SUMMARY
A causal role of gene amplification in tumorigenesis is well-known, while amplification of DNA regulatory elements as an oncogenic driver remains unclear. In this study, we integrated next-generation sequencing approaches to map distant estrogen response elements (DEREs) that remotely control transcription of target genes through chromatin proximity. Two densely mapped DERE regions located on chromosomes 17q23 and 20q13 were frequently amplified in ERα-positive luminal breast cancer. These aberrantly amplified DEREs deregulated target gene expression potentially linked to cancer development and tamoxifen resistance. Progressive accumulation of DERE copies was observed in normal breast progenitor cells chronically exposed to estrogenic chemicals. These findings may extend to other DNA regulatory elements, the amplification of which can profoundly alter target transcriptome during tumorigenesis.
INTRODCUTION
For more than two decades, considerable efforts aimed at identifying oncogenes located near or within amplified genomic regions in cancer have been at the forefront of cancer research (Albertson 2006; Lupski et al., 2005; Stephens et al., 2011). A longstanding view is that these amplification events contribute to increased expression dosages of genes located in the regions for oncogenesis. The first oncogene, MYCN, mapped to chromosome 2p24 is amplified up to 300 copies in homogeneously staining regions of neuroblastomas (Schwab et al., 1983). High MYCN transcription levels typically accompanied with this genomic copy-number gain (Schwab 1999). ERBB2, localized on chromosome 17q21, is another well-characterized gene frequently amplified and overexpressed in breast cancer (Santarius et al., 2010; Slamon et al., 1987). These findings indicate that genomic amplification is a major mechanism underlying the activation of oncogenes during tumor development.
Intensive efforts have recently been undertaken to globally screen for and analyze genomic amplicons in breast cancer (Curtis et al., 2012; Nikolsky et al., 2008; Pollack et al., 2002; Santarius et al., 2010). In a study of 2,000 primary breast tumors, copy-number gains were frequently found on chromosomes 1q, 7p, 8, 11q, 14q, 16, 17q and 20q (Curtis et al., 2012). Oncogenes including PIK3CA, EGFR, ERBB2, and FOXA1 located in these amplicons were overexpressed in specific breast cancer subtypes. However, other copy-number gains were not necessarily correlated with up-regulation of candidate loci found in amplicons (Cancer Genome Atlas Network 2012; Curtis et al., 2012; Santarius et al., 2010). In other global surveys, >80% of amplified or structurally rearranged regions did not harbor protein-coding or microRNA loci aberrantly expressed in cancer (Hampton et al., 2009; Stephens et al., 2011). In those cases, copy-number gains of DNA regulatory elements with the strong potential to impact target gene transcription have often been overlooked. Transcription repressors or activators usually bind to these regulatory regions located near transcription start sites (TSSs) of genes. Furthermore, increasing evidence has recently indicated that DNA regulatory regions are remotely located downstream or far upstream from TSSs or even on different chromosomes. Through intra- or inter-chromatin looping, these regulatory elements are brought near TSSs for transcriptional repression or activation of target genes (Carroll et al., 2006; Fullwood et al., 2009; Hsu et al., 2010; Hu et al., 2008; Visel et al., 2009). Frequent chromatin interactions may increase DNA breaks at or nearby the looping sites, contributing to genomic instability in cancer cells. Inappropriate DNA repair may lead to genomic fusion and duplication of regulatory elements and gene promoters (Berger et al., 2011; Lin et al., 2009; Mani et al., 2009). In addition to oncogene amplification, we hypothesize that genomic amplification of DNA regulatory elements offers another mechanistic explanation for extensive transcription alterations during tumorigenesis.
Our goal in this study was to comprehensively map distant estrogen response elements (DEREs) in breast cancer that regulate the transcription of remote target genes using integrated next-generation sequencing approaches. We also examined the role of estrogen receptor activation through its ligand estradiol in enhancing DERE-mediated transcriptional control and aberrant gains of DERE copy. Furthermore, we addressed whether DERE amplification can be associated with endocrine resistance in breast cancer
RESULTS
Integrative Analyses Identify Densely Mapped DERE Regions
To globally survey estrogen-induced chromatin interaction events, we performed chromosome conformation capture (3C) coupled with paired-end sequencing in MCF-7 cells stimulated with E2 (17β-estradiol, 70 nM) for 24 hr (Dekker et al., 2002; Hagège et al., 2007). Based on our previous study, progressive increases in chromatin interaction frequency occurred during this stimulation period (Hsu et al., 2010). Cross-linked chromatin was digested with BamHI for subsequent diluted ligation (Figure 1A, STEP 1). Ligated DNA fragments attributed to different chromosome regions coming into close proximity through chromatin looping were subjected to paired-end sequencing (total 0.3 billion 51-bp sequence reads; Table S1). Poisson regression modeling was used to determine true chromatin-looping events (FDR=8.35%). To demonstrate the reliability of this 3C assay, reproducibility, saturation, and sensitivity analyses were also conducted on two biological replicates (Figure S1A–B and Table S2).
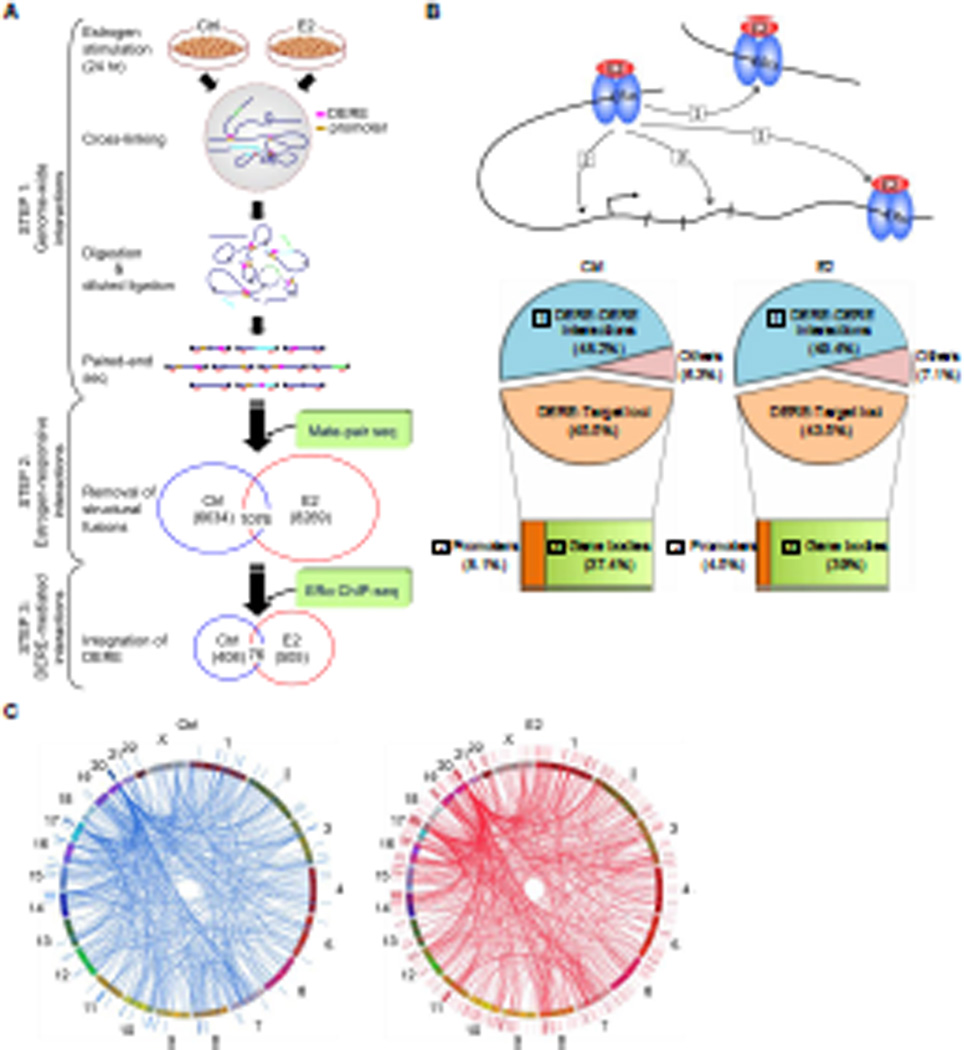
Figure 1. Integrative Mapping of ERα-mediated Chromatin Interactions Based on Next-generation Sequencing Approaches.
(A) Integrative scheme of identifying ERα/DERE-mediated chromatin interaction sites in E2-stimulated MCF-7 human breast cancer cells. Chromosome conformation capture (3C) assay coupled with paired-end sequencing was performed on both untreated (Ctrl) and estrogen-treated (E2, 70 nM) MCF-7 cells to survey chromatin interaction events in a genome-wide manner (STEP 1) (see also Figure S1A–B and Tables S1–2). To identify genuine interaction sites, genomic fusions and self-ligated fragments mapped by mate-pair sequencing were filtered-out from the 3C-seq dataset (STEP 2) (see also Tables S3–6). The filtered data were then integrated with ERα ChIP-seq datasets (0 and 24 hr, respectively) and distant estrogen response elements (DEREs) were mapped to define DERE-associated chromatin interaction events (STEP 3) (see also Tables S7).
(B) Genomic distribution of ERα-mediated chromatin interaction sites. ERα-mediated interaction sites mapped within 10-kb regions of DEREs, which have no known target genes, were defined as DERE-DERE interactions. In the target loci category, the regions within 10-kb upstream and 1-kb downstream of the transcription start site (TSS) of a gene were defined as promoters. “Others” were defined as ERα-mediated interaction sites mapped in gene-desert regions.
(C) Circular visualization of ERα-mediated interactions upon E2 stimulation. Circular plots depict interactive loci of different chromatin loops using the Circos software (http://mkweb,bcgsc.ca/circos/). Chromosomes are individually colored. The locations of DEREs are represented as lines outside the chromosomes “circle”. Four clustered DEREs were identified in 1p13, 3p14, 17q23, and 20q13 regions (see also Figure S1C).
As the above sequencing approach might capture structural fusions, we comprehensively mapped all putative translocation sites in MCF-7 cells by mate-pair sequencing with ~31X converge of the genome (total 2.4 billion 51-bp sequence reads;Table S3). All potential false-positive fusions produced by random ligations were eliminated using Poisson regression modeling (FDR<0.000001). A total of 429 translocation fusions were identified in the MCF-7 genome (Table S4). By excluding these translocation events from the 3C dataset, 6,634 and 8,269 chromatin interaction events were unique to the control and E2-stimulated cells, respectively (Figure 1A, STEP 2; Tables S5–6).
To further define DERE-associated chromatin interactions, we performed a time-course chromatin immunoprecipitation using an ERα antibody coupled with single-end sequencing (ERα ChIP-seq) in MCF-7 cells stimulated with E2 for 0, 0.5, 1 and 24 hr (Figure 1A, STEP3). A total of 0.3 billion 51-bp sequence reads were processed and 58.9% of those reads mapped to unique chromosome locations (Table S7). The wBELT Peak Calling program was applied to identify specifically ERα-bound DEREs upon E2 stimulation (Hsu et al., 2010). De novo binding events (n=11,397) accumulated by 1 hr and decreased by 24 hr. Integration with the aforementioned chromatin looping data, we found 505 DERE-associated chromatin interaction events after E2 treatment for a 24-hr period (Figure 1A, STEP3). Notably, 408 pre-existing interactions simultaneously disappeared, consistently supporting our previous finding that ERα binding also results in chromatin dissociation in target sites (Hsu et al., 2010). In addition, 76 of these pre-existing interaction events remained unchanged upon E2 stimulation. When categorizing these interaction sites based on their genomic locations, we found that DEREs from either the same or different chromosomes frequently interacted with each other, accounting for ~49% of all interaction events (Figure 1B). As expected, DEREs were found to interact with an overall large number of target genes through intra- and inter-chromatin looping (n=467). However, only 4–8% of all DERE interactions occurred near promoter regions while the majority (37–39%) of these interaction sites occurred within gene bodies (Figure 1B).
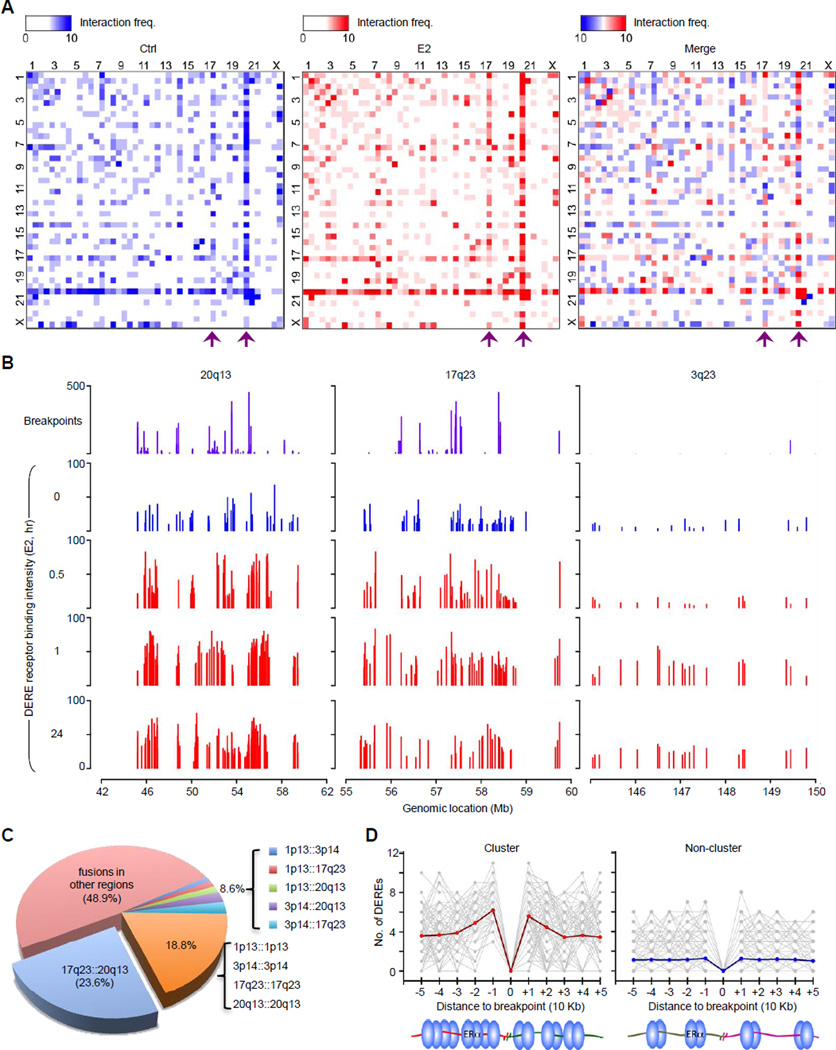
Interestingly, DERE interaction sites were frequently clustered in specific chromosome regions, such as 1p13, 3p14, 17q23, and 20q13 (Figures 1C and S1C). Pair-wise heat maps depicting interaction frequencies between different DEREs showed that chromatin segments on 20q13 or 17q23 most frequently interacted with other segments on different chromosomes (see purple arrows in Figure 2A). In addition, these two regions displayed the highest density (46–51 sites per megabase) of DEREs bound by ERα compared to other genomic regions (e.g., 3q23, 2–5 sites per megabase) after E2 treatment (Figure 2B, blue and red landscape maps, and Table S8). We also found that these two regions contained hot spots for genomic translocations (Figure 2B, purple maps; Figure S2A–B). When surveying the entire MCF-7 genome, we observed that 23.6% of all fusion events occurred between 17q23 and 20q13 (Figure 2C). Moreover, these densely localized DEREs were frequently mapped near or at clustered breakpoints in 17q23 and 20q13 regions (5–10 breakpoints per megabase) compared to other non-clustered regions (p<0.001; Figure 2D). Additional data indicate that estrogenic stimulation leads to distant chromatin interactions involving ERα-bound DEREs, most notably in 17q23 and 20q13 (Table S8).
Figure 2. Co-localization of Densely ERα-bound DEREs and Translocation-susceptible Regions.
(A) Heat maps of DERE-DERE chromatin interactions. Frequencies of DERE-DERE interactions in p and q arms of individual chromosomes were plotted. Purple arrows indicate two major sites of DERE-DERE interactions on 20q13 and 17q23, respectively. See also Figure S2C–E for a proposed model of DERE-DERE fusions and amplification.
(B) Genomic maps of translocation breakpoints and ERα-bound DEREs in three representative regions (3q23, 17q23, and 20q13) of MCF-7 cells. MCF-7 cells stimulated with E2 (70 nM) in a time-dependent manner (0, 0.5, 1, and 24 hr) were subjected to ChIP-seq for defining ERα-bound DEREs. Fusion frequencies of breakpoint sites are plotted in purple and binding intensities of ERα-bound DEREs in blue (untreated) and red (E2-treated). See also Figure S2A–B for whole-genome and individual chromosome maps, respectively.
(C) Pie chart summarizing all fusion events in MCF-7 cells based on mate-pair sequencing data.
(D) Distribution of DEREs nearby translocation sites in clustered and non-clustered breakpoints. The number of DEREs per 10-kb span is calculated in each direction relative to each breakpoint site. The red and blue lines are the average value of DEREs.
Amplified DERE Copies are Linked to Adverse Outcome of ERα-positive Luminal Breast Cancer
In addition to chromosomal fusions, interphase fluorescence in situ hybridization (FISH) analysis indicated that DERE copies in 20q13 were increased with time in E2-treated MCF-7 cells (Figure 3A; statistics of 20q13 DERE copies shown in right-upper scatter plot). Notably, long-term E2 exposure also triggered the formation of DERE clusters (see inserted squares and right-lower scatter plot). Quantitative PCR analyses further confirmed that estrogen stimulation significantly led to DERE amplification on 17q23 and 20q13 regions in MCF-7 cells (p<0.001; Figure 3B). The ERα antagonist- ICI 182,780 (ICI, 100 nM) effectively inhibited DERE copy increases elicited by E2 (see E2+ICI group).
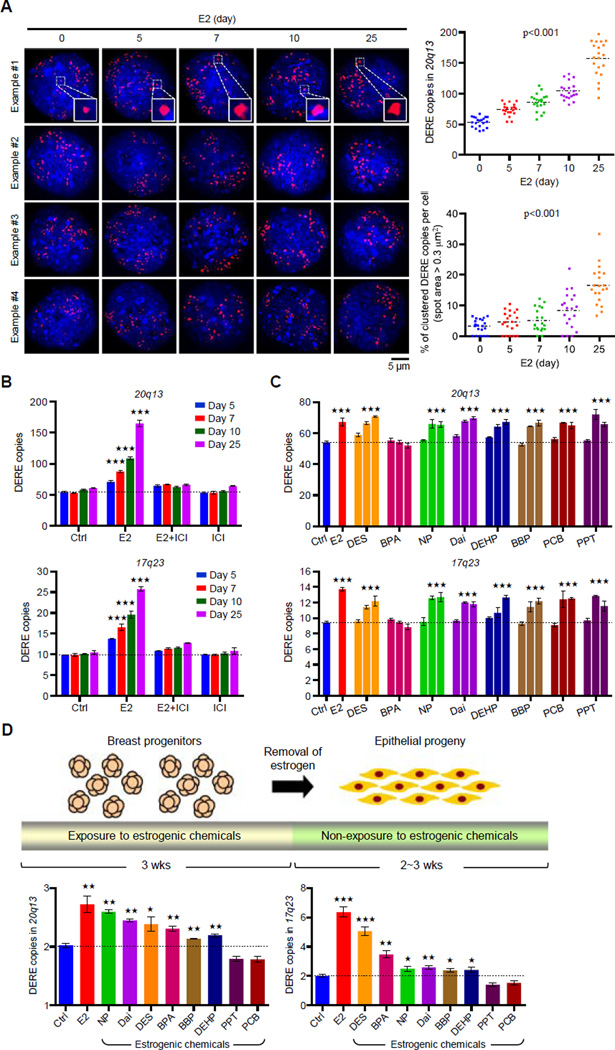
Figure 3. Prolonged Estrogen Exposure Leads to Amplification of DERE Copies.
(A) Interphase fluorescence in situ hybridization (FISH) analysis of amplified 20q13 DERE copies in E2-treated (70 nM) MCF-7 cells for different time periods (0, 5, 7, 10, and 25 days). Representative four images in each condition were shown. Inserted squares: clustered DEREs. Quantification of DERE copies per cell was performed by CellSens software and presented in the scatter plot (n=20). A spot with area size over 0.3 µm2 was counted as the clustered DERE region. p<0.001 (two way ANOVA test), compared to “0” group.
(B) Quantitative PCR analysis of two amplified DERE copies located in 20q13 (upper) and 17q23 (lower). MCF-7 cells were continuously exposed to E2 (70 nM) and/or ICI 182,780 (100 nM) for different time periods (5, 7, 10, and 25 days) in charcoal-stripped conditions (n=6 replicates in two biological batches of treatment). Mean ± SD. ***, p<0.001 (Student’s t test), compared to “Ctrl” cells.
(C) Dose-dependent gains of 20q13 and 17q23 DERE copy in MCF-7 cells exposed to different estrogenic chemicals. MCF-7 cells were cultured in charcoal-stripped conditions and exposed to ethanol (Ctrl), E2 (70 nM), or estrogenic chemicals with 5-fold different dose, including diethylstilbestrol (DES, 14-70-140 nM), bisphenol A (BPA, 0.5-4–20 nM), 4-nonylphenol (NP, 0.2-1–5 µM), daidzein (Dai, 2-10–50 µM), N-butyl-benzyl phthalate (BBP, 2-10-50 µM), di(2-ethylhexyl)-phthalate (DEHP, 2-10–50 µM), 4,4’-dichloro-biphnyl (PCB, 0.02-0.1–0.5 nM), and 1,3,5-tris(4-hydroxyphenyl)- 4-propyl-1H-pyrazole (PPT, 0.02-0.1–0.5 nM), respectively, for 5 days. These treatment doses were selected and modified based on our previous findings (Hsu et al., 2009 and 2010). Genomic DNA from treated cells was collected for quantitative PCR analysis of 17q23 and 20q13 DERE copies. Mean ± SD (n=6 replicates in two biological batches of treatment). ***, p<0.001 (Student’s t test), compared to “Ctrl” cells.
(D) Differential copy changes of 20q13 and 17q23 DEREs in normal epithelial cells preexposed to estrogenic chemicals. Experimental scheme of an in vitro exposure system is shown in the upper panel. Floating mammospheres containing breast progenitor cells were preexposed to dimethyl sulfoxide (DMSO as control, Ctrl), E2 (70 nM), or estrogenic chemicals, including DES (70 nM), BPA (4 nM), NP (1 µM), Dai (10µM), BBP (10µM), DEHP (10µM), PCB (0.1 nM), and PPT (0.1 nM), respectively, for 3 weeks. Differentiated epithelial cells were then subjected to quantitative PCR analysis (lower) of 17q23 and 20q13 DERE copies. Mean ± SD (n=6 replicates in two batches of treatment). ***, p<0.001 (Student’s t test), compared to “Ctrl” cells.
Based on the above results, we hypothesized that prolonged exposure to estrogenic chemicals contributes to DERE amplification. MCF-7 cells were used to determine whether other estrogenic chemicals recapitulate estrogen-mediated gains of DERE copy (Hsu et al., 2009 and 2010). Seven of the eight compounds examined, except bisphenol A (BPA), demonstrated increases in DERE copies on 17q23 and 20q13 in a dose-dependent manner (p<0.001; Figure 3C).
To determine whether this estrogen-driven DERE amplification occurs in breast stem/progenitor cells, normal mammospheres harboring self-regenerating cells were exposed to E2 or other aforementioned estrogens (Figure 3D, upper). We then measured changes of DERE copy located on 17q23 and 20q13 in differentiated progeny without additional estrogenic exposure. In two biological replicates, increased DERE copies were consistently observed in epithelial cells pre-exposed to E2 or six of the eight compounds tested (p<0.01; Figure 3D, lower). One possible explanation as to differences in drug response between normal and cancer cells could be due to “U-shaped” dose-response curves in reaction to particular drugs (Almstrup et al., 2002; Li et al., 2007; Vandenberg et al., 2006).
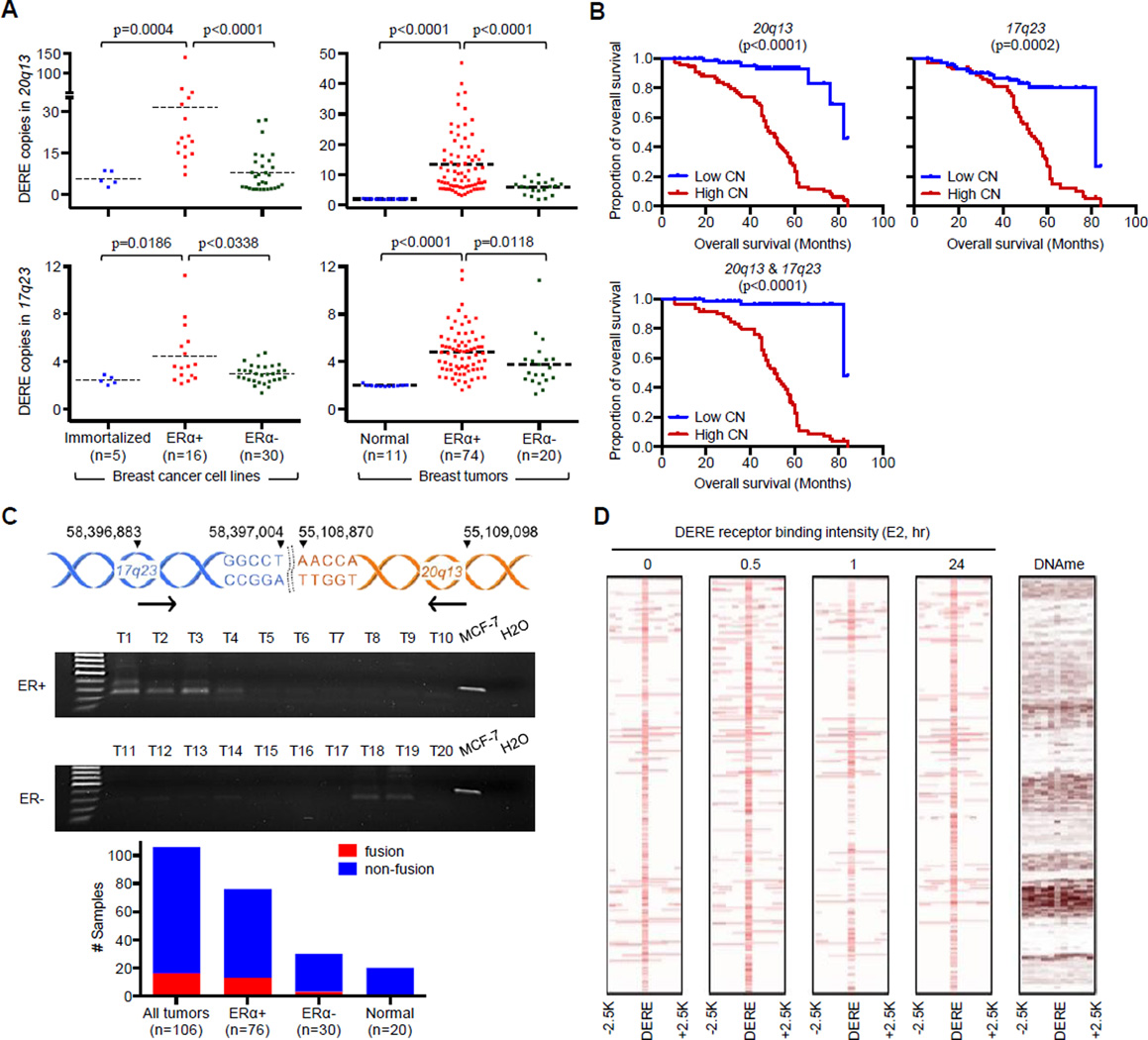
Examining DERE copy number in the Integrative Cancer Biology Program (ICBP) panel of 46 breast cancer and 5 immortalized cell lines revealed that DERE copies were significantly increased in ERα-positive breast cancer cell lines compared to ERα-negative cancer and immortalized lines (p<0.0004 in 20q13 and p<0.0186 in 17q23; Figure 4A, left). Furthermore, ERα-positive luminal-A cell lines (Figure S3A–B, dashed red lines) appeared to have on average higher DERE copies than those of luminal-B subtypes (dashed blue lines). Quantitative PCR analysis of 105 clinical samples showed a similar DERE amplification event in ERα-positive breast tumors (p<0.0118, Figure 4A, right). The clinical relevance of these observations illustrated by a Kaplan-Meier analysis demonstrated that gains of 17q23 and/or 20q13 DERE copy were positively associated with reduced overall survival in patients with ERα-positive breast cancer (p<0.0002, Figure 4B). In contrast, no association between DERE amplification and survival was apparent in ERα-negative tumors (p<0.6452; Figure S3C).
Figure 4. Amplification of DERE Copies Preferentially Occurs in ERα-positive Breast Cancers.
(A) Quantitative analysis of 17q23 and 20q13 DERE copies in 51 immortalized and breast cancer cell lines (left) and 105 clinical samples, including 94 breast tumors and 11 normal tissues (right). See also Figure S3A–B for copy number of 20q13 and 17q23 DEREs in luminal breast cancer cell lines; Figure S3D–F and Table S9 for association of TP53-involved signaling network in 20q13 DERE amplification.
(B) Kaplan-Meier survival curves of ERα-positive breast cancer patients (n=74) harboring either high (n>2) or low copy (n<2) of the 20q13 (left) or 17q23 (middle) or both (right) DEREs. Wilcoxon test was used to determine statistical significance. See also Figure S3C for overall survival curves of ERα-negative breast cancer patients (n=19).
(C) PCR analysis of 17q23∷20q13 fusion fragment in 106 primary breast tumors and 20 normal tissues. Genomic location of interrogated fusion is shown in upper panel. Gel pictures of PCR results from ten representative ER-positive and -negative tumors, respectively, are shown, plus an MCF-7 positive control and H2O negative control.
(D) Intensity maps of DERE receptor binding upon different time periods of E2 treatment and DNA methylation. The flanking regions of DERE (centered) from −2.5-Kb to +2.5-Kb were shown. The heat map of DNA methylation in untreated MCF-7 cells was generated using MeDIP-seq data from our previous study (Hsu et al., 2010).
It was next of interest to investigate the association of genomic rearrangement with DERE amplification in patient samples. We examined 17q23∷20q13, the most frequent structural fusion in the MCF-7 genome, in 126 primary tissues using PCR analysis (Figure 4C, upper; Table S4). This fusion fragment was detected in 15.1% (16 out of 106) of tumors, but not in adjacent normal tissues (Figure 4C, see “All tumors” in bar chart). The majority of fusions were found in ERα-positive tumors (13 out of 16), suggesting that ERα signaling activation may contribute to both DERE amplification and the occurrence of 17q23∷20q13 structural fusions. To determine whether aberrant epigenetic changes of DERE regions influence these structural fusions in MCF-7 cells, we generated heat maps of DERE receptor binding occupancy and DNA methylation in DERE receptor binding regions using the aforementioned ChIP-seq data and our previously published MeDIP-seq data (Figure 4D) (Hsu et al., 2010). We observed that DEREs occupied by ERα were frequently hypomethylated compared with their flanking regions, which were hypermethylated. Hypomethylated regions are known to be structurally unstable and prone to develop breakage and fusion in cancer cells (You and Jones 2012).
Further in silico analysis identified 133 differentially expressed genes in three ERα-positive cell lines (600MPE, MDA-MB-361, and ZR-7530) having fewer 20q13 DERE copies (n<15) relative to three other cell lines (LY2, BT474, and MCF-7) with the highest copy-number gains (n>50) (Figure S3D and Table S9) (Heiser et al., 2009). Ingenuity Pathways Analysis (IPA) suggested that TP53/BRCA1-related DNA damage response in LY2, BT474, and MCF-7 cells enhanced this aberrant DERE copy increases (Figure S3E–F) (Mohr et al., 2011). Up-regulated (e.g., NBN, DNMT3B, HLTF and NCOA3) and down-regulated (e.g., CYP286, SNCG, and RRAS) loci involved in this damage response pathway likely promote DNA break-repair functions, leading to amplification of 20q13 DEREs in breast cancer cell lines. This finding, therefore, provides a future opportunity to investigate the relationship between DNA-damage regulatory pathways and DERE amplification in breast cancer.
Take together, these integrative results strongly indicated that 1) amplified DERE copies are associated with the development of ERα-positive luminal breast cancer and poorer survival in patients; and 2) this process is a general phenomenon in both normal and cancer cells exposed to different estrogenic chemicals. We further suggest that ERα binding sites in 17q23 and 20q13 regions are highly susceptible to breakage and fusion, contributing to genomic instability in cancer.
Amplified DERE Copies Regulate Target Genes through Long-range Chromatin Interactions
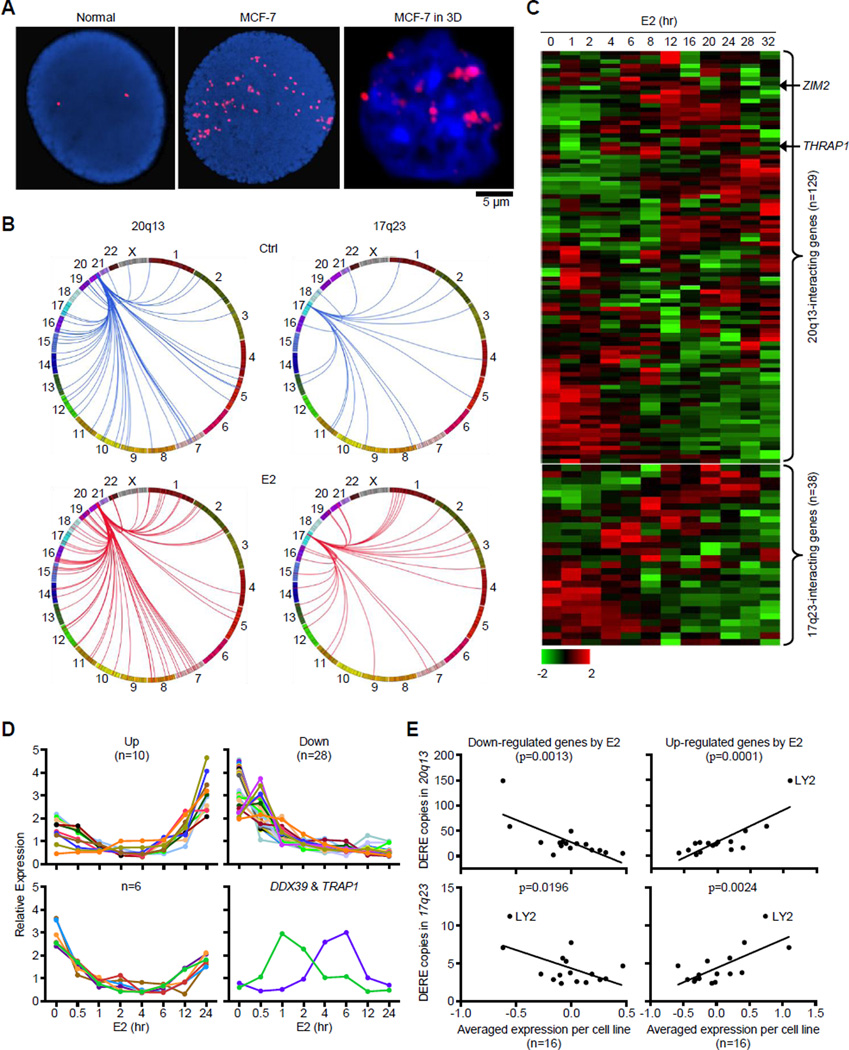
To address the biological consequence of DERE amplification, we investigated the effect of increased DERE copies on transcription of potentially oncogenic target genes using three-dimensional interphase fluorescence in situ hybridization (3D-FISH) analysis. A 3D-FISH approach preserving nuclear architecture of MCF-7 cells showed that DEREs tended to aggregate in 10–12 sub-nuclear locations (Figure 5A, right). In contrast, DEREs were scattered into 50–60 spots in a “compressed” nucleus by traditional 2D-FISH (middle). Consistent with the “transcription factory” concept (Lieberman-Aiden et al., 2009; Mitchell and Fraser 2008; Osborne et al., 2007), DEREs located on different chromosomes might be brought together to “sub-nuclear depots” for coordinate transcriptional control of multiple genes. Upon binding by ERα, DEREs (Figure 5B, red lines) might reach out through chromatin movement to interact with target genes while existing contacts (blue lines) dissociate from target loci in E2-stimulated MCF-7 cells. Integrating our 3C dataset with a published time-course study of gene expression in MCF-7cells (Cicatiello et al., 2010), we mapped two groups of target genes, including 95 loci remotely interacting with 20q13 DEREs and 38 genes with 17q23 DEREs (Figure 5C). A time-course analysis independently confirmed that 46 of these estrogen-responsive targets displayed down-regulated, up-regulated, or cycling patterns of gene expression over a 24-hr period of E2 stimulation (Figures 5D and S4A–B). These differential expression profiles regulated by DEREs, however, were less obvious in normal breast epithelial cells stimulated with E2 (see THRAP1 and ZIM2 as two examples in Figure 6B, blue line). Integration of our qPCR results of DERE copy with a published expression microarray data (i.e., ICBP breast cancer cell line panel; Heiser et al., 2009) further demonstrated that increased DERE copies preferentially correlated with increased or decreased expression levels of E2-responsive genes in 16 ERα-positive but not 30 ERα-negative cell lines (Figures 5E and S4C). Among these ICBP cell lines, the LY2 cell line was an outlier with the greatest number of 17q23 and 20q13 DERE copies, which seemed to have little effect on target gene regulation. These results indicate that ERα-bound DEREs remotely modulate transcriptional control of distant genes through long-range chromatin interactions in estrogen/ERα-driven tumorigenesis.
Figure 5. Amplified DERE Copies Regulate Target Genes through Estrogen-induced Chromatin Interactions.
(A) Interphase fluorescence in situ hybridization (FISH) analysis of amplified DERE copies in compressed (left and middle) and intact (right) nuclei.
(B) Circos plots depict chromatin interactions of two amplified DERE (20q13 and 17q23) with their respective target genes in untreated (Ctrl) and E2-treated MCF-7.
(C) Time-course analysis of gene expression synchronously regulated by either 20q13 (upper) or 17q23 (lower) DEREs. Heat maps generated using a published dataset (Cicatiello et al., 2010) show expression patterns of 20q13- or 17q23-interacting genes in response to E2 stimulation.
(D) Independent time-course analysis of 46 estrogen-responsive targets regulated by 20q13 DEREs. Total RNA isolated from E2-treated (70 nM) MCF-7 cells at different time-points was subjected to quantitative RT-PCR analysis. Based on the data obtained from two independent sets of experiments, four different patterns of gene expression were identified in E2-treated MCF-7 cells. Data were summarized in a heat map and shown in 46 bar charts with individual genes (see Figure S4A–B).
(E) Correlation analysis of DERE copy changes and DERE-regulated target gene expression in ERα-positive breast cancer cell lines (n=16). Expression microarray data of the ICBP cell lines (Heiser et al., 2009) were integrated with experimental copy-number results to interrogate the correlation between DERE amplification and transcriptional regulation. Down- and up-regulated genes were identified from Figure 5C. See also Figure S4C for correlation analysis in ERα-negative breast cancer cell lines (n=30).
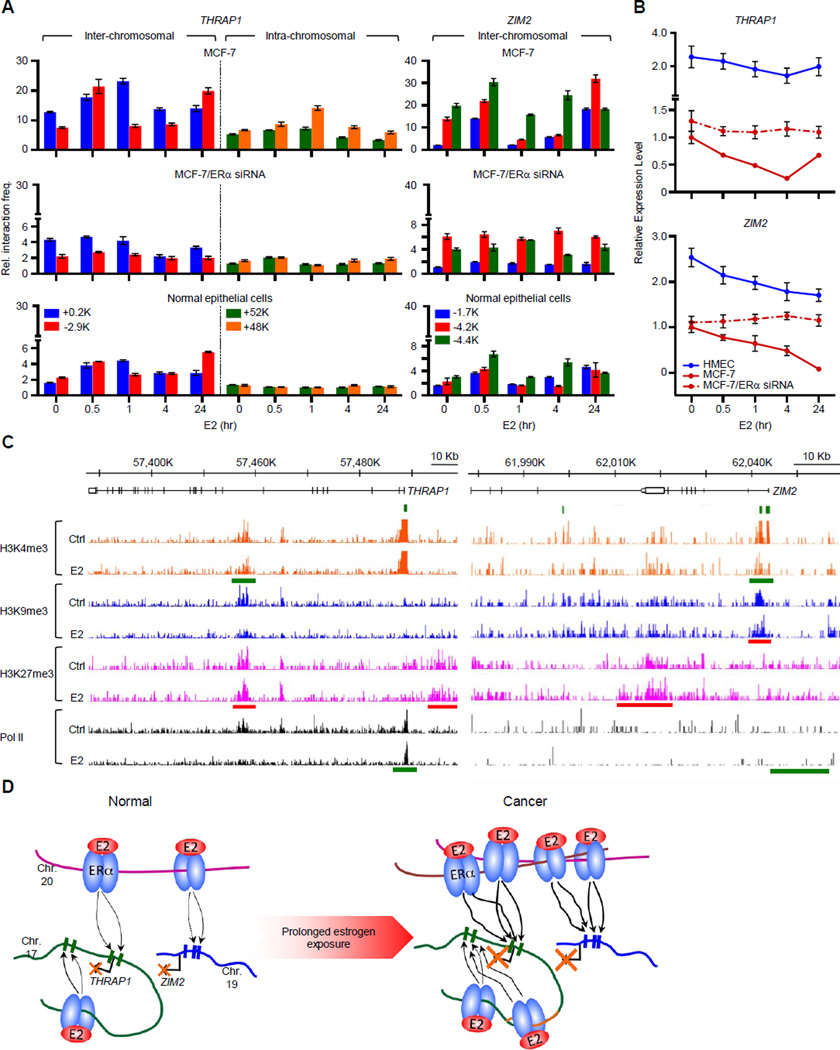
Figure 6. Two Representative Examples of Amplified DERE-regulated Target Genes through Long-range Chromatin Interactions.
(A) Chromosome conformation capture coupled with quantitative PCR (3C-qPCR) analysis of two target loci, ZIM2 and THRAP1. Cross-linked chromatin from E2-treated MCF-7 and normal epithelial cells was digested with either BamHI or HindIII and then ligated under diluted conditions. DEREs at 20q13 were designated as “baits”, and digested areas of two candidate loci, ZIM2 and THRAP1, were “interrogated fragments”. Ligated DNA was subjected to the 3C-qPCR. Data are shown as relative interaction frequencies compared to those of GAPDH as an internal control. Mean ± SD (n=6).
(B) Expression analyses of THRAP1 and ZIM2 in normal breast epithelial cells (HMEC) or cancer cells (MCF-7) in response to E2 (70 nM) determining using quantitative RT-PCR. Cells with or without ESR1 (ERα) knockdown by siRNA were treated with E2 for the indicated times. See also Figure S5 for the effect of siRNA on ESR1 expression.
(C) Genomic landscapes of histone modifications and Pol II occupancy on THRAP1 and ZIM2 loci upon estrogen stimulation. A published ChIP-seq data including three histone marks (H3K4me3, H3K9me3, and H3K27me3) and Pol II was used to investigate the occupancy of epigenetic marks on the two genes from −20-Kb upstream region of transcription start sites to transcription termination sites (Joseph et al., 2010). Red bars, increased occupancy; green bars, reduced occupancy.
(D) Proposed coordinate model of DERE-modulated chromatin interactions for transcriptional regulation in response to estrogen. In normal cells, DEREs at 20q13 are brought to THRAP1 (at 17q23.2) and ZIM2 (at 19q13.43), respectively, through chromatin movement to repress expression. During tumor progression under continuous estrogen exposure, genomic fusions and amplifications occur in the 20q13 DEREs attributed to prolonged physical contact between DEREs in an unstable cancer genome. The DEREs from 20q13 are replicated and inserted into different chromosomes, leading to increased interaction frequencies between DEREs and the respective gene loci, which profoundly alter their transcriptional regulation.
Amplified DERE Copies Repress Candidate Tumor-suppressor Loci and Drive Cell Proliferation of ERα-positive Luminal Cancers
To confirm long-range transcription regulation by DEREs, we conducted functional analyses of two selected target loci, THRAP1 (or MED13) and ZIM2. The rationale for choosing THRAP1 was that its gene product is an integral component of the repressive Mediator complex recently implicated in tumor growth (Monni et al., 2001; Knuesel et al., 2009). ZIM2 was also selected because its biological functions have not been explored in breast cancer. As a zinc-finger imprinted gene localized on 19q13, ZIM2 plays a regulatory role for axon guidance signaling involved in pancreatic cancer development (Biankin et al., 2012; Kim et al., 2004). We conducted chromosome conformation capture (3C)-qPCR assay to examine whether DEREs located on 17q23 and 20q13 can remotely regulate THRAP1 and ZIM2. In MCF-7 cells stimulated with E2 over a 24-hr period, inter-chromosomal interactions were more frequently observed between DEREs located on 20q13 and the THRAP1 gene (+0.2- and −2.9-kb flanking regions of TSS) and the ZIM2 promoter regions (−1.7-, −4.2-, and −4.4-kb) (Figure 6A, left and right). Intra-chromosomal interactions were also observed between 17q23 DEREs and THRAP1 gene-body regions (i.e., exon 13–14 regions located +52 and +48-kb from TSS), indicating cooperative efforts between different DEREs in regulating gene expression (Figure 6A, left and middle). Inter- and intra-chromosomal interactions associated with progressive down-regulation of the two loci in E2-stimulated MCF-7 cells (Figure 6B, red line). However, siRNA knockdown of ERα (Figure S5) markedly attenuated these inter- and intra-chromosomal interactions, abrogating the time-course repression of these genes (Figure 6A–B, dashed-dotted red lines). Compared to MCF-7 cells with 50~60 copies of amplified 20q13 segment (Figures 3A–B and 5A), the effect of this chromatin-mediated repression was substantially less in normal breast epithelial cells with only two copies (Figure 5A, normal epithelial cells). Upon E2 stimulation, chromatin interactions in normal cells were less frequent with only a slight effect on gene repression (Figure 6B, blue line).
Since epigenetic changes are known to contribute to transcription repression and these long-range chromosomal interactions may lead to histone modifications in target regions (You and Jones 2012), we determined genomic landscapes of three histone marks- H3K4me3, H3K9me3, and H3K27me3 and RNA polymerase II (Pol II) in the two loci using a published dataset for MCF-7 cells (Joseph et al., 2010). Using THRAP1 as an example (Figure 6C, left), E2 stimulation resulted in an increase in the H3K27me3 (red bar) repressive mark occupancy on promoter region and simultaneously decreased Pol II binding (green bar) at its transcription start site (TSS). A similar response was also observed at the ZIM2 locus (Figure 6C, right). Upon E2 treatment, increased recruitment of repressive H3K9me3 (red bar) to the TSS region was accompanied reduced H3K4me3, an active histone mark, and less Pol II binding (green bars).
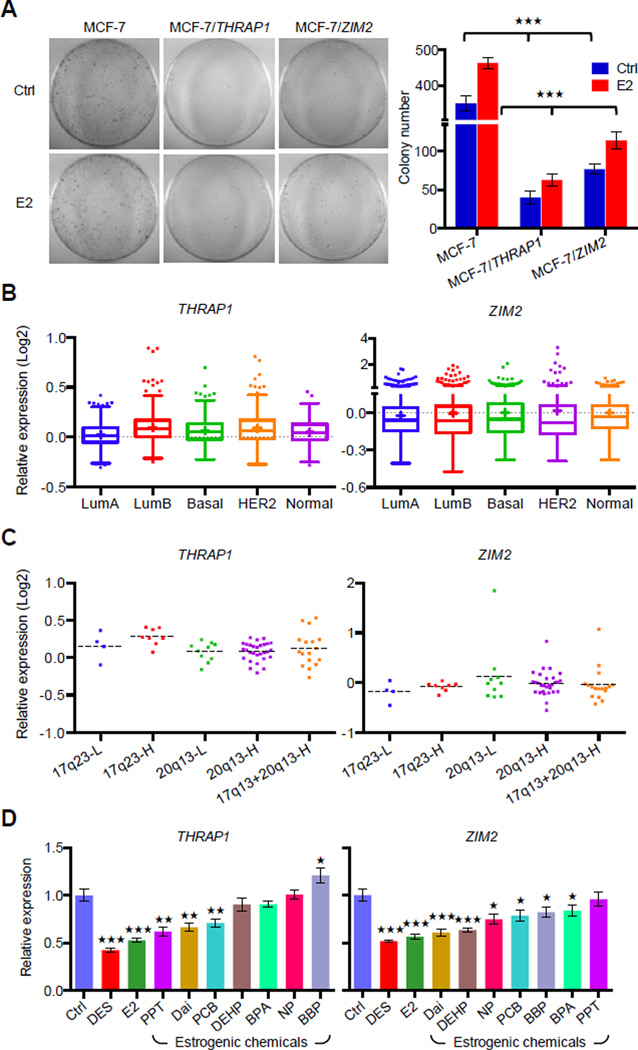
To further investigate the biological significance of THRAP1 and ZIM2 in breast tumorigenesis, proliferation of MCF-7 cells transiently transfected with either THRAP1 or ZIM2 was monitored using the foci formation assay. THRAP1 or ZIM2 expression levels in the transfected cells were confirmed using RT-qPCR analysis (Figure S6). Repressed THRAP1 and ZIM2 by DEREs likely contributed to the observed E2-stimulated MCF-7 cell proliferation (Figure 7A, MCF-7). This proliferation was greatly reduced by transient expression of THRAP1 or ZIM2, suggesting that these two genes play tumor-suppressor roles in ERα-positive luminal cancer cells (p<0.001; Figure 7A). In silico analysis of an expression microarray dataset (Curtis et al., 2012) further confirmed down-regulation of THRAP1 or ZIM2 in ERα-positive luminal-A tumors relative to other breast cancer subtypes (p<0.05, Figure 7B). An inverse correlation between increased DERE copies on 20q13 and down-regulation of these two genes was further observed (p<0.01; Figure 7C). Consistent with these findings, estrogen-induced DERE copy gains possibly initiated in progenitors chronically preexposed to estrogenic chemicals resulted in permanent expression changes of THRAP1 or ZIM2 in the progeny (Figure 7D). Taken together, these results indicate that DERE-mediated transcriptional regulation may result in repression of tumor suppressor genes such as THRAP1 and ZIM2, leading to aberrant cell proliferation and tumor progression.
Figure 7. Amplified DEREs Repress Candidate Tumor Suppressor Expression for ERα-positive Luminal Cancer Proliferation.
(A) Determining tumor-suppressor features of two DERE-regulated loci, THRAP1 and ZIM2, in MCF-7 cells. To determine cell proliferation rate, colony formation assays were conducted in cells transiently expressing THRAP1 or ZIM2, respectively, upon E2 stimulation (70 nM) (see also Figure S6 for expression levels of THRAP1 and ZIM2 in MCF-7 transfectants). Colony numbers of two biological replicates scored by three independent researchers are shown (right).
(B–C) In silico analysis of THRAP1 and ZIM2 mRNA expression levels in breast cancer subgroups (B) and ERα-positive breast tumors within differential copies of 20q13 and 17q23 DEREs (C) using a published microarray breast cancer cohort (Curtis et al., 2012). A total of 1986 breast tumors with subgroup information were analyzed in (B); 68 ERα-positive breast tumors within amplification of either 17q23 or 20q13 DEREs were applied in the study.
(D) Expression levels of THRAP1 and ZIM2 in normal breast epithelia preexposed to estrogenic chemicals as shown in Figure 3D. THRAP1 and ZIM2 expression was determined by RT-qPCR of differentiated epithelial progeny after preexposure to estrogenic chemicals (see exposure scheme in Figure 3D, upper). Mean ± SD (n=6 replicates in two batches of treatment). ***, p<0.001; **, p<0.01; *, p<0.05 (Student’s t test), comparing to “Ctrl” cells.
Amplified DERE Copies Deregulate Anti-proliferation and Apoptosis Signaling Networks Associated with Tamoxifen Resistance in Breast Cancer
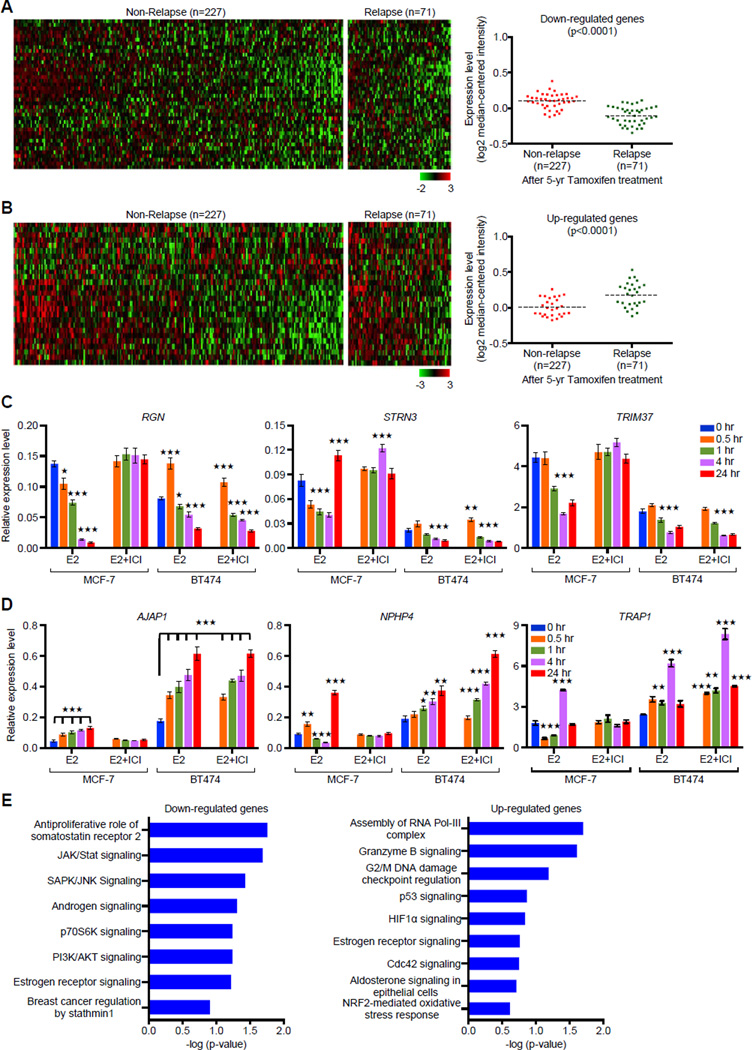
We further examined whether altered expression of DERE-regulated genes associates with adverse treatment outcome of ERα-positive breast cancer by surveying published expression microarray data of multiple breast cancer cohorts (Hatzis et al., 2011; Symmans et al., 2010). We found that differential expression of a subset (n=67) of DERE-regulated loci is associated with patient relapse after endocrine therapies (Figures 8A–B and S7A–B). Comparing gene expression profiles in a 298 patients cohort (Symmans et al., 2010), 40 of the 67 DERE-interacting loci were significantly repressed in primary tumors from 71 patients, who subsequently developed local recurrence or distant metastasis after 5 years of tamoxifen therapy (p<0.0001, Figure 8A). In contrast, only 27 of the 67 DERE-interacting loci were up-regulated in those same patients (p<0.0001, Figure 8B). To validate the above findings, 26 of the 67 DERE-regulated loci were randomly selected for gene expression analysis in an ERα-positive BT474 breast cancer cell line, which is HER2-overexpressing and resistant to tamoxifen (Wang et al., 2006). Increased crosstalk between ERα and HER2 pathways may contribute to endocrine resistance in this cell line. Estrogen stimulation significantly induced differential expression of those genes in both MCF-7 and BT474 cells while ICI attenuated E2-driven up- and/or down-regulation only in MCF-7, but not BT474 cells (Figures 8C–D and S7C–D). One possible explanation for this observation is that HER2 may bypass the ERα blockade to regulate DERE-interacting loci in BT474 cells. An alternative explanation is that despite the ICI treatment, high copy-number DEREs in BT474 cells may provide an additional ERα binding reservoir for partial transcription regulation of these target genes (Figures S3A–B and S7C–D). An IPA approach further demonstrated that these differentially expressed genes were mapped to several biologic pathways, including cell cycle control (e.g., G2/M DNA damage checkpoint regulation and CDC42 signaling) pathways (Figure 8E). Down-regulated genes belonged to signaling pathways such as anti-proliferative somatosatin receptor 2 and SAPK/JNK signaling, which has been associated with tamoxifen-induced apoptosis (Mandlekar and Kong 2001). Signaling cascades modified by up-regulated genes included attenuation of Granzyme B-associated apoptosis, leading to advanced luminal cancer development (Jiang et al., 2006).
Figure 8. Expression Signature of DERE-interacting Genes Correlates with Relapse after Tamoxifen Therapy.
(A–B) In silico analysis of DERE-regulated genes in an ERα-positive breast tumor cohort is associated with relapse. A breast cancer cohort including a total of 298 patients with ERα-positive breast tumors and 5-year tamoxifen treatment was used to study the clinical significance of DERE-regulated genes (Symmans 2010). Forty down-regulated (A) and twenty-seven up-regulated (B) genes were significantly associated with relapse after tamoxifen treatment (p<0.0001) (see also Figure S7A–B for validation analysis using another independent cohort within endocrine therapy history).
(C–D) Expression analysis of 26 DERE-interacting genes Involved in tamoxifen resistance. Quantitative RT-PCR analysis was performed on MCF-7 and BT474 cells treated with E2 (70 nM) alone/and ERα antagonist- ICI 182,780 (ICI, 1 µM) in five time periods (0, 0.5, 1, 4, and 24 hr). Expression data were shown in 26 bar charts with individual genes (see also Figure S7C–D). Mean ± SD (n=6 replicates in two biological batches). (C) Down-regulated genes; (D) Up-regulated genes.
(E) Signaling pathways associated with DERE-regulated genes in tamoxifen resistance. Ingenuity Pathway Analysis was used to determine signaling pathways associated with down- and up-regulated genes (identified from A and B) in tamoxifen resistance. Mean ± SD (n=6 replicates in two batches of treatment). ***, p<0.001; **, p<0.01; *, p<0.05 (Student’s t test), comparing to control cells (time point “0 hr”).
DISCUSSION
Genomic amplification in 17q23 and 20q13 regions is commonly observed in breast cancer with poor prognosis (Andersen et al., 2002; Bilal et al., 2012; Ginestier et al., 2006). Moreover, these amplification events have been implicated in endocrine therapy (Bilal et al., 2012; Han et al., 2006; Symmans et al., 2010). Specifically, Han et al. (2006) reported that chromosome 17q23 is among the most commonly amplified regions in breast cancer patients whose tumors recurred within 5 years of post-tamoxifen therapy. It was previously suggested that oncogenes located within these amplicons express high levels of corresponding mRNA and proteins involved in cancer development and tamoxifen resistance. As a result, identifying driver genes located within these large amplified regions (4.5-Mb for 17q23 and 19-Mb for 20q13) was subject to intense investigations (Collins et al., 2001; Han et al., 2006; Monni et al., 2001; Sinclair et al., 2003). However, systematic screening of a total of ~148 genes in these regions identified only a handful of putative oncogenes. For example, overexpression of ZNF217 and MAP3K3 located on 20q13 can attenuate apopototic signaling, leading to chemotherapy resistance (Ginestier et al., 2006; Santarius et al., 2010). In the 17q23 region, overexpression of RPS6KB1, TBX2, PPM1D, RAD51C, and APPBP2 frequently associates with tumors displaying luminal or HER2 phenotypes (Bilal et al., 2012; Monni et al., 2001; Sinclair et al., 2003). One proposed explanation for the low number of oncogenes associated with these amplified regions is that selection of only a few oncogenic drivers is sufficient to drive tumorigenesis in these cell types. This process may be accompanied by amplification of neighboring genes as neutral passengers (Ginestier et al., 2006; Sinclair et al., 2003).
In the present study, we offer an additional explanation that DNA regulatory elements located within these genomic regions, such as DEREs, are co-amplified during breast tumorigenesis. We suggest that increases in DERE copy number enhance ERα-modulated transcriptional activity activated by E2, which is associated with induction of pro-growth factors as well as repression of growth suppressing genes (Carroll et al., 2006; Fullwood et al., 2009; Welboren et al., 2009). An integrative analysis of next-generation sequencing data indicated that densely ERα-bound DEREs (46–51 per megabase) are localized in 17q23 and 20q13 regions. These ERα-bound DEREs interact with target genes outside the amplified regions through long-range chromosomal interactions. Our experimental evidence further reveals that the amplified DERE copies enhance the frequency of chromatin interactions in breast cancer cells, markedly altering transcriptomic profiles of target genes involved in cancer development. In silico analysis shows that these genes exist in a network associated with apoptotic signaling pathways, such as the serine proteases, Granzyme B (Han et al., 2005). The observed increase in the frequency of chromatin interactions between amplified DEREs and target genes likely attenuate Granzyme B-induced apoptosis, thereby contributing to advanced luminal cancer development (Jiang et al., 2006). Moreover, based on differential expression patterns in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant BT474 cells (Figures 8C–D and S7C–D), we propose that amplified DEREs may deregulate gene expression associated with anti-proliferation and apoptosis signaling networks that are implicated in tamoxifen resistance. Although the estrogen-dependent transcription mechanism may be blocked by tamoxifen, tumors containing high copy-number DEREs may still have residual responses to estrogen-dependent growth compared to tumors with low copy-number DEREs. As such, it will be interesting to interrogate in the future whether high copy-number DEREs can be used as putative biomarkers for predicting patients’ intrinsic resistance to endocrine therapy.
In addition to the aforementioned findings, this study further demonstrates that amplified DNA regulatory elements are caused by sustained stimulation of DERE-DERE interactions (Figures 1, 2, and S2). In tumorigenesis, these amplified events may intensify chromatin interactions (see two examples in Figure 6D), leading to inter- and intra-chromosomal rearrangements. Although the molecular mechanisms underlying these chromosomal amplifications in cancer remain to be fully explored, we anticipate that E2 stimulated, DERE-mediated chromatin interactions may be a driving force of genomic instability. It is well established that loss of genome stability and integrity is a hallmark of cancers, leading to somatic copy-number alterations and non-random distributions of breakpoints (De and Mochor 2011a; Hampton et al., 2009). Our results suggest that DNA breakpoints preferentially cluster together at specific chromosomal regions previously implicated in breast tumorigenesis, indicating that those regions harboring clustered breakpoints are exquisitely prone to genomic rearrangements (De and Mochor 2011b; Hampton et al., 2009). Furthermore, during neoplastic transformation, defects in double-strand break repair can occur (Hampton et al., 2009), destabilizing these physical interactions and promoting insertions and self-duplication of 20q13 DERE clusters, for example, into regions of seven derivative chromosomes (Figure S2C–D).
While our ChIP-seq and 3C-seq analyses point to amplification DERE-mediated interactions, normalization to copy-number variation of DEREs is not presently conducted for several reasons. First, the occurrence of ERα-binding events or the frequency of chromatin interactions may not linearly correspond to increased DERE copies. Second, while applying normalization to DERE copies relies on an assumption that all amplified copies in a given DERE locus participate evenly in ERα-binding and associated chromatin interaction, not all copies are expected to contribute equally to these events. Lastly, DERE-mediated chromatin interactions are highly dynamic upon estrogen stimulation, irrespective of their copy-number effect. Our ongoing study using Hi-C (an improved version of 3C-seq) suggests that estrogen stimulation may lead to progressive increases and then decreases in DERE interactions between 17q23 and 20q13 over a 24-hr period (data not shown) (Lieberman-Aiden et al., 2009). Therefore, the present analysis supports the concept that estrogenic exposure drives genomic alterations involving chromosomal amplification of distant estrogen-responsive regulatory elements. These genetic lesions persist in breast progenitor cells, and when accompanied by altered gene expression profiles, possibly lead to tumorigenesis. In addition, our data suggest that amplified regulatory elements in 17q23 and 20q13 can be used as potential prognostic markers for anti-estrogen resistance.
EXPERIMENTAL PROCEDURES
A more detailed description of the experimental procedures and reagents used in this study can be found in the Supplemental Experimental Procedures.
Breast Tissue Collection and In Vitro Exposure Model
Non-cancerous and cancer samples were collected by the tissue procurement service in accordance with the protocols approved by the Institutional Review Board of The Ohio State University and had been de-identified prior to the analysis. For isolation of breast progenitor cells, non-cancerous sections were obtained from individuals undergoing reduction mammoplasties mainly due to macromastia. These tissues were dissociated mechanically and enzymatically, and single cells were isolated and grown into progenitor-containing cells, called mammospheres (2,000–10,000 cells per mammosphere), in ultra-low attachment plates (Corning) with serum-free mammary epithelial growth medium (Cambrex) as described (Dontu et al., 2003). Progenitor cells, repopulated in the suspension culture up to six passages were then exposed to DMSO (dimethyl sulfoxide, Control), 17β-estradiol (E2), or estrogenic chemicals in phenol red–free medium for 3 weeks (medium changed twice weekly). After preexposure, progenitor-containing cells were placed on a collagen substratum (BD Biosciences) in phenol red-free medium for 2–3 weeks without additional exposure. Under these culture conditions, progenitors were differentiated into mature breast epithelial cells as described previously (Hsu et al., 2009 and 2010). Six sets of biological replicates were interrogated in this study. The MCF-7 and BT474 breast cancer cell line, obtained from the American Type Culture Collection, were maintained in phenol red-free medium. To minimize the effect of endogenous estrogen on each experiment, MCF-7 and BT474 cells were cultured in media containing charcoal-stripped serum for 24 hr prior to any short-term (a 24-hr period) treatment. For long-term exposure, cells were cultured in charcoal-stripped serum condition with any treatment.
Library Preparation and Illumina Sequencing Processing of ChIP-seq, 3C-seq, and Mate-pair Seq
ChIP-seq, 3C-seq, and mate-pair seq libraries were prepared using the Illumina ChIP-seq, paired-end, and mate-pair sample preparation kits, respectively, according to the manufacturer. Libraries were validated using Agilent Technologies 2100 Bioanalyzer and then sequenced on Genome Analyzer IIx sequencer as follows: 3C-seq, 2 × 36-bp; mate-pair seq, 2 × 51-bp; ChIP-seq, 1 × 51-bp. Detailed experimental description and data analyses are provided in the Supplemental Experimental Procedures.
Statistical Analyses
All quantitative PCR results were presented as the mean ± SD of n independent measurements. Statistical comparisons between the two groups were made by Student’s t test using SigmaPlot 11, and multiple groups were determined by ANOVA test. For samples with equal variance, the paired Student’s t test was used. For samples with unequal variance, the Mann-Whitney Rank Sum test was utilized.
Supplementary Material
HIGHLIGHTS.
Densely ERα-bound DEREs co-localize in translocation-susceptible regions.
Estrogen exposure contributes to amplification of 20q13 and 17q23 DEREs.
Coordinate model of DERE-mediated looping on regulating target gene transcription.
Amplified DEREs deregulates target genes correlated to tamoxifen resistance in breast cancer.
Significance.
Genomic amplification of 17q23 and 20q13 is a known prognostic factor in breast cancer. Despite exhaustive search for oncogenic drivers within these genomic regions, only a handful of putative oncogenes have been identified. By conducting integrative analyses, we discover that densely ERα-bound DEREs localized in these regions may contribute to breast tumorigenesis. Increased DERE copies enhance chromatin interactions, leading to transcriptional repression of genes associated with tumor-suppressor and apoptosis pathways. The increased frequency of chromatin interactions may be a relevant factor in the development of a previously undescribed subtype of tamoxifen-resistant breast cancer.
ACKNOWLEDGEMENTS
We would like to thank the Genomic Sequencing Facility of the Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio (UTHSCSA) for next-generation sequencing. This work was supported by R01 CA069065, R01 ES017594, U01 ES015986 (Gene and Environment Initiative), U54 CA113001 (Integrative Cancer Biology Program), P30 CA054174 (Cancer Center Support Grant) at the U.S. National Institutes of Health and by generous gifts from the Cancer Therapy and Research Center Foundation and the Voelcker Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The Sequence Read Archive accession number for the 3C-seq, mate-pair seq, and ChIP-seq reported in this paper is SRA091617.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, nine tables, and Supplemental Experimental Procedures and can be found with this article online.
REFERENCES
- Andersen CL, Monni O, Wagner U, Kononen J, Barlund M, Bucher C, Haas P, Nocito A, Bissig H, Sauter G, et al. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am. J. Pathol. 2002;161:73–79. doi: 10.1016/S0002-9440(10)64158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Fernandez MF, Petersen JH, Olea N, Skakkebaek NE, Leffers H. Dual effects of phytoestrogens result in u-shaped dose-response curves. Environ. Health Persepct. 2002;110:743–748. doi: 10.1289/ehp.02110743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esqueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreastic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal E, Vassallo K, Toppmeyer D, Barnard N, Rye IH, Almendro V, Russnes H, Borresen-Dale AL, Levine AJ, Bhanot G, et al. Amplified loci on chromosome 8 and 17 predict early relapse in ER-positive breast cacners. PLoS One. 2012;7:e38575. doi: 10.1371/journal.pone.0038575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Mutarelli M, Grober OM, Paris O, Ferraro L, Ravo M, Tarallo R, Luo S, Schroth GP, Seifert M, et al. Estrogen receptor alpha controls a gene network in luminal-like breast cacner cells comprising multiple trasncription factors and microRNAs. Am. J. Pathol. 2010;176:2113–2130. doi: 10.2353/ajpath.2010.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Volik S, Kowbel D, Ginzinger D, Yistra B, Cloutier T, Hawkins T, Predki P, Martin C, Wernick M, et al. Comprehensive genomc sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landsapes of cancer genomes. Nat. Biotechnol. 2011a;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Mochor F. DNA secondary structures and epigenetic determination of cancer genome evolution. Nat. Struct. Mol. Biol. 2011b;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Cervera N, Finetti P, Esteyries S, Estein B, Adelaide J, Xerri L, Viens P, Jacquemier J, Charafe-Jauffret E, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin. Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. A sequence-level map of chormosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–177. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Goldstein LA, Gastman BR, Rabinovitz A, Rabinowich H. Disruption of Mcl-1.Bim complex in granzyme B-mediated mitochondrial apoptosis. J. Biol. Chem. 2005;280:16383–16392. doi: 10.1074/jbc.M411377200. [DOI] [PubMed] [Google Scholar]
- Han W, Han WR, Kang JJ, Bae JY, Lee JH, Bae YJ, Lee JE, Shin HJ, Hwang KT, Hwang SE, et al. Genomic alerations identified by array comparartive genomic hybridization as prognostic narkers in tamoxifen-treated estrogen receptor-positive breast cacner. BMC Cancer. 2006;6:92. doi: 10.1186/1471-2407-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang J, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. Subtype and pathway specific responses to anticancer compounds in breast cacner. Proc. Natl. Acad. Sci. U. S. A. 2009;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Deatherage DE, Rodriguze BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Xenoestrogen-induced epigenetic repression of microRNA-9–3 in breast epithelial cellsp. Cancer Res. 2009;69:5936–5945. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–744. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc. Natl. Acad. Sci. USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Orr BA, Kranz DM, Shapiro DJ. Estrogen induciton of the granzyme B inhibitor, proteinase inhibitor 9, protects cells against apoptosis ediated by cytotoxic T lymphocytes and nature killer cells. Endocrinology. 2006;147:1419–1426. doi: 10.1210/en.2005-0996. [DOI] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim G, Thomsen JS, et al. Integrative model of genomic factors for determining binidng site selection by estrogen receptor-α. Mol. Syst. Biol. 2010;6:456–468. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bergmann A, Lucas S, Stone S, Stubbs L. Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics. 2004;84:47–58. doi: 10.1016/j.ygeno.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Andersen ME, Heber S, Zhang Q. Non-monotoic dose-repsonse relationship in steriod hormone receptor-mediated gene expression. J. Mol. Endocrinol. 2007;38:569–585. doi: 10.1677/JME-07-0003. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and coveyed phenotypes. PLos Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlekar S, Kong AN. Mechanisms of tamoxifen-induced apoptosis. Apoptosis. 2001;6:469–477. doi: 10.1023/a:1012437607881. [DOI] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P. Transcriotion factories are unclear sub-compartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr J, Helfrich H, Fuge M, Eldering E, Buhler A, Wrinkler D, Volden M, Kater AP, Mertens D, Te Ras D, et al. DNA damage-induced transcriptional program in CLL: biological and diagnostic implications for functional p53 testing. Blood. 2011;117:1622–1632. doi: 10.1182/blood-2010-08-300160. [DOI] [PubMed] [Google Scholar]
- Monni O, Bariund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolsky Y, Sviridov E, Dosymbekov D, Ustyansky V, Kaznacheev V, Dezso Z, Mulvey L, Macconaill LE, Winckler W, Serebryiskaya T, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancerp. Cancer Res. 2008;68:9532–9540. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferenitally relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Schwab M. Oncogene amplification in solid tumors. Semin. Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- Sinclair CS, Rowley M, Naderi A, Cough FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res. Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Biqnell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmans WF, Hatzis C, Sotiriou C, Andre F, Peintinger F, Regitnig P, Daxenbichler G, Desmedt C, Domont J, Marth C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Wadia PR, Schaeberie CM, Rubin BS, Sonnenschein C, Soto AM. The mammary gland response to estradiol: monotonic at the cellular level, non-monotonic at the tissue-level of organiztion? J. Steriod. Biohem. Mol. Biol. 2006;101:263–274. doi: 10.1016/j.jsbmb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Yang XY, Zhang X, An P, Kim HJ, Huang J, Clarke R, Osborne CK, Inman JK, Appella E, Farrar WL. Disruption of estrogen receptor DNA-binding domain and related intramolecular communication restores tamoxifen sensivity in resistant breast cancer. Cancer Cell. 2006;10:487–499. doi: 10.1016/j.ccr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Meqens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERαlpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer Genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.