Abstract
The poly(A) tail of mRNA has an important influence on the dynamics of gene expression. On one hand, it promotes enhanced mRNA stability to allow production of the protein, even after inactivation of transcription. On the other hand, shortening of the poly(A) tail (deadenylation) slows down translation of the mRNA, or prevents it entirely, by inducing mRNA decay. Thus deadenylation plays a crucial role in the post-transcriptional regulation of gene expression, deciding the fate of individual mRNAs. It acts both in basal mRNA turnover, as well as temporally- and spatially-regulated translation and decay of specific mRNAs. Here, we discuss mRNA deadenylation in eukaryotes, focusing on the main deadenylase, the Ccr4-Not complex, including its composition, regulation and functional roles.
Keywords: mRNA, deadenylation, poly(A) tail, Ccr4-Not, gene expression
INTRODUCTION
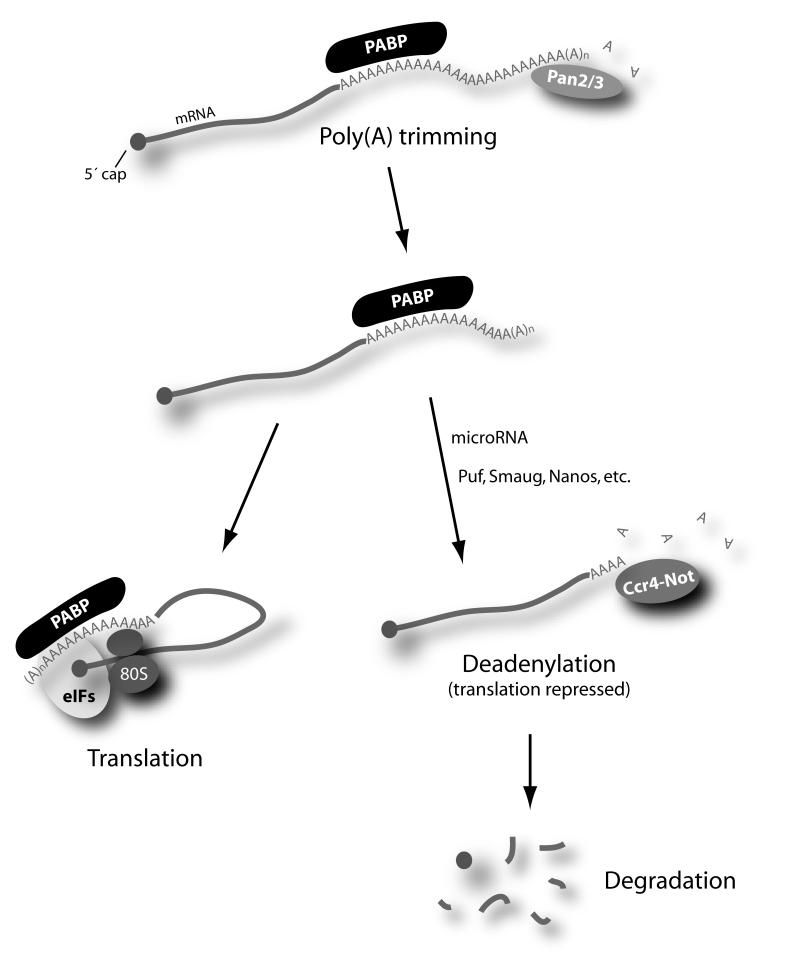
Regulated transcription is often the focus of studies on gene expression where polymerases, transcription factors and chromatin structure play important roles. However, post-transcriptional regulation is also a key factor in modulating gene expression [1], and much of this is controlled through the mRNA poly(A) tail. With the exception of some histone mRNAs, all protein coding mRNAs in eukaryotes contain a 3′ poly(A) tail. mRNAs exit the nucleus with a uniform poly(A) tail length (~70 nucleotides in yeast, ~250 nucleotides in mammals) but deadenylases act in the cytoplasm to shorten or remove it. Deadenylation occurs in both gene- and context-specific manners to allow differential control of poly(A) tail lengths [2]. Importantly, poly(A) tail length affects gene expression in two major ways – it controls mRNA stability and translational efficiency (Figure 1). This is essential for many processes including: periodic expression of cell cycle-related genes, microRNA-mediated gene silencing, and expression of maternal, masked mRNAs during oogenesis and early development [3-7]. By regulating poly(A) tail length, eukaryotes possess a highly sophisticated mechanism to allow exquisite control of gene expression.
Figure 1. Deadenylation of mRNAs in eukaryotes.
Poly(A) binding proteins (PABP) protect the 3′ end of mRNA from exonucleases and stimulate translation. The Pan2/3 complex trims poly(A) tails, while the Ccr4-Not complex removes them. Regulatory signals (e.g. microRNAs and Puf proteins) accelerate deadenylation by recruiting Ccr4-Not to target mRNAs.
In this review, we aim to give an overview of deadenylation, with a focus on the Ccr4-Not complex – the major deadenylase in eukaryotes. We emphasise proteins from the budding yeast Saccharomyces cerevisiae but also cover mechanisms in other organisms.
POLY(A) TAILS AND mRNA STABILITY
Poly(A) binding proteins (PABPs – Pab1p in yeast) protect the 3′-ends of mRNAs from exonucleases [8]. Indeed, the first step of mRNA degradation is normally the shortening or removal of the poly(A) tail [9, 10]. Computational studies have shown that the most significant factor determining mRNA degradation is the rate of deadenylation [9]. Deadenylation is followed by one of two alternate degradation pathways: 3′→5′ degradation by the cytoplasmic exosome, or removal of the 5′ cap (decapping) by Dcp1p/Dcp2p and 5′→3′ degradation by the Xrn1p exonuclease [8]. Recently, it was shown that this latter type of degradation can occur co-translationally, allowing the ribosome to complete the current round of translation before the mRNA is completely degraded [11].
POLY(A) TAILS AND TRANSLATION
Poly(A) tails also function during initiation of translation. Poly(A) binding proteins interact with the eukaryotic translation initiation factor 4F (eIF4F) that is bound to the 5′ cap [12]. These interactions effectively circularise the mRNA, acting as a control measure to ensure that only intact mRNAs are translated (Figure 1). Importantly, poly(A) binding proteins and eIF4F enhance recruitment of the small ribosomal subunit to promote translation initiation. Thus, a longer poly(A) tail can result in stimulation of translation whereas deadenylation decreases the efficiency of translation initiation [3]. Experiments showing co-regulation of poly(A) tail lengths of functionally related genes supports this [3, 4]. For example, in yeast the lengths of mRNA poly(A) tails of cell-cycle regulated genes are generally short suggesting that the strict temporal regulation of their expression depends, not only on transcription, but also on deadenylation [3]. In higher eukaryotes, cycles of deadenylation and cytoplasmic polyadenylation act in a general mechanism to control mitosis by regulating translation of cell cycle mRNAs [4].
THE EUKARYOTIC DEADENYLASES
Ccr4-Not complex
A major deadenylase activity involved in both basal mRNA decay and regulated deadenylation of specific transcripts is found within the evolutionarily conserved Ccr4-Not complex [10, 13]. Ccr4-Not is composed of nine different proteins subunits (including the deadenylase subunits Ccr4p and Pop2p/Caf1p) and will be discussed in detail below.
Pan complex
In ccr4 mutant yeast, a complex containing the Pan2p and Pan3p proteins is responsible for mRNA deadenylation, suggesting functional redundancy with Ccr4-Not [14]. Pan2p is the deadenylase subunit, containing a nuclease domain belonging to the DEDD superfamily. Pan3p acts as a regulator, binding to both Pan2p and poly(A) binding proteins. A ccr4/pan2 double mutant yeast strain grows very slowly and demonstrates no deadenylation, indicating that Ccr4p and Pan2p are responsible for most deadenylation [14]. The precise role of the Pan complex is unclear but it is thought to “trim” poly(A) tails to mature lengths, dependent on PABPs and on mRNA-specific signals (Figure 1) [13, 15]. In agreement with this, deletion of either pan2 or pan3 results in longer poly(A) tails but only causes a minor deadenylation defect [15]. There may be a complex interplay between the Ccr4-Not and Pan complexes, such that each contributes to cytoplasmic deadenylation. For example, Pan2/3 may shorten the poly(A) tail to a certain length, allowing Ccr4-Not to complete the deadenylation [13]. Further research is needed to clarify this.
PARN
The PARN nuclease (previously known as DAN) is found only in vertebrates. It forms a homodimer and differs from the other deadenylases because it binds to and is stimulated by the mRNA 5′ cap [13, 16]. Deadenylase activity is inhibited by poly(A)- and cap-binding proteins. PARN is responsible for deadenylation of a number of mRNAs, and often these are subject to intricate regulation. For example, PARN acts on specific mRNAs to repress their translation (until cytoplasmic poly(A) polymerases once again lengthen the poly(A) tail). This occurs during mitosis, with maternal mRNA in oocytes and in plant embryogenesis [4, 17, 18].
Nocturnin and Ngl
Two further deadenylases, Nocturnin and Ngl/ANGEL, share nuclease domain similarity with Ccr4 (i.e. they are members of the exonuclease/endonuclease/phosphatase, or EEP, superfamily). Nocturnin has been identified as a rhythmically expressed deadenylase in the cytoplasm of the retinal photoreceptor cells of Xenopus laevis and is thought to play a role in post-transcriptional regulation of circadian-related mRNAs [19]. Ngl is a deadenylase involved in 5.8S rRNA processing [20].
REGULATED REMOVAL OF THE POLY(A) TAIL BY THE CCR4-NOT COMPLEX
The Ccr4-Not complex is a 3′→5′ exoribonuclease with a preference for poly(A). Many of its subunits were first identified as transcriptional regulators. The Ccr4-Not complex localises to promoter regions, interacts with Transcription Factor IID (TFIID) and SAGA, and facilitates chromatin modifications such as trimethylation of histone H3 lysine 4 (H3K4me3) [21-23]. Thus, the Ccr4-Not complex may regulate gene expression not only through post-transcriptional deadenylation, but also at a transcriptional level. Both functions respond to extra- or intra-cellular stimuli. For example, the complex plays key roles in the cellular response to DNA replication stress and DNA damage [24], cell stress [25] and cell cycle control [26]. A recent microarray-based study suggested that the Ccr4-Not complex is important for the expression of most (>85%) of the yeast genome [27].
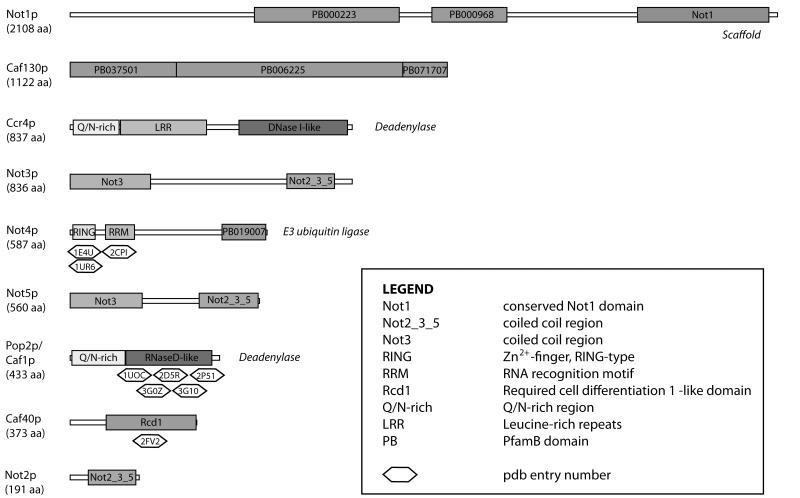
The Ccr4-Not complex is conserved across eukaryotes. In yeast, it is composed of nine subunits that can be purified as a 1 MDa particle [21, 22, 28]. The core complex consists of: two deadenylases (Ccr4p and Pop2p/Caf1p); five Not proteins (Not1p – Not5p); Caf40p; and Caf130p (Figure 2) [29]. At least two “modules” appear to exist within the Ccr4-Not complex: genetic and biochemical experiments have suggested that the Not proteins associate in one complex while Ccr4p, Pop2p, Caf40p and Caf130p exist in a second subcomplex. The Not and deadenylase modules are joined through the 240 kDa Not1p protein which acts as a scaffold and is necessary for yeast viability [30].
Figure 2. Schematic representation of yeast Ccr4-Not complex subunits.
Domains are depicted according to known structures and predictions using the Pfam database. PfamB domains are computationally generated and unannotated but can give insights into functional conserved regions. Functional roles are given on the left of each subunit, in italics. PDB accession codes are given underneath each protein for any available structures. It should be noted that in higher eukaryotes the composition of the Ccr4-Not complex varies slightly. Not3p and Not5p are very similar proteins and in humans they have only one ortholog, CNOT3. In contrast, the deadenylase subunits Ccr4p and Pop2p have two human orthologues (CNOT6/hCcr4a and CNOT6L/hCcr4b; CNOT7/hCaf1 and CNOT8/hPop2/CALIF).
The two deadenylase subunits are unrelated: Ccr4p contains a DNaseI-like domain of the EEP superfamily while Pop2p contains a RNaseD-like domain belonging to the DEDD family. It is unclear why both are present. Deletion of either ccr4 or pop2 results in defects in the rate and extent of deadenylation [14]. Experiments so far have indicated that Ccr4p activity is predominant [10, 31]. For example, overexpression of Ccr4p overcomes the deadenylation defects in a yeast strain with a pop2 deletion, and point mutations in catalytic residues of Ccr4p, but not Pop2p, inhibits deadenylation in vivo and in vitro [31, 32]. Correspondingly, overexpression of Ccr4p, but not Pop2p, in a wild type yeast strain increases mRNA deadenylation [31].
Although it appears to have lower activity, Pop2p is also a functional deadenylase and may be responsible for regulated deadenylation of specific transcripts, with Ccr4p being responsible for basal mRNA degradation [32, 33]. In addition, Pop2p likely plays a role to stabilise the complex and/or activate Ccr4p. Indeed, Ccr4p associates with the Ccr4-Not complex primarily though Pop2, and catalytically inactive Pop2p can complement a yeast strain containing a pop2 deletion [34].
In human cells, there are two isoforms of both Ccr4 (CNOT6/hCcr4a and CNOT6L/hCcr4b) and Pop2 (CNOT7/hCaf1 and CNOT8/hPop2). Since each isoform may have different substrate specificities, this allows formation of multiple types of Ccr4-Not deadenylase complexes.
In addition to its deadenylase activity, the Ccr4-Not complex possesses E3 ubiquitin ligase activity in the RING finger of its Not4p subunit. The substrates of this activity remain poorly defined but include the heterodimeric nascent polypeptide-associated complex (NAC), or EGD complex [35]. NAC/EGD binds to the ribosome, near the polypeptide exit tunnel to protect nascent chains as they emerge, participating in protein folding and ribosome biogenesis [36]. Its ubiquitination promotes ribosome association but does not affect protein stability [37]. Not4p also ubiquitinates translational arrest products that occur after translation of poly(A) [38]. Poly(A) tails are not normally translated and therefore, by recognising arrested ribosomes Not4p promotes degradation of aberrant proteins that have been produced from mRNAs lacking a stop codon (nonstop mRNAs). Finally, Not4p ubiquitinates the histone H3 Lys 4 (H3K4) demethylase Jhd2p/JARID1C, targeting it for degradation by the proteasome to regulate H3K4 trimethylation and gene expression [23].
It was recently shown that the Ccr4-Not complex (and Not3 in particular) is a conserved regulator of heart function [39]. Neely et al. (2010) performed a RNAi screen in Drosophila and identified a number of Ccr4-Not components as important for cardiac function. These include Not1, Not2, Not3, Not4, the Drosophila homolog of NAC and the E2 Ubc4. Importantly, not3 −/− knockout mice are embryonic lethal while not3 +/− mice have heart defects (spontaneous impairment of cardiac contractility and increased susceptibility to heart failure). Moreover, the Ccr4-Not complex is likely involved in heart function in humans since SNPs in Ccr4-Not genes are correlated with altered cardiac QT intervals [39].
Together, the Ccr4-Not complex probably plays an integrated role in transcription, mRNA stability, and translational regulation. Its two catalytic activities (deadenylation and E3 ubiquitin ligase) could mediate most of the functions ascribed to the Ccr4-Not complex. However, the extent and relevance of the E3 ubiquitin ligase activity, and how this is linked to deadenylation, is still unclear. Further research will be needed to clarify this.
microRNA-TARGETED DEADENYLATION
microRNAs (miRNAs) are short RNAs that bind to sequences in the 3′ untranslated regions (UTRs) of their target mRNAs with imperfect complementarity. This recruits Argonaute (Ago) proteins and GW182/TNRC6 to form a miRNA-loaded RNA-induced silencing complex (miRISC) that silences expression of the target mRNA (reviewed in [40] and [41]). At least part of the silencing mechanism involves mRNA deadenylation. Specifically, miRISC recruits the Ccr4-Not complex, triggering deadenylation of target mRNAs and leading to mRNA decay and/or translational repression [5, 42, 43].
miRNA-induced deadenylation requires the deadenylase activity of Pop2/Caf1 and an interaction between Ago and GW182 [5, 42, 44]. GW182 is thought to recruit the Ccr4-Not complex, although no direct interactions have been shown. Tethering of GW182 to mRNA results in deadenylation, independent of miRNA or Ago [7]. The details of this process remain unclear, including whether Ccr4 activity is also required and how the recruitment is mediated.
Interestingly, PABP is also necessary for miRNA-induced deadenylation [5], even though in vitro assays have demonstrated that PABP inhibits deadenylation by Ccr4-Not complexes [31]. A possible explanation is given by recent studies showing that the C-terminal domain of GW182 interacts with PABP (reviewed in [7]). Thus, GW182 might antagonize PABP function and thereby destabilize the PABP-poly(A) interaction. This, in turn, could increase the accessibility of poly(A)-tails for deadenylases.
The temporal order of miRNA-mediated repression is heavily debated. On one hand it has been suggested that repression of translation initiation occurs first, followed by mRNA deadenylation and degradation [5]. On the other hand, deadenylation may take place first and consequently mRNAs with shorter poly(A)-tails are destabilised and translation initiation becomes less efficient [45].
PROTEIN-TARGETED DEADENYLATION
The Ccr4-Not complex can be recruited to mRNA by conserved regulatory proteins (e.g. Pumilio/FBF (Puf) proteins) bound to specific sequences in the 3′ UTR [46]. Indeed, tethering of either Pop2 or specific 3′ UTR-binding proteins to a model mRNA is sufficient to trigger rapid deadenylation and mRNA decay [47-49].
Puf proteins are found in all eukaryotes and they control mRNA stability and translation, for example in development, memory and stem cell maintenance. Puf proteins increase the efficiency of Ccr4-Not-mediated deadenylation of specific mRNAs by interacting with both binding elements in the 3′ UTR and Pop2p, thereby directly recruiting the complex to target mRNAs [50]. They bind to large groups of specific mRNAs (e.g. they bind 10-15% of yeast transcripts) and each of the six yeast Puf protein seems to regulate a different set of functionally related genes [51].
Similarly, other evolutionarily conserved proteins bind sequences in the 3′ UTR and recruit the Ccr4-Not complex: Nanos binds to Not4 [49, 52]; Bicaudal-C/Bic-C binds Not3/5 [53] and Smaug/Vts1p recruits the Ccr4-Not complex, although its direct interaction partner has not been characterised [48]. These conserved recruitment factors all play crucial roles early development and have been characterised in the context of Drosophila embryonic development. However, this fine-tuning of translational control plays a general role wherever spatial or temporal regulation of protein synthesis occurs, for example in mitosis and in synapses [4, 54]. Future work should address exactly how these proteins control gene expression.
PERSPECTIVES
Many questions remain unanswered about the mechanisms of regulated deadenylation of mRNAs in eukaryotes. For example, what is the relationship between different deadenylases, specifically the Ccr4-Not and Pan complexes? How is deadenylation coupled to other mechanisms of gene regulation? What are the precise mechanisms of recruitment factors? It has been suggested that deadenylation requires remodelling of poly(A)-binding proteins on the poly(A) tail to make it more accessible [55, 56]. However, it is unclear what promotes this reorganisation – perhaps additional regulatory proteins assist in releasing or altering the structure of PABPs. Structural models of intact deadenylation complexes as well as detailed biochemical characterisations will begin to answer these questions and explain the cellular determinants of poly(A) tail length.
Acknowledgements
Work in the authors’ laboratory is funded by the Medical Research Council (MRC).
Abbreviations used
- DAN
deadenylating nuclease
- eIF
eukaryotic translation initiation factor
- miRISC
miRNA-induced silencing complex
- miRNA
microRNA
- NAC
nascent polypeptide-associated complex
- PABP
poly(A) binding protein
- PAN
Pab1-dependent poly(A) nuclease
- PARN
poly(A) ribonuclease
- RNAi
RNA interference
- SNP
single nucleotide polymorphism
- TFIID
Transcription Factor IID
- UTR
untranslated region
REFERENCES
- 1.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 2.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 3.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 5.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, 3rd, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat Rev Mol Cell Biol. 2010;11:379–384. doi: 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 8.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 9.Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 11.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 14.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 15.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H. Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease. Structure. 2009;17:276–286. doi: 10.1016/j.str.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–1214. doi: 10.1261/rna.7540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 20.Faber AW, Van Dijk M, Raue HA, Vos JC. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA. 2002;8:1095–1101. doi: 10.1017/s1355838202021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 22.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 23.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolstencroft RN, Beilharz TH, Cook MA, Preiss T, Durocher D, Tyers M. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J Cell Sci. 2006;119:5178–5192. doi: 10.1242/jcs.03221. [DOI] [PubMed] [Google Scholar]
- 25.Mulder KW, Inagaki A, Cameroni E, Mousson F, Winkler GS, De Virgilio C, Collart MA, Timmers HT. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics. 2007;176:181–192. doi: 10.1534/genetics.106.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS. The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell. 2009;20:3840–3850. doi: 10.1091/mbc.E09-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, Collart MA. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 30.Maillet L, Tu C, Hong YK, Shuster EO, Collart MA. The essential function of Not1 lies within the Ccr4-Not complex. J Mol Biol. 2000;303:131–143. doi: 10.1006/jmbi.2000.4131. [DOI] [PubMed] [Google Scholar]
- 31.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 33.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol Cell Biol. 2010;30:460–469. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J Biol Chem. 2006;281:31389–31398. doi: 10.1074/jbc.M604986200. [DOI] [PubMed] [Google Scholar]
- 36.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panasenko OO, David FP, Collart MA. Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination. Genetics. 2009;181:447–460. doi: 10.1534/genetics.108.095422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmen L, Gupta V, Arora S, Sarangi R, Dan D, Fujisawa S, Usami T, Xia CP, Keene AC, Alayari NN, Yamakawa H, Elling U, Berger C, Novatchkova M, Koglgruber R, Fukuda K, Nishina H, Isobe M, Pospisilik JA, Imai Y, Pfeufer A, Hicks AA, Pramstaller PP, Subramaniam S, Kimura A, Ocorr K, Bodmer R, Penninger JM. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 41.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 47.Finoux AL, Seraphin B. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J Biol Chem. 2006;281:25940–25947. doi: 10.1074/jbc.M600132200. [DOI] [PubMed] [Google Scholar]
- 48.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 49.Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- 50.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 51.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci U S A. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci U S A. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon E, Seraphin B. A specific role for the C-terminal region of the Poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 2007;35:6017–6028. doi: 10.1093/nar/gkm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]