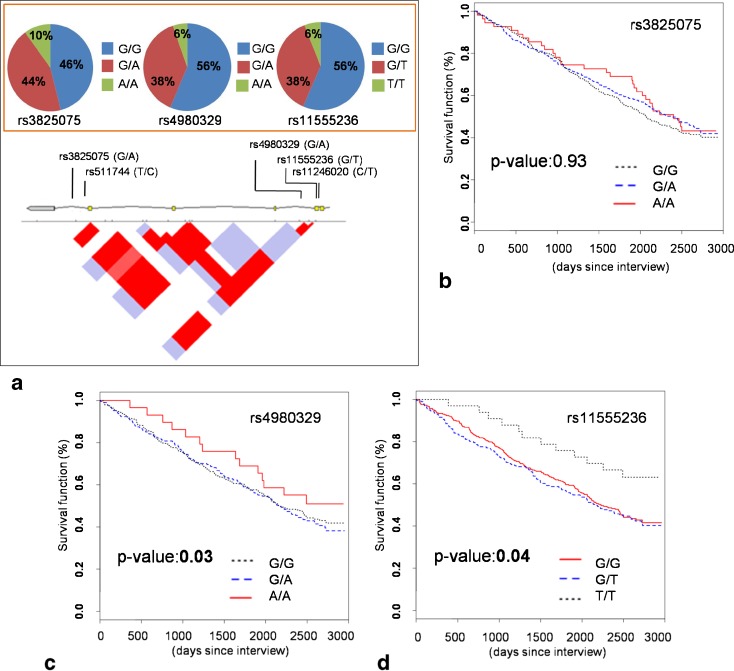
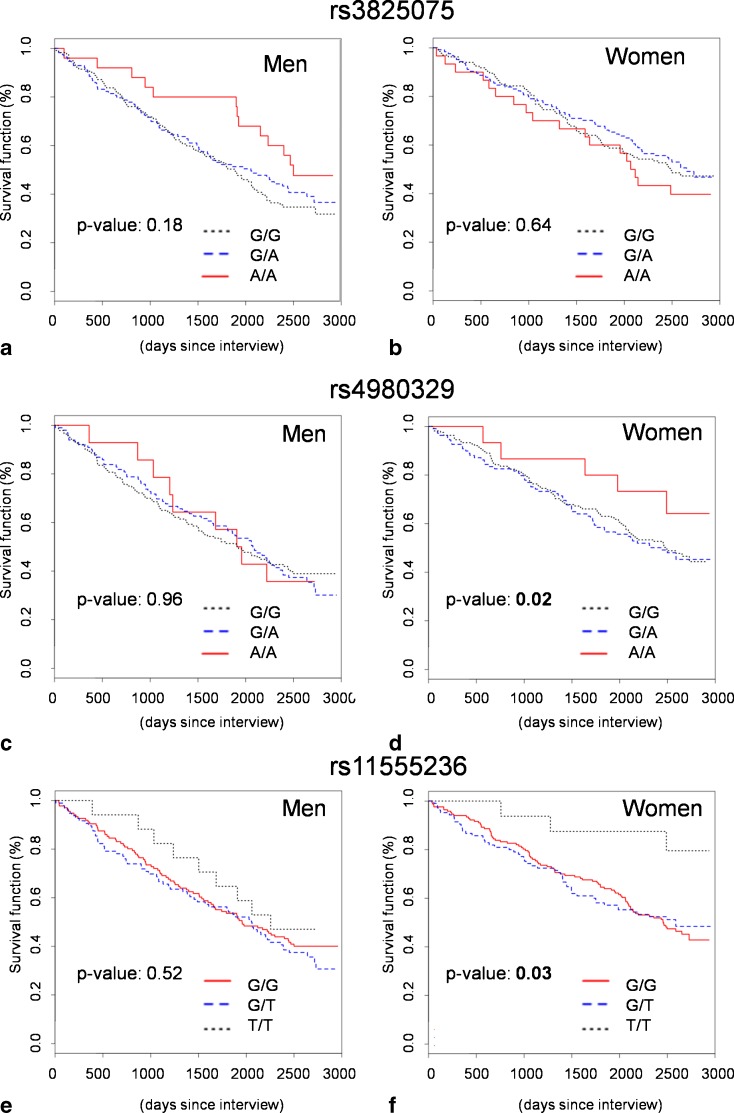
As the TRELONG study features a longitudinal follow-up with mortality data, we decided to reanalyze SIRT3 genetic variability considering mortality as a dependent variable and plotting survival curves according to each SNP genotype (Fig. 1). As mortality might be influenced by several other variables, we controlled for TRELONG prevalent disorders (diabetes, cardiovascular diseases, cerebral vasculopathies, and cancer), risk factors (cholesterol level, scholarity), age, and gender. We found an association between rs4980329 and survival, with the minor allele (A) giving an advantage for longevity. The calculated HRs, considering the major allele homozygous genotype as reference, were 1.08 and 0.66 for (GA) and (AA) genotypes, respectively. A second association was found for rs11555236, with the minor allele (T) conferring an advantage. For this polymorphism, the HRs using the major allele homozygous genotype as references were 1.18 and 0.58, respectively. No evidence was found for rs3825075. To investigate whether these associations had a different outcome in males or females, we performed a gender-stratified analysis, correcting for all the other variables influencing survival listed above. Survival plots of the TRELONG population according to gender are shown in Fig. 2. We found a significant effect of rs4980329 and rs11555236 minor alleles for women only (corrected p values and HR: p value, 0.02; HR, 0.54 and p value, 0.03; HR, 0.27, respectively), while the association was no longer present in males (corrected p values, 0.96 and 0.52, respectively).
Fig. 1.
Genotyping and survival curves of the TRELONG population. a Graphical representation of the SIRT3 gene portion encompassing the single nucleotide polymorphisms genotyped in the TRELONG population. The genotypic frequency is graphically indicated for rs3825075, rs4980329, and rs11555236, while just the position is shown for the tagged SNPs rs511744 and rs11246020. Below the gene structure, where exons are in yellow, a linkage disequilibrium graphical representation is reported; red diamonds are suggestive of high LD (D' >80 %). b Survival plot of the whole TRELONG population stratified according to rs3825075 genotype. The reported p value is corrected for possible confounding factors affecting longevity (age, gender, scholarity, cholesterol level, cardiovascular disease, vascular cerebropathies, diabetes, and cancer). c Survival plot of the whole TRELONG population stratified according to rs4980329 genotype. The p value was corrected as above. d Survival plot of the whole TRELONG population stratified according to rs11555236 genotype. This SNP was reported to be in linkage disequilibrium to a putative enhancer variant within SIRT3 intron 5 (Bellizzi et al. 2005). The p value was corrected for the same variables as previously detailed
Fig. 2.
Longitudinal survival analysis in the TRELONG population after gender stratification. Survival curves were plotted according to genotype and gender to search for a gender-specific effect of the investigated SIRT3 SNPs. a–b rs3825075 survival curve in males and females, respectively. c–d rs4980329 survival curve in males and females, respectively. e–f rs11555236 survival curve in males and females, respectively. The reported p values are corrected for potential confounders (age, cholesterol level, scholarity, cardiovascular disease, vascular cerebropathies, diabetes, and cancer)
Functional correlation of rs11555236
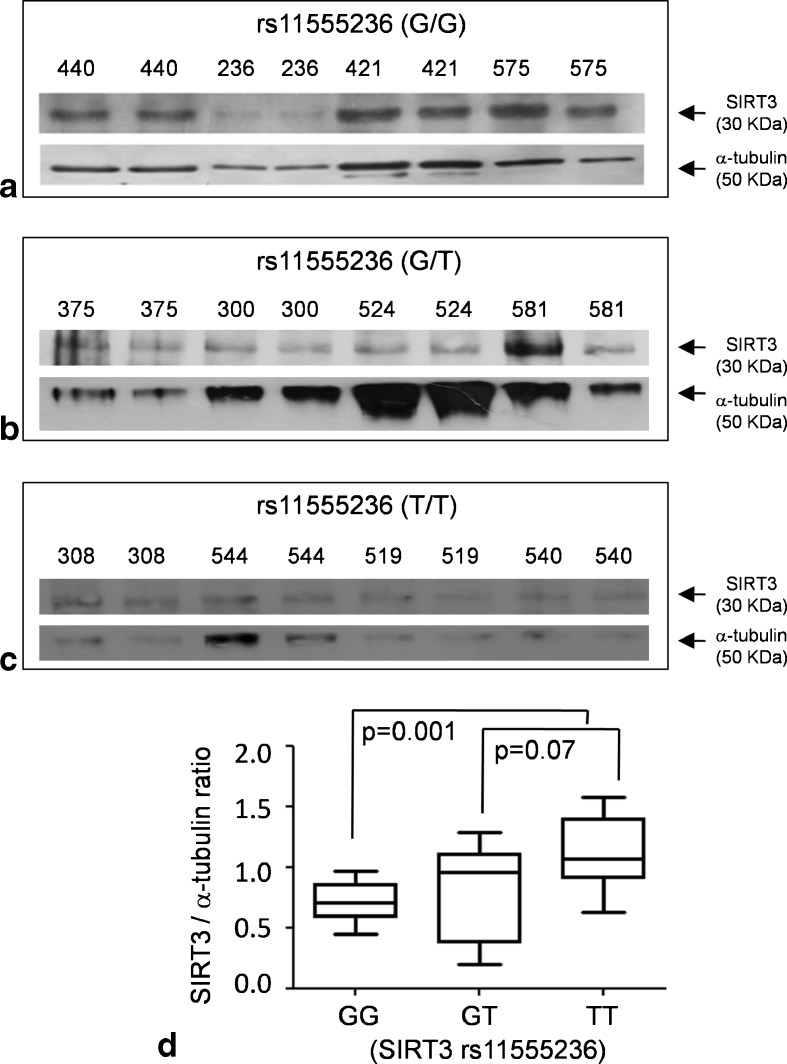
As it was hypothesized that rs11555236 might be a genetic marker of longevity in linkage disequilibrium with a putative functional enhancer within intron 5 of SIRT3 (Bellizzi et al. 2005), we decided to assess whether the rs11555236 genotype correlated with a modulation of SIRT3 expression level. We focused on the SIRT3 isoform having a molecular weight of around 30 kDa that is considered the fully active protein relevant for mitochondrial metabolic homeostasis (Hallows et al. 2008). We analyzed SIRT3 protein level by Western blotting in PBMCs from the TRELONG population, building up three groups (n = 10 subjects each) balanced for age and gender, stratified according to rs11555236 genotype (GG, GT, or TT). Figure 3 shows SIRT3 protein immunoreactivity and the densitometric quantification. In the group homozygous for the rs11555236 minor allele (T), SIRT3 reactivity was significantly enhanced in comparison to the (GG) carriers, using α-tubulin as normalization internal control. The normalized mean densitometric values for the three groups were as follows: (GG) = 0.71 ± 0.08; (GT) = 0.80 ± 0.4; and (TT) = 1.11 ± 0.3. These values fitted an additive model of allelic effect (linear correlation coefficient r2 = 0.892).
Fig. 3.
SIRT3 Western blotting in peripheral blood mononucleate cells. To assess a possible functional correlation of rs11555236 variants with SIRT3 expression, we selected a group of subjects from the TRELONG study, balanced for gender, and evaluated SIRT3 protein immunoreactivity. Each subject, identified in the figure by the number above the corresponding gel lane, was assessed in duplicate. a–c Representative Western blotting where subjects were divided according to rs11555236 genotype (GG, GT, or TT). A total of ten subjects for each genotype were analyzed. Alpha-tubulin immunoreactivity is also shown as loading control for normalization. Variation among subjects of α-tubulin reactivity was due to different cellularity of the corresponding PBMC fraction. d Box plot showing the densitometric analysis of the Western blotting data. Optical density values of SIRT3 band were quantified and normalized to α-tubulin. Values were compared by nonparametric Mann–Whitney U test
Discussion
Longevity is a complex phenotype arising from multiple variables, and genetic background plays a pivotal role not only for a deterministic control on hereditary disorders, but also in modulating the individual response to environmental hits and drug therapies (Eaton et al. 2012; Murabito et al. 2012; Chung et al. 2010; Seripa et al. 2010). Genes with involvement in basic physiologic processes are rational candidates to search for variability linked to increased survival at oldest ages (Tazearslan et al. 2012). To this respect, SIRT3 deserves attention both for its role in mitochondrial homeostasis, a fundamental aspect of cell physiology, and for the available data associating SIRT3 to survival in animal models or in the elderly (Benigni et al. 2009; Halaschek-Wiener et al. 2009; Rose et al. 2003; Bellizzi et al. 2005). We have decided to analyze an informative set of SNPs of SIRT3 gene in a prospective study (TRELONG) allowing us to have access to survival data over a 7-year follow-up. This work has been carried out in a unique group of subjects: a community-based population with the highest longevity rates in Italy (Gallucci et al. 2007). To this respect, our analysis reflects closely the correlation between SIRT3 and longevity in the Italian population in the real world. As in a previous genetic study performed in our laboratory where we had already investigated a panel of SIRT3 SNPs (Polito et al. 2013), we decided to select from this panel those matching or tagging a list of SIRT3 genetic variants whose presence was highlighted in a population of Caucasian elderly, suggesting their involvement in a prosurvival background (Halaschek-Wiener et al. 2009). Thanks to the existence of linkage disequilibrium blocks within SIRT3, we were able to choose a couple of SNPs from Polito et al. (2012) (rs3825075 and rs4980329) that were linked to the SNPs rs511744 and rs11246020 reported by Halaschek-Wiener et al. (2009; Fig. 1). The third SNP, rs11555236, was linked to longevity in an Italian study and supposed to be in linkage disequilibrium with a putative functional enhancer (a variable number of tandem repeat (VNTR) element) located within SIRT3 intron 5 (Bellizzi et al. 2005). The cross-sectional association study was performed by splitting the TRELONG population around 85 years, a critical point discriminating long-living individuals that has allowed us to find other informative genetic variability in the same population (Albani et al. 2009). However, no association was found with this experimental design. To reanalyze data taking into account the real survival curve of each subject, a prospective analysis was conducted by inputting mortality data of the TRELONG study over a 7-year follow-up. The main result was the association between rs11555236 and survival in the whole population, having the minor allele (T) carriers a more favorable survival outcome. This is in agreement with the available data in the Italian population by Rose et al. (2003), with our added value of a more sound longitudinal design. At difference from the above-cited study, where the association was significant in males only, we found a detectable effect in the whole population and by splitting according to gender the association kept significant in women only. Importantly, we had also association between rs4980329 and whole population survival, with a gender effect significant for women. From the genetic point of view, the parallelism between rs4980329 and rs11555236 results was expected due to the predicted location of these SNPs in a linkage disequilibrium block according to the available HapMap data (www.hapmap.org) (see Fig. 1). Our experimental confirmation allows to consider these SNPs as alternative markers to be genotyped in association to longevity. The fact that these polymorphisms were associated to longevity in the whole population but under gender stratification in females only might tune down their impact to survival, but we and others have already reported several cases of analogous outcome after gender stratification (Albani et al. 2009; Napolioni et al. 2011; Anselmi et al. 2009; Cederholm et al. 2007). This might suggest that SIRT3 genetic elements play a gender-selective role in modulating longevity, even though our results are not deciding to this respect, as we found association only after correction for several other variables affecting longevity, while the uncorrected interaction (gender × polymorphism) was not significant (data not shown).
Also in reason of its complex DNA sequence structure, we have not performed in the present study the assessment of the putative VNTR enhancer element of SIRT3 gene described by Bellizzi et al. (2005), but taking advantage from its reported linkage disequilibrium with rs11555236, we decided to further investigate the functional action of the VNTR element in a group of TRELONG subjects bearing the rs11555236 variants. We hypothesized that some sequences of the VNTR element acted as a cis-enhancer on SIRT3 gene, leading to an increase of SIRT3 protein level. We verified this by measuring SIRT3 protein expression in PBMCs, as this was the only available tissue specimen from the TRELONG study. Our observation that the (T) allele, that correlated with longevity, was also associated to an increased level of SIRT3 protein is intriguing, as it might suggest that increased SIRT3 level is beneficial, as expected from the current knowledge on this protein functions, dealing with response to oxidative stress and control of the mitochondrial acetylome (Bause and Haigis 2012; Fritz et al. 2012). We acknowledge that this assessment suffers from limitations; first of all, the lack of independent tissue source to confirm a widespread and not tissue-specific effect. Moreover, we are not able to rule out the possibility that other genetic or epigenetic modulations occurring in PBMC might have affected the transcription rate of the SIRT3 gene. Finally, the exact VNTR sequence linked to rs11555236 (T) allele should be further investigated.
In summary, this study points to an involvement of SIRT3 variability in human longevity, with a particular emphasis on the role of the rs11555236 SIRT3 polymorphic site as a genetic and functional marker.
Electronic Supplementary Material
(JPEG 71 kb)
(JPEG 109 kb)
Acknowledgments
This work was supported by “Fondazione Italo Monzino,” Milan, Italy and by grants from “Interdisciplinary Geriatric Research Foundation (FORGEI)” and from “Fondazione Veneto Banca.”
Contributor Information
Diego Albani, Phone: +39-2-39014594, FAX: +39-2-3546277, Email: diego.albani@marionegri.it.
Eleonora Ateri, Email: elly.a86@gmail.com.
Stefano Mazzuco, Email: mazzuco@stat.unipd.it.
Alice Ghilardi, Email: alice.ghilardi@marionegri.it.
Serena Rodilossi, Email: serena.rodilossi@marionegri.it.
Gloria Biella, Email: gloria.biella@marionegri.it.
Fausta Ongaro, Email: ongaro@stat.unipd.it.
Piero Antuono, Email: PAntuono@mcw.edu.
Paolo Boldrini, Email: pboldrini@ulss.tv.it.
Enrico Di Giorgi, Email: edigiorgi@ulss.tv.it.
Andrea Frigato, Email: afrigato@ulss.tv.it.
Elisabetta Durante, Email: edurante@ulss.tv.it.
Livio Caberlotto, Email: lcaberlotto@ulss.tv.it.
Andrea Zanardo, Email: azanardo@ulss.tv.it.
Marinella Siculi, Email: msiculi@ulss.tv.it.
Maurizio Gallucci, Email: mgallucci@ulss.tv.it.
Gianluigi Forloni, Email: gianluigi.forloni@marionegri.it.
References
- Albani D, Batelli S, Polito L, Prato F, Pesaresi M, Gajo GB, De Angeli S, Zanardo A, Galimberti D, Scarpini E, Gallucci M, Forloni G. Interleukin-6 plasma level increases with age in an Italian elderly population ("The Treviso Longeva"-Trelong-study) with a sex-specific contribution of rs1800795 polymorphism. Age Dordr. 2009;31:155–162. doi: 10.1007/s11357-009-9092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani D, Mazzuco S, Polito L, Batelli S, Biella G, Ongaro F, Gustafson DR, Antuono P, Gajo G, Durante E, Caberlotto L, Zanardo A, Siculi M, Gallucci M, Forloni G. Insulin-like growth factor 1 receptor polymorphism rs2229765 and circulating interleukin-6 level affect male longevity in a population-based prospective study (Treviso Longeva-TRELONG) Aging Male. 2011;14:257–264. doi: 10.3109/13685538.2011.607521. [DOI] [PubMed] [Google Scholar]
- Albani D, Polito L, Forloni G. Sirtuins as novel targets for Alzheimer's disease and other neurodegenerative disorders: experimental and genetic evidence. J Alzheimers Dis. 2010;19:11–26. doi: 10.3233/JAD-2010-1215. [DOI] [PubMed] [Google Scholar]
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Bause AS, Haigis MC. SIRT3 regulation of mitochondrial oxidative stress. Exp Gerontol. 2012 doi: 10.1016/j.exger.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G. Disruption of the AngII type 1 receptor promotes longevity in mice. J Clin Invest. 2009;119:524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederholm T, Persson M, Andersson P, Stenvinkel P, Nordfors L, Madden J, Vedin I, Wretlind B, Grimble RF, Palmblad J. Polymorphisms in cytokine genes influence long-term survival differently in elderly male and female patients. J Intern Med. 2007;262:215–223. doi: 10.1111/j.1365-2796.2007.01803.x. [DOI] [PubMed] [Google Scholar]
- Chung WH, Dao RL, Chen LK, Hung SI. The role of genetic variants in human longevity. Ageing Res Rev. 2010;9(Suppl 1):S67–S78. doi: 10.1016/j.arr.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton NR, Krueger RF, South SC, Gruenewald TL, Seeman TE, Roberts BW. Genes, environments, personality, and successful aging: toward a comprehensive developmental model in later life. J Gerontol A Biol Sci Med Sci. 2012;67:480–488. doi: 10.1093/gerona/gls090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci M, Ongaro F, Bresolin F, Bernardi U, Salvato C, Minello A, Amici GP, Barasciutti E, Mazzuco S, Gajo GB, De Angeli S, Forloni GL, Albani D, Zanardo A, Regini C. The Treviso Longeva (Trelong) study: a biomedical, demographic, economic and social investigation on people 70 years and over in a typical town of North-East of Italy. Arch Gerontol Geriatr. 2007;44(Suppl 1):173–192. doi: 10.1016/j.archger.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Amirabbasi-Beik M, Monfared N, Pieczyk M, Sailer C, Kollar A, Thomas R, Agalaridis G, Yamada S, Oliveira L, Collins JA, Meneilly G, Marra MA, Madden KM, Le ND, Connors JM, Brooks-Wilson AR. Genetic variation in healthy oldest-old. PLoS One. 2009;4(8):e6641. doi: 10.1371/journal.pone.0006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?—functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411:e11–e13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Bonasio R, Narendra V, Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol. 2012 doi: 10.1128/MCB.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescai F, Blanché H, Nebel A, Beekman M, Sahbatou M, Flachsbart F, Slagboom E, Schreiber S, Sorbi S, Passarino G, Franceschi C. Human longevity and 11p15.5: a study in 1321 centenarians. Eur J Hum Genet. 2009;17:1515–1519. doi: 10.1038/ejhg.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murabito JM, Yuan R, Lunetta KL. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J Gerontol A Biol Sci Med Sci. 2012;67:470–479. doi: 10.1093/gerona/gls089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolioni V, Carpi FM, Giannì P, Sacco R, Di Blasio L, Mignini F, Lucarini N, Persico AM. Age- and gender-specific epistasis between ADA and TNF-α influences human life-expectancy. Cytokine. 2011;56:481–488. doi: 10.1016/j.cyto.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Polito L, Kehoe PG, Davin A, Benussi L, Ghidoni R, Binetti G, Quadri P, Lucca U, Tettamanti M, Clerici F, Bagnoli S, Galimberti D, Nacmias B, Sorbi S, Guaita A, Scarpini E, Mariani C, Forloni G, Albani D (2013) The SIRT2 polymorphism rs10410544 and risk of Alzheimer's disease in two Caucasian case–control cohorts. Alzheimers Dement 9(4):392–399 [DOI] [PubMed]
- Polito L, Kehoe PG, Forloni G, Albani D. The molecular genetics of sirtuins: association with human longevity and age-related diseases. Int J Mol Epidemiol Genet. 2010;1:214–225. [PMC free article] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, Bonafe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/S0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- Seripa D, Pilotto A, Panza F, Matera MG, Pilotto A. Pharmacogenetics of cytochrome P450 (CYP) in the elderly. Ageing Res Rev. 2010;9:457–474. doi: 10.1016/j.arr.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Tazearslan C, Cho M, Suh Y. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J Gerontol A Biol Sci Med Sci. 2012;67:376–383. doi: 10.1093/gerona/glr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPEG 71 kb)
(JPEG 109 kb)