Abstract
In preparation for a space flight on STS-126, two in vitro culture systems were used to investigate macrophage colony stimulating factor-dependent macrophage differentiation from mouse primary bone marrow cells. The patented Techshot Cell Cult Bioreactor and the BioServe Fluid Processing Apparatus (FPA)1 were operated in different orientations to determine their impact on macrophage growth and differentiation. Bone marrow cell parameters were determined after cells were grown in FPAs incubated at 37°C in vertical or horizontal orientations, and macrophage cell recovery was significantly higher from FPAs that were incubated in the horizontal orientation compared to “vertical” FPAs. Similarly, when bone marrow cells were grown in the Techshot bioreactor, there were significant differences in the numbers of macrophages recovered after 7 days, depending on movement and orientation of the bioreactor. Macrophage recovery was highest when the patented bioreactor was rotated in the horizontal, x-axis plane (merry-go-round fashion) compared to static and vertically, y-axis plane rotated (Ferris wheel fashion) bioreactors. In addition, the expression of F4/80 and other differentiation markers varied depending on whether macrophages differentiated in FPAs or in bioreactors. After 7 days, significant differences in size, granularity and molecule expression were seen even when the same primary bone marrow cells were used to seed the cultures. These data show that culture outcomes are highly dependent on the culture device and device orientation. Moreover, the impact of the culture system needs to be understood in order to interpret space flight data.
Keywords: cell differentiation, macrophage, fluid processing apparatus, space flight hardware
Introduction
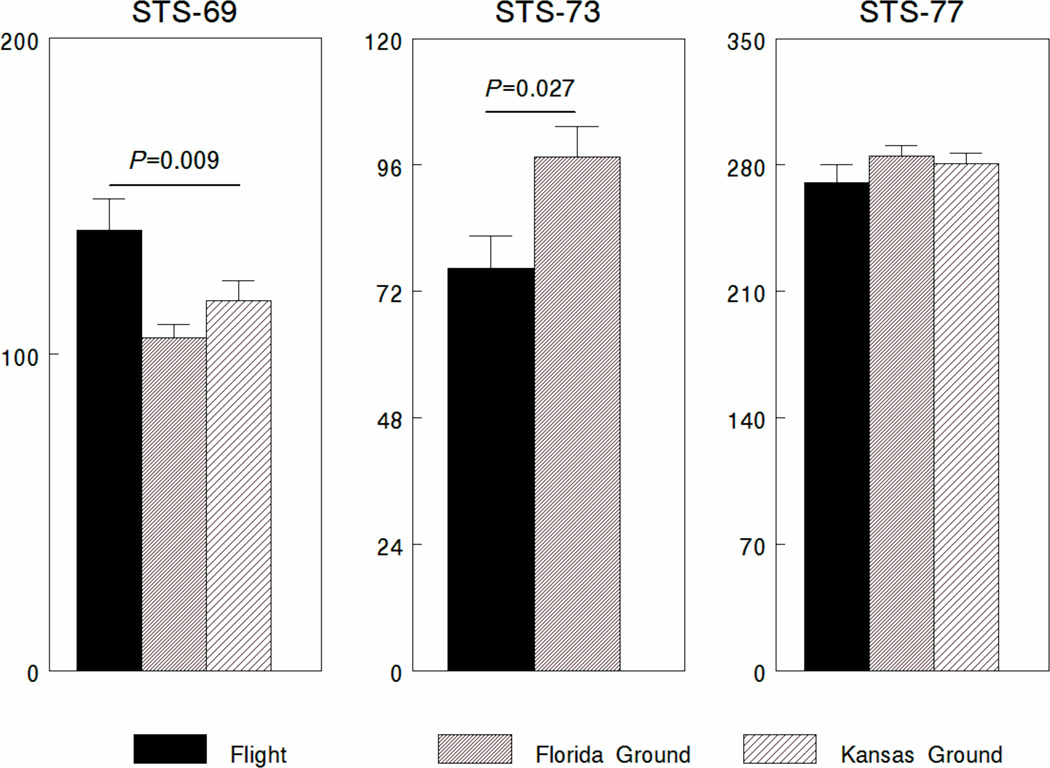
Spaceflight and/or microgravity can alter the immune system at organismic levels [1, 2]as well as cellular levels [3–5]. In particular, the hematopoietic system that generates cells in the monocyte/macrophage lineage may be a target. In 1986, Taylor et al. [6]detected monocytopenia post flight in seven out of 11 astronauts that flew on the space shuttle. In addition, some astronauts exhibited changes in insulin-like growth factor-1 receptor expression on monocytes post flight [7]. Changes in colony forming units-granulocyte macrophage (CFU-GM) and colony forming units-macrophage (CFU-M) were seen in rats, post flight on biosatellites [8, 9]and during the Spacelab Life Sciences (SLS)-2 space lab flight on space transportation system (STS)-58. [10]. However, comparison of CFU-M after two 7-day space shuttle flights found that there were inconsistencies in CFU-M in the bone marrow of rats post flight [11]. We also found different outcomes when we measured the development of CFU-M in vitro after three different shuttle missions (Figure 1). During STS-69, a 260 hr. mission, we found that significantly more CFU-M developed in cultures grown in space compared to ground controls. In contrast, during STS-73, a 382 hr space-flight mission, we found the opposite result with inhibited CFU-M generation in space-flight cultures compared to ground controls (Figure 1). There was no impact of space flight on CFU-M generation during the STS-77 shuttle mission, a 240 hr. mission.
Figure 1. CFU-M development on STS-69, 73 and 77.
Soft agar colony assays to detect CFU-M (macrophage colony forming units) were conducted in FPAs as described [12]. Colony assays were set up for shuttle flight on STS-69 and 73 (and the associated ground controls) approximately 48 hours prior to turnover to the shuttle loading crew. The FPAs were equilibrated for 12 hours at 37° C and 8% CO2capped with rubber septa to trap equilibrated air, and shipped at ambient temperature to Kennedy Space Center (KSC) for integration into Group Activation Packs (GAPs) [21]. To control for transportation of the FPAs to KSC, ground controls were conducted at KSC and in our laboratory at Kansas State University (KSU). Colony assays set up for shuttle flight and ground controls for STS-73 were set by harvesting bone marrow cells at KSU which were immediately shipped in RPMI10 (on ice) to KSC. Ammonium Chloride lysis of red blood cells and all subsequent assay procedures were performed in the laboratory facilities at KSC Space Life Science Support Facility (SLSL). These assays were set up approximately 20 hours prior to turnover to the shuttle loading crew. As a result of the assay setup being conducted at KSC there were no ground controls conducted at KSU.
Space flight experiments are extraordinarily challenging. Not only do you have to modify experimental conditions and equipment for the space-flight environment, the ability to reproduce the exact flight conditions from flight-to-flight is next to impossible. Occasionally, one finds that equipment used for space flight is an advance on the technique used for normal ground experiments [12], but this is rare. Frequently, secondary payloads are at the mercy of the requirements of the primary payloads. Therefore, variables such as temperature, astronaut time for manipulations and time-lines are not optimal.
The assay of macrophage hematopoietic precursors can be done two ways. To identify the number of precursors, bone marrow cells are isolated and cultured in soft agar containing growth factors such as macrophage colony stimulating factor (M-CSF) or granulocyte-macrophage colony stimulating factor (GM-CSF) at appropriate concentrations [13]. Assessment of how many colonies grow provides an estimate about the number of CFU-M or CFU-GM that are present in the stem cell population [14]. Alternatively, macrophages can be induced to differentiate in M-CSF and the cells that differentiate into macrophages in 6–8 days can be phenotyped based on their cell-surface molecules, assayed for functional assays, or used for other kinds of analyses [15].
Our group had an opportunity to determine if macrophage development was affected in vitro on the space shuttle flights STS-57, STS-60 and STS-62 [16]. Mouse bone marrow stem cells were incubated in liquid culture in medium containing M-CSF during the space flights. The differentiated macrophages were fixed during flight and phenotyped by flow cytometry and the supernatants were collected from viable cells and were assayed for interleukin-6 (IL-6) post flight. However, because these experiments were secondary payloads, the temperature conditions were not physiological and some questions about the results remained[16]. Therefore, we explored additional opportunities to attempt to resolve the issue about the impacts of space flight on macrophage development at the cellular level.
Preflight Verification Tests
Although space flight offers the opportunity to do research in microgravity, there is also the challenge of adapting tissue culture methods for the space environment. For our experiments, two possible hardware configurations were tested. One was the Cell Cult Bioreactor from Techshot (Formerly Space Hardware Optimization Technology, Inc. Greenville, IN) [17–19]. The patented bioreactor vessel [19] holds approximately 50 ml of culture medium and is approximately 6 cm (diameter) × 2.7 cm (height) in size (Figure 2). The bioreactor in its final assembled configuration was computer controlled with flexible feeding and collection schedules and volumes. The bioreactor could be rotated at 1 revolution per minute (RPM) during flight to allow for adequate mixing of fresh culture medium or fixative. We also tested the BioServe Fluid Processing Apparatus (FPA)[16, 20–22]. These culture vessels are 1.35 cm diameter (1.31 mm thickness) glass barrels that are 11.70 cm long (Figure 3 A-C). Transfer of the medium from one chamber to another is accomplished through a bypass on one side of the tube (Figure 3A). These culture devices allow the incubation of cells in a volume of 3.0 ml which can be supplemented with one or two additional 1.5 ml medium supplements or one feeding and the addition of fixatives/preservatives. Detailed set up of the equipment for bone marrow cells has been described by our group previously [5, 16].
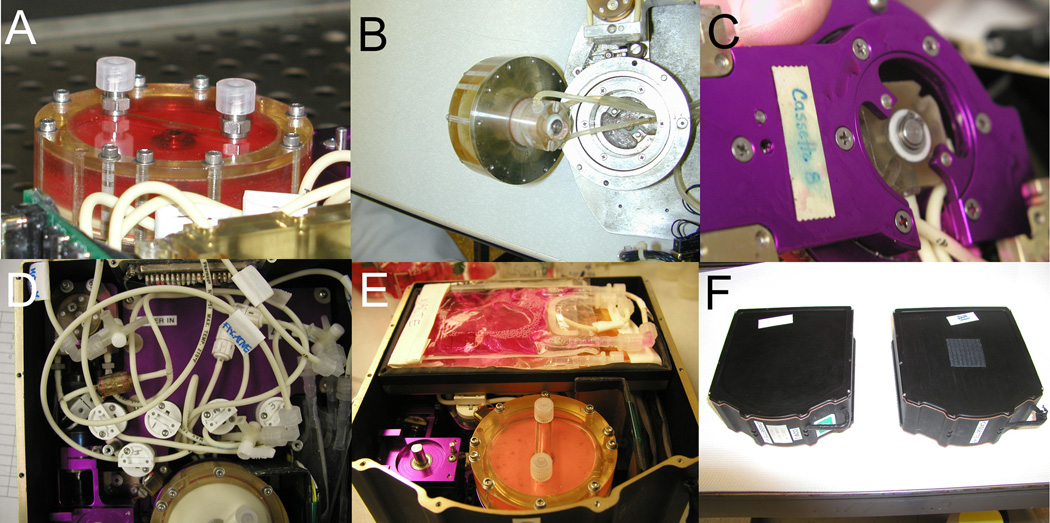
Figure 2. Bioreactor configuration.
The Techshot bioreactor vessel is a 6 cm (diameter) × 2.7 cm (height) culture vessel (Panel A) that is connected via rotary unions to medium feeding lines (Panel B, removed and viewed from below). The bioreactor is served by peristaltic pumps (Panel C) and computer controlled solenoid valves (Panel D) that control fresh medium or waste flow from or into Teflon bags (Panel E). The completed assembly (Panel F) can be inserted into a middeck-sized locker for use in space flight [17, 18].
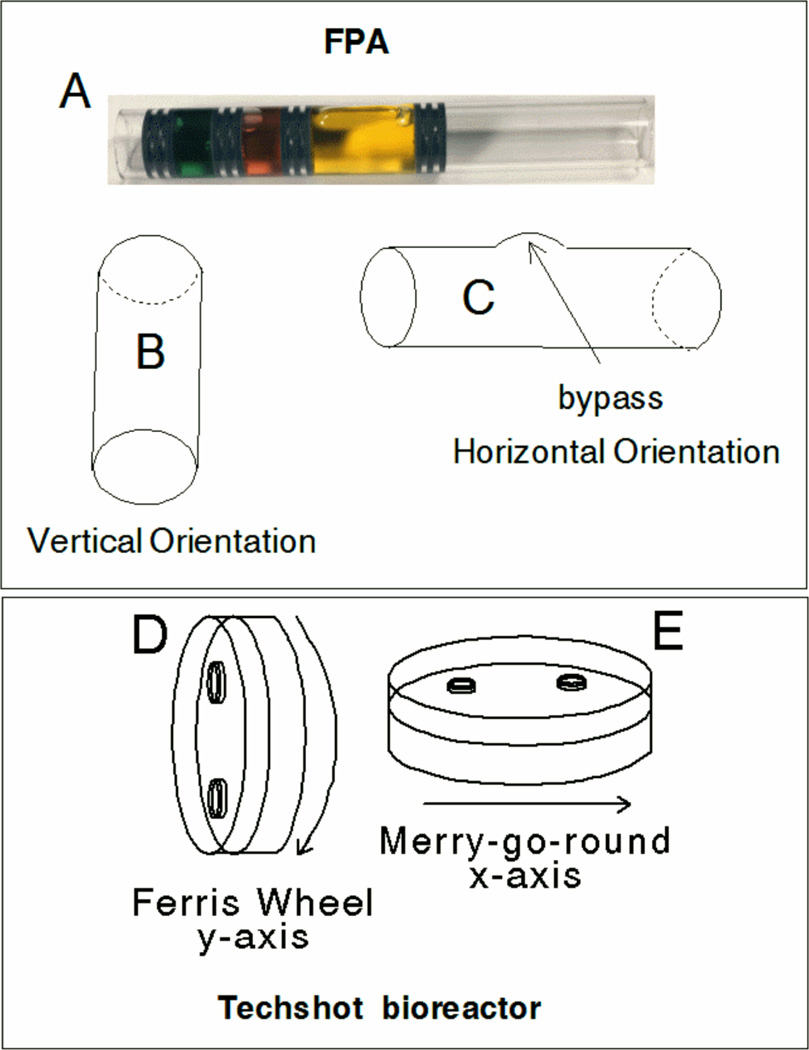
Figure 3. Fluid Processing Apparatus and Techshot bioreactor orientations.
FPA diagram (A) shows a loaded example in which 1×107 cells were placed in three ml of medium in chamber 1 (yellow) separated by two rubber septa. The FPA is engineered with a bypass so that when the second septum from the right crosses the bypass during motion from left to right the medium in the second chamber (red, 1.5 ml) will mix with the material in chamber 1 and the first two septa from the right will compress together. The next two septa also compress with additional movement to mix the contents of chamber 3 (green, 1.5 ml) with the contents previously mixed; showing vertical (B) or horizontal (C) orientation. The Techshot bioreactor was incubated in a vertical (Ferris wheel) or (E) horizontal (merry-go-round) configurations and rotated at 1 RPM.
There were significant differences in the complexity and capabilities of the culture systems available to us which presented some questions about the data each system was capable of providing. This is not a new concept. Others have compared hardware in ground based experiments to determine strengths and weaknesses of systems like rapidly rotating clinostats and random positioning machines [23]. For shuttle-flight experiments, most comparisons are made to samples incubated on Earth, although some space-flight experiments have been developed with controls in 1xg centrifuges that were also in space [24–26]. Comparisons have also been made with animals subjected to space flight and antiorthostatic suspension on Earth [9]. However, for our in vitro experiments, the orientation of the culture devices became a concern to us, especially for our Earth controls. In particular, the bioreactor could be oriented in “Ferris wheel” (Figure 3D) or “merry-go-round”(Figure 3E) orientations. We compared cell recovery {% cell recovery = (# viable cells recovered/ # viable cells added to the culture vessel)x100}from bioreactors that were rotated (at 1 RPM) or kept stationary that were incubated in both horizontal and vertical orientations (Table 1). One-third or fewer viable cells were recovered from bioreactors that were not rotated compared to bioreactors that were rotated at 1 RPM. In addition, approximately 80% more viable cells were obtained from bioreactors incubated in the horizontal orientation compared to the vertical orientation (Table 1). When we differentiated macrophages in FPAs incubated in vertical (Figure 3B) or horizontal orientations (Figure 3C), we also found that there were differences in the numbers of cells recovered. We averaged 3.7±0.9×106 cells per FPA after 7 days in M-CSF (15 ng/ml) when FPAs were stored in the vertical position. In contrast, we averaged 1.2±0.3×107 cells per FPA when the FPAs were stored in a horizontal position when bone marrow cells were differentiated into macrophages in medium containing M-CSF. The cell input recovered was 120% vs. 37% for horizontal and vertical FPAs, respectively. In general, the surface area that cells had to spread on was greater in both the bioreactor and the FPAs when the culture units were in the horizontal compared to the vertical orientation. It suggests that cell recovery is improved with greater surface area, independent of fluid volume. FPA’s were generally more efficient in viable cell numbers recovered compared to the larger bioreactors.
Table 1.
Recovery of cells from bioreactor
| Horizontal1 | Vertical1 | |||
|---|---|---|---|---|
| Rotation | Stationary | Rotation | Stationary | |
| Viable Cells Recovered2 | 1.6±0.3 × 107 | 3.6±0.5 × 106 | 3.2±0.5 × 106 | 1.1±0 × 106 |
| % Cell input Recovered3 | 5.3% | 1.2% | 1.1% | 0.4% |
3×108 primary mouse bone marrow cells were seeded in a chamber volume of 50 ml containing 15ng/ml mCSF.
Cells were incubated for 7 days. They were fed with 15 ml fresh medium on days 3 and 5.
% Cell input Recovered=(# of viable cells recovered/# viable cells added to the culture vessel)×100
We examined the cell surface phenotype of macrophages that differentiated in FPAs or the bioreactor. Cells were incubated in 15 ng/ml of M-CSF for 7 days and assessed for the expression of several cell surface molecules commonly found on macrophages. Both the FPAs and bioreactors were oriented in the horizontal positions and the bioreactors were rotated at 1 RPM. The bioreactors were fed with 15 ml of medium (with M-CSF) on days 3 and 5. The FPAs were stationary except when they were fed with 1.5 ml of medium (with M-CSF) on days 3 and 5 of the 7 day culture period. Cells were stained with fluorochrome-labelled antibodies specific for macrophage surface proteins c-fms [27–29], Mac-2 [30, 31], F4/80 [32, 33] and Ly6G [34–36]. Freshly harvested bone marrow cells are generally smaller with several subpopulations of cells, reflecting the large number of neutrophils in mouse bone marrow [37] and other populations of cells including undiffererentiated stem cells [38–41]. In FPAs the cells expressing Ly6G fall into two subpopulations. There is a small, less granular subpopulation and a large, granular Ly6G-negative subpopulation (Figure 4B and E). This contrasts bone marrow macrophages that differentiate in bioreactors which were mostly Ly6G positive and had continuums of sizes and granularities (Figure4C and F). For F4/80 expression, most of the cells recovered from FPAs and bioreactors were F4/80 positive (Figure 5). However, the cells in FPAs were smaller and agranular (Figure 5B and E) compared to cells collected from bioreactors after 7 days of differentiation, which expressed a continuum of granularities and sizes (Figure 5C and F). We saw similar trends for Mac-2 (Figure 6) and c-fms (Figure 7) expression. Macrophages were clearly differentiating in both culture devices over the 7-day differentiation period. However, the data clearly demonstrate that the properties of the cells were very distinct based on size, granularity and antibody staining. This is an important insight because it illustrates that just like the number of macrophages that develop in each type of culture device, the cell phenotype is hardware dependent. It is not clear why the cell phenotypes were so distinct when bone marrow cells were incubated in the two different hardware. The smaller fluid volume of the FPA may have had an impact. Although the numbers of cells in each of the hardware (1×107 vs. 3×108) was adjusted, the final cell concentration in an FPA was 1.7×106 cells/ml (after all septa movements) compared to 6×106 cells per ml in the Techshot bioreactor. Perhaps the increased cell concentration induced more culture stress. Alternatively, the distinct shapes of the devices may have influenced the data. This hypothesis is supported by the experiments presented here. When we changed the orientations of FPAs or the Techshot bioreactor, there were distinct changes in the culture outcomes. Additional experiments will be needed to test these hypotheses.
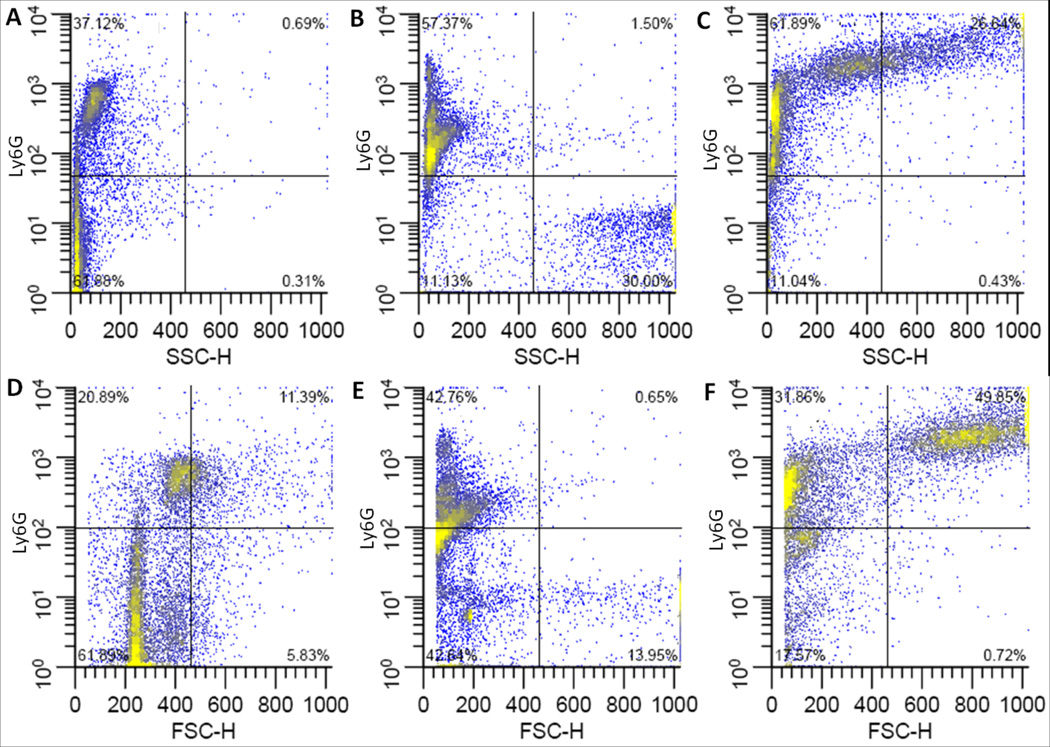
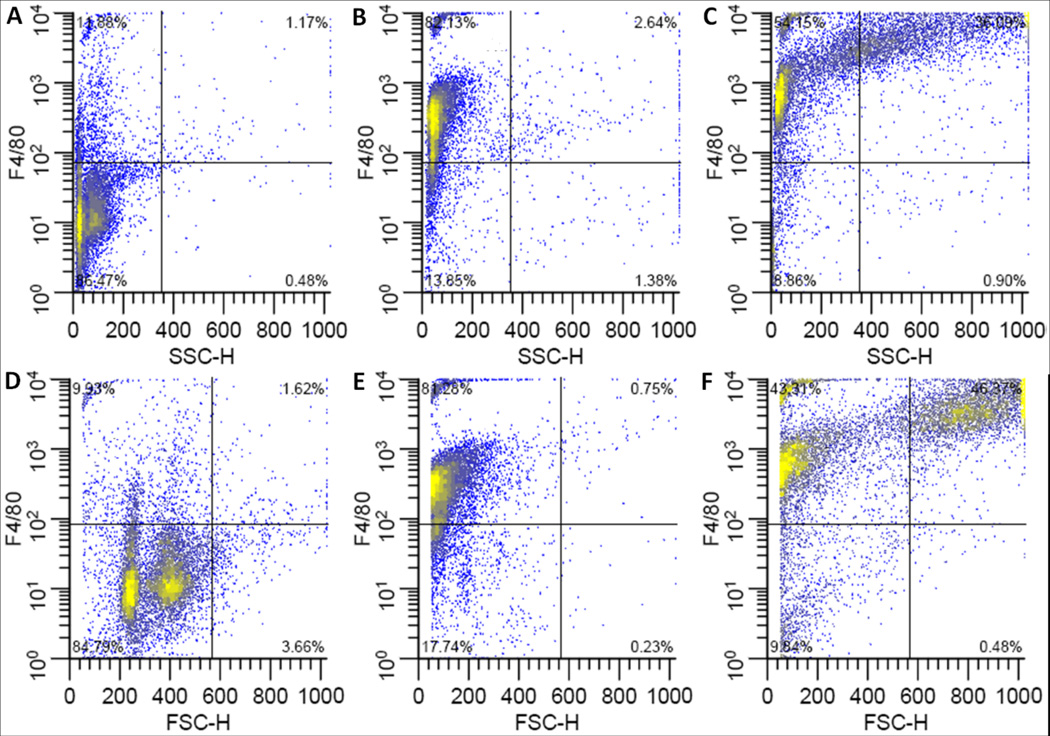
Figure 4. Flow cytometric analysis of Ly6G on bone marrow derived macrophages.
Representative plots of Ly6G fluorescence vs. side scatter (top row) or forward scatter (bottom row). Undifferentiated, freshly harvested bone marrow cells (A, D), or cells differentiated in medium containing 15 ng/ml M-CSF in FPAs (B, E) or Bioreactors (C,F). Culture vessels were incubated in horizontal orientations.
Figure 5. Flow cytometric analysis of F4/80 on bone marrow derived macrophages.
Representative plots of F4/80 fluorescence vs. side scatter (top row) or forward scatter (bottom row). Undifferentiated, freshly harvested bone marrow cells(A, D), or cells differentiated in medium containing 15 ng/ml M-CSF in FPAs (B, E) or Bioreactors (C,F). Culture vessels were incubated in horizontal orientations.
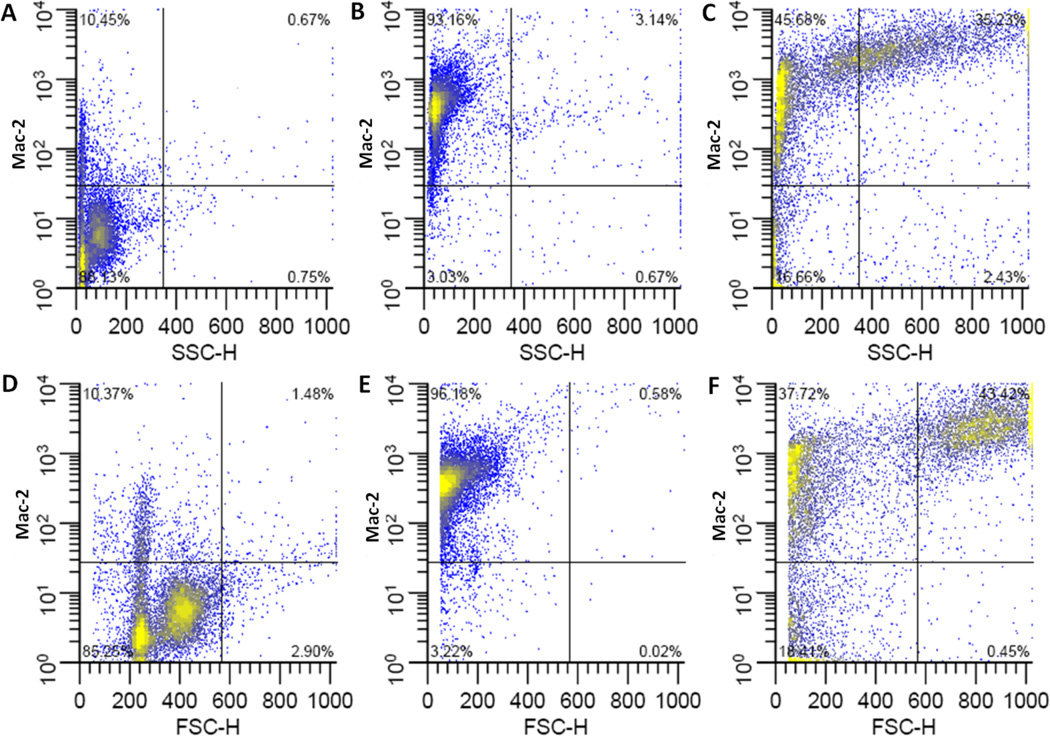
Figure 6. Flow cytometric analysis of Mac-2 on bone marrow derived macrophages.
Representative plots of Mac-2 fluorescence vs. side scatter (top row) or forward scatter (bottom row). Undifferentiated, freshly harvested bone marrow cells (A, D), or cells differentiated in medium containing 15 ng/ml M-CSF in FPAs (B, E) or Bioreactors (C,F). Culture vessels were incubated in horizontal orientations.
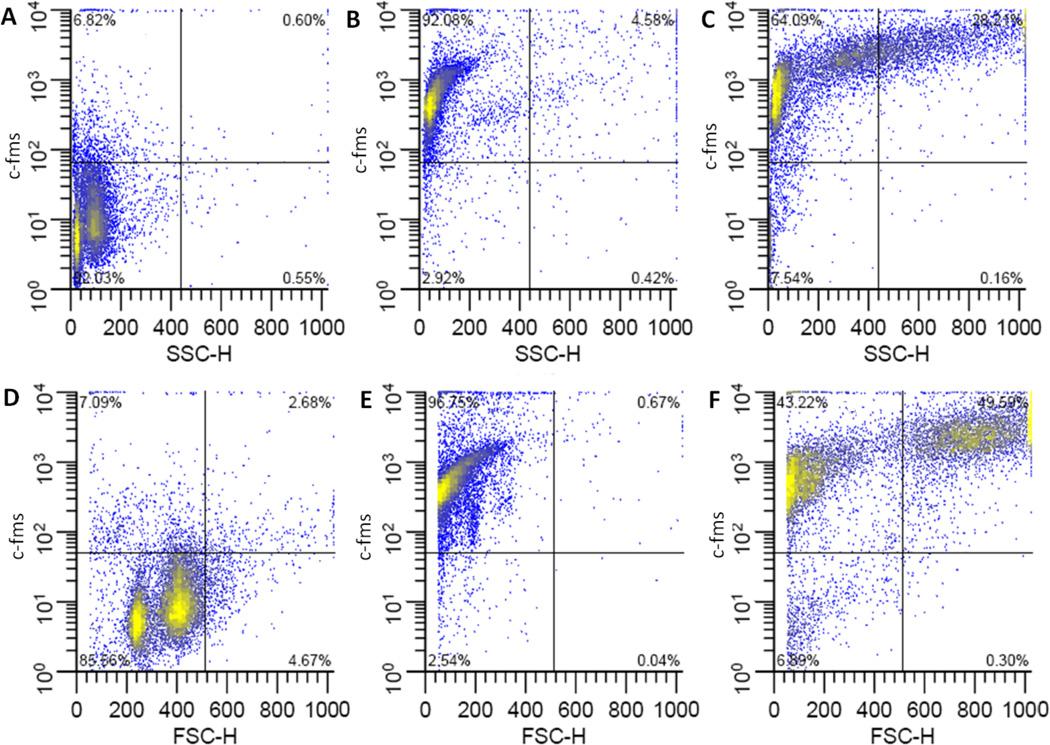
Figure 7. Flow cytometric analysis of c-fms on bone marrow derived macrophages.
Representative plots of c-fms fluorescence vs. side scatter (top row) or forward scatter (bottom row). Undifferentiated, freshly harvested bone marrow cells(A, D), or cells differentiated in medium containing 15 ng/ml M-CSF in FPAs (B, E) or Bioreactors (C,F). Culture vessels were incubated in horizontal orientations.
STS-126
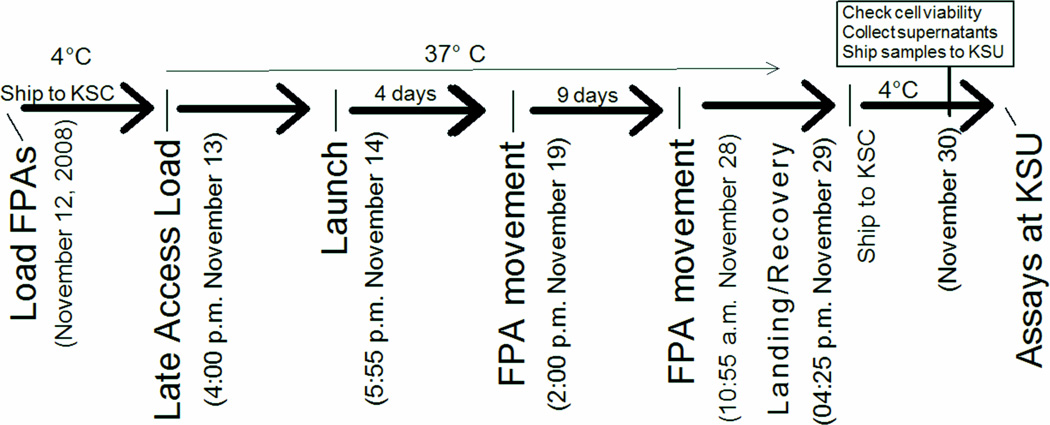
We initially developed our experiment for the Techshot bioreactor. However, the loss of the space shuttle Columbia on February 1, 2003 and the announcement on April 4, 2006 of a change in the NASA strategic plan[42], restricted access to the space shuttle laboratory. In 2008, we secured another opportunity to fly an experiment on the space shuttle Endeavour, STS-126. This 14-day flight provided the opportunity to assess bone marrow macrophage differentiation in vitro in response to recombinant mouse M-CSF using FPA hardware[5]. Cells were fixed with formalin and RNA was preserved at day 14 of the space flight (Figure 8). Cells were returned to Earth for cell surface molecule assessments and transcriptional arrays were performed using the Affymetrix Mouse Genome 430 2.0 array [5].
Figure 8. Time-line of activities for bone marrow macrophage differentiation on STS-126.
The FPAs were prepared as previously described [5] and were shipped to the NASA SLSL at KSC 2 days before launch. The FPAs were loaded into GAPs and placed at 37°C in the Commercial Generic Bioprocessing Apparatus (CGBA). Parallel temperature and activation profile conditions were maintained on GAPs kept at the SLSL. The cells in the FPAs were re-fed with 1.5 ml of medium supplemented with rmM-CSF on day 5 of the spaceflight by combining chambers 1 and 2 (See Figure 3). On day 14 of spaceflight, the content of the FPAs’ third chamber were mixed with the cell suspension in the previously merged chambers to preserve RNA or to fix or maintain cells until processing after landing. STS-126 landing occurred at the Edwards Air Force Base in California after 16 days of space flight. Samples were placed at 4° C and transported to the SLSL in Florida.
We discovered that cells proliferated more in space compared to ground-incubated cells. There were also several changes in cell surface molecules on macrophages differentiated in space compared to ground controls [5]. We saw significant shifts in the distribution of cells expressing CD31, Ly6C, CD11b, F4/80, Mac-2 and c-fos in cells differentiated in space flight compared to ground controls [5].
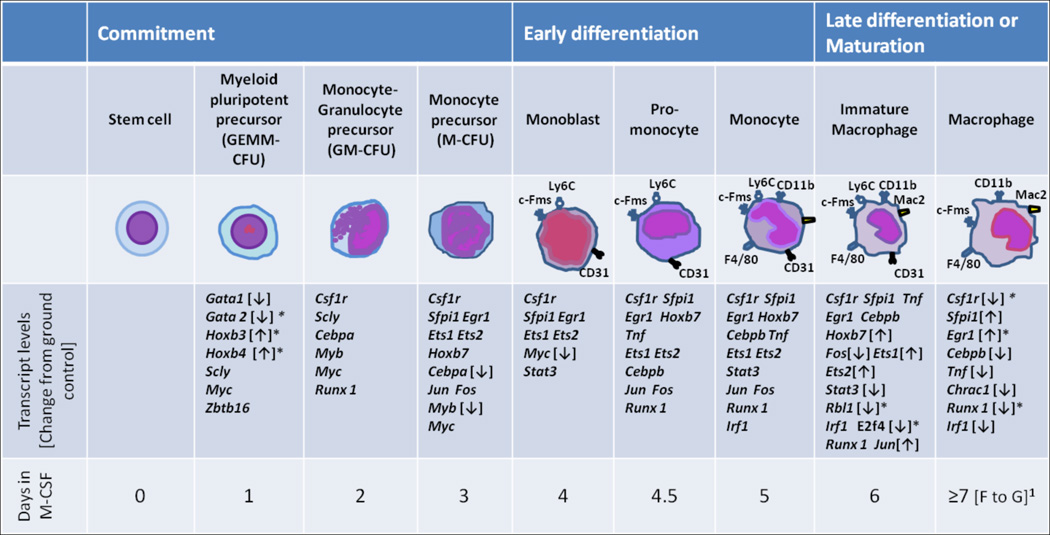
The transcript levels in macrophages differentiated in space were also different from those inground controls. Six hundred and seven genes had transcript levels >1.5 fold higher for flight samples than ground controls. In contrast, we found that 1071 genes had gene transcript levels >1.5 fold lower than ground controls (P < 0.05)[5]. The genes were sorted into biological function categories using Ingenuity Pathway Analysis (IPA) software and 33 genes were identifiedin the M-CSF/c-fms pathway. Bone marrow stem cells differentiate into macrophages in the presence of M-CSF in an ordered fashion from myeloid pluripotent precursors to monoblasts to macrophages (Figure 9). Gene expression occurs concomitantly during this process [43–46]. When we compared transcript levels of genes that were higher or lower in cells differentiated during space flight compared to ground controls, we found several genes that were significantly up or down regulated in the M-CSF/c-fms pathway (Figure 9). Moreover, the down regulation of Rbl-1 and E2f4 transcripts would be expected because of their roles in proliferation [47, 48] and cell differentiation [49]. Hoxb3 and Hoxb4 molecules are necessary for myeloid cell proliferation early in differentiation [44, 45] and the patterns of the transcripts seen in the space-flight samples were consistent with the increased proliferation we detected in space flight compared to ground controls [5].
Figure 9. Transcriptional profile of bone marrow cells as they differentiate into macrophages.
Correlation of macrophage differentiation stage (top 3 rows) with known transcriptional activation during differentiation (4thand 5th row).1Arrows [↑ or ↓] indicate transcript levels of macrophages differentiated for 14 days in space during STS-126 compared to ground controls. ↑ or ↓ trends are noted only at the last known stage of transcriptional activity during differentiation [43]. * indicates significantly different from ground controls (≥1.5 fold) after Benjamini-Hochberg correction [51], P < 0.05.
Current and Future Developments
The space flight experiment on STS-126 was a last-minute opportunity. The preflight verification tests comparing the Techshot bioreactor to FPAs prepared us for the flight. In our proposal to NASA, we showed that we could obtain meaningful data using the FPA hardware and that we understood the culture system. Results from STS-126 were validated by the consistency between the transcript expression data and the functional cell proliferation data [5]. This also provides strong justification for NASA to provide investigators ample opportunities to validate their culture systems because the acquired data may be dependent on the culture system. This will be especially important because of the retirement of the space shuttle and the readjustment of cell culture systems to new space platforms [50].
Acknowledgements
We thank Dr. Alan Forsman for help with STS-69, -73 and -77, Michelle Pate for her help with the preflight verification tests and Nanyan Lu for help with the array data from STS-126. We thank engineers Eric Taylor and Ken Barton of Techshot, Inc. for extensive support of the Cell Cult bioreactor system. Our work has been supported by NASA grants NAG2-1274 and NNX08BA91G, the NASA space grant consortium, the American Heart Association grant 0950036G, NIH grants AI55052, AI052206, AI088070, RR16475 and RR17686, the Terry C. Johnson Center for Basic Cancer Research and the Kansas Agriculture Experiment Station. This is Kansas Agriculture Experiment Station publication13-172-J.
Footnotes
List of abbreviations. Colony forming units-granulocyte macrophage (CFU-GM); Commercial Generic Bioprocessing Apparatus (CGBA); Fluid Processing Apparatus (FPA); Granulocyte-macrophage colony stimulating factor (GM-CSF); Group Activation Packs (GAPs); Ingenuity Pathway Analysis (IPA); Interleukin-6 (IL-6); Kansas State University (KSU); Kennedy Space Center (KSC); Macrophage colony forming units (CFU-M); Macrophage colony stimulating factor (M-CSF); Revolution per minute (RPM); Space Life Science Support Facility (SLSL); Space transportation system (STS).
References Cited
- 1.Sonnenfeld G. The immune system in space, including Earth-based benefits of space-based research. Curr Pharm Biotechnol. 2005 Aug;vol. 6:343–349. doi: 10.2174/1389201054553699. [DOI] [PubMed] [Google Scholar]
- 2.Chapes SK, Ganta RR. Mouse infection models for space flight immunology. Adv. Space Biol. Med. 2005;vol. 10:81–104. doi: 10.1016/s1569-2574(05)10004-5. [DOI] [PubMed] [Google Scholar]
- 3.Fleming SD, Edelman L, Chapes SK. Effects of corticosterone and microgravity on inflammatory cell production of superoxide. J. Leukoc. Biol. 1991;vol. 50:69–76. doi: 10.1002/jlb.50.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Woods K, Chapes S. Abrogation of TNF-mediated cytotoxicity by space flight involves protein kinase c. Exp. Cell Res. 1994 Mar;vol. 211:171–174. doi: 10.1006/excr.1994.1074. [DOI] [PubMed] [Google Scholar]
- 5.Ortega MT, Lu N, Chapes SK. Evaluation of in vitro macrophage differentiation during space flight. Advances in Space Research. 2012 May 15;vol. 49:1441–1455. doi: 10.1016/j.asr.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor G, Neale L, Dardano J. Immunological analyses of U.S. Space Shuttle crew members. Aviat. Space Environ. Med. 1986;vol. 57:213–217. [PubMed] [Google Scholar]
- 7.Meehan R, Neale L, Kraus E, Stuart C, Smith M, Cintron N, Sams C. Alteration in human mononuclear leucocytes following space flight. Immunol. 1992;vol. 76:491–497. [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenfeld G, Mandel AD, Konstantinova IV, Taylor GR, Berry WD, Wellhausen SR, Lesnyak AT, Fuchs BB. Effects of spaceflight on levels and activity of immune cells. Aviat Space Environ Med. 1990 Jul;vol. 61:648–653. [PubMed] [Google Scholar]
- 9.Sonnenfeld G, Mandel A, Konstantinova I, Berry W, Taylor G, Lesnyak A, Fuchs B, Rakhmilevich A. Space flight alters immune cell function and distribution. J. Appl. Physiol. 1992;vol. 73:191S–195S. doi: 10.1152/jappl.1992.73.2.S191. [DOI] [PubMed] [Google Scholar]
- 10.Ichiki A, Gibson L, Jago T, Strickland K, Johnson D, Lange R, Allebban Z. Effects of spaceflight on rat peripheral blood leukocytes and bone marrow progenitor cells. J. Leukoc. Biol. 1996 Jul;vol. 60:37–43. doi: 10.1002/jlb.60.1.37. [DOI] [PubMed] [Google Scholar]
- 11.Chapes SK, Simske SJ, Sonnenfeld G, Miller ES, Zimmerman RJ. Effects of spaceflight and PEG-IL-2 on rat physiological and immunological responses. J. Applied Physiol. 1999;vol. 86:2065–2076. doi: 10.1152/jappl.1999.86.6.2065. [DOI] [PubMed] [Google Scholar]
- 12.Forsman AD, Herpich AR, Chapes SK. Improved soft-agar colony assay in a fluid processing apparatus. In Vitro Cell Dev Biol Anim. 1999 Jan;vol. 35:55–60. doi: 10.1007/s11626-999-0044-2. [DOI] [PubMed] [Google Scholar]
- 13.Golde DW, Gasson JC. Cytokines: myeloid growth factors. In: Gallin JI, Goldstein IM, Snyderman R, editors. Inflammation: Basic Principles and Clinical Correlates. New York: Raven Press; 1988. pp. 253–261. 1988. [Google Scholar]
- 14.Metcalf D. The molecular contol of cell division, differentiation commitmant and maturation in haemopoietic cells. Nature. 1989;vol. 339:27–30. doi: 10.1038/339027a0. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford M, Witsell A, Schook L. Mechanisms generating functionally heterogeneous macrophages: chaos revisited. J. Leukoc. Biol. 1993 May;vol. 53:602–618. doi: 10.1002/jlb.53.5.602. [DOI] [PubMed] [Google Scholar]
- 16.Armstrong JW, Gerren RA, Chapes SK. The effect of space and parabolic flight on macrophage hematopoiesis and function. Exp Cell Res. 1995 Jan;vol. 216:160–168. doi: 10.1006/excr.1995.1020. [DOI] [PubMed] [Google Scholar]
- 17.Vellinger JC, Doyle JF, Hill S, Todd P, Wells ME, Chapes SK. Multiple robotic bioreactor systems for cell and tissue research in low gravity. In: American Institute of Aeronautics and Astronautics, editor. Archive Set 809. vol. AIAA Paper 2001–5012. 2001. [Google Scholar]
- 18.Vellinger J, Todd P, Taylor E, Kennedy DJ, Jones A, Boling RE. Overview of SHOT hardware capabiliites and range of cell biology experiment designs. J. Grav. Physiol. . 2004;vol. 11:51–56. [PubMed] [Google Scholar]
- 19.Vellinger JC, Barton KW, Wells ME, Deuser MS. Bioreactor apparatus and cell culturing system. United States Patent U. S. Patent No. 7,198,940. 2007 Apr 3;
- 20.Luttges MW. Recognizing and optimizing flight opportunities with hardware and life sciences limitations. Trans Kans Acad Sci. 1992;vol. 95:76–86. [PubMed] [Google Scholar]
- 21.Hoehn A, Klaus DM, Stodieck LS. A modular suite of hardware enabling spaceflight cell culture research. J Gravit Physiol. 2004 Mar;vol. 11:39–49. [PubMed] [Google Scholar]
- 22.Wilson JW, Ott CM, zu Bentrup KH, Ramamurthy R, Quick L, Porwollik S, Cheng P, McClelland M, Tsaprailis G, Radabaugh T, Hunt A, Fernandez D, Richter E, Shah M, Kilcoyne M, Joshi L, Nelman-Gonzalez M, Hing S, Parra M, Dumars P, Norwood K, Bober R, Devich J, Ruggles A, Goulart C, Rupert M, Stodieck L, Stafford P, Catella L, Schurr MJ, Buchanan K, Morici L, McCracken J, Allen P, Baker-Coleman C, Hammond T, Vogel J, Nelson R, Pierson DL, Stefanyshyn-Piper HM, Nickerson CA. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. U S A. 2007 Oct 9;vol. 104:16299–16304. doi: 10.1073/pnas.0707155104. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzenberg M, Pippia P, Meloni MA, Cossu G, Cogoli-Greuter M, Cogoli A. Microgravity simulations with human lymphocytes in the free fall machine and in the random positioning machine. J Gravit Physiol. 1998 Jul;vol. 5:P23–P26. [PubMed] [Google Scholar]
- 24.Hashemi BB, Penkala JE, Vens C, Huls H, Cubbage M, Sams CF. T cell activation responses are differentially regulated during clinorotation and in spaceflight. FASEB J. 1999 Nov;vol. 13:2071–2082. doi: 10.1096/fasebj.13.14.2071. [DOI] [PubMed] [Google Scholar]
- 25.Vellinger J, Ormsby R, Thomas N, Kennedy D, Hudson D, Todd P. Providing an optimal environment utilizing the Avian Development Facility for research in microgravity. Warrendale, PA: SAE International; 2001. Paper 2001-01-2289. [Google Scholar]
- 26.Chang TT, Walther I, Li C-F, Boonyaratanakornkit J, Galleri G, Meloni MA, Pippia P, Cogoli A, Hughes-Fulford M. The Rel/NF-κB pathway and transcription of immediate early genes in T cell activation are inhibited by microgravity. J. Leukoc. Biol. 2012 Jul 2; doi: 10.1189/jlb.0312157. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan J, Leenen PJM, Bertoncello I, Nishikawa S, Hamilton JA. Macrophage lineage cellls in inflammation: characterization by colony-stimulating factor-1 (CSF-1) receptor (c-fms), ER-MP58, and ER-MP20 (Ly-6C) expression. Blood. 1998;vol. 92:1423–1431. [PubMed] [Google Scholar]
- 28.Guilbert L, Stanley E. The interaction of 125I-colony-stimulating factor-1 with bone marrow-derived macrophages. J. Biol. Chem. 1986;vol. 261(9):4024–4032. [PubMed] [Google Scholar]
- 29.Rettenmier CW, Roussel MF, Ashmun RA, Ralph P, Price K, Sherr CJ. Synthesis of membrane-bound colony-stimulating factor 1 (CSF-1) and down modulation of CSF-1 receptors in NIH 3T3 cells transformed by cotransfection of the human CSF-1 and c-fms (CSF-1 receptor) genes. Mol. Cell. Biol. 1987 Jul 1;vol. 7:2378–2387. doi: 10.1128/mcb.7.7.2378. 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong S, Hughes RC. Macrophage surface glycoproteins binding to galectin-3 (Mac-2-antigen) Glycoconj J. 1997 Feb;vol. 14:267–274. doi: 10.1023/a:1018554124545. [DOI] [PubMed] [Google Scholar]
- 31.Ho MK, Springer TA. Mac-2, a novel 32,000 Mr mouse macrophage subpopulation-specific antigen defined by monoclonal antibodies. J Immunol. 1982 Mar;vol. 128:1221–1228. [PubMed] [Google Scholar]
- 32.Caminschi I, Lucas KM, O'Keeffe MA, Hochrein H, Laabi Y, Kontgen F, Lew AM, Shortman K, Wright MD. Molecular cloning of F4/80-like-receptor, a seven-span membrane protein expressed differentially by dendritic cell and monocyte-macrophage subpopulations. J Immunol. 2001;vol. 167:3570–3576. doi: 10.4049/jimmunol.167.7.3570. [DOI] [PubMed] [Google Scholar]
- 33.Hume DA, Ross IL, Himes SR, Sasmono RT, Wells CA, Ravasi T. The mononuclear phagocyte system revisited. J. Leukoc. Biol. 2002;vol. 72:621–627. [PubMed] [Google Scholar]
- 34.Ferret-Bernard S, Sai P, Bach JM. In vitro induction of inhibitory macrophage differentiation by granulocyte-macrophage colony-stimulating factor, stem cell factor and interferon-gamma from lineage phenotypes-negative c-kit-positive murine hematopoietic progenitor cells. Immunol. Lett. 2004 Feb 15;vol. 91:221–227. doi: 10.1016/j.imlet.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Leenen PJ, Melis M, Slieker WA, Van Ewijk W. Murine macrophage precursor characterization. II. Monoclonal antibodies against macrophage precursor antigens. Eur. J. Immunol. 1990 Jan;vol. 20:27–34. doi: 10.1002/eji.1830200105. [DOI] [PubMed] [Google Scholar]
- 36.Ammon C, Meyer SP, Schwarzfischer L, Krause SW, Andreesen R, Kreutz M. Comparative analysis of integrin expression on monocyte-derived macrophages and monocyte-derived dendritic cells. Immunology. 2000 Jul;vol. 100:364–369. doi: 10.1046/j.1365-2567.2000.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boxio R, Bossenmeyer-Pourie C, Steinckwich N, Dournon C, Nusse O. Mouse bone marrow contains large numbers of functionally competent neutrophils. J. Leukoc. Biol. . 2004 Apr;vol. 75:604–611. doi: 10.1189/jlb.0703340. [DOI] [PubMed] [Google Scholar]
- 38.Watt SM, Williamson J, Genevier H, Fawcett J, Simmons DL, Hatzfeld A, Nesbitt SA, Coombe DR. The heparin binding PECAM-1 adhesion molecule is expressed by CD34+ hematopoietic precursor cells with early myeloid and B-lymphoid cell phenotypes. Blood. 1993 Nov 1;vol. 82:2649–2663. [PubMed] [Google Scholar]
- 39.Corsi MM, Sandberg JK, Wasserman K, Maes HH, Kiessling R. Generation and function of bone marrow-derived dendritic cells from CD4/CD8(−/−) double-knockout mice. Immunol Lett. 1999;vol. 67:243–249. doi: 10.1016/s0165-2478(99)00018-8. [DOI] [PubMed] [Google Scholar]
- 40.Mazo IB, Honczarenko M, Leung H, Cavanagh LL, Bonasio R, Weninger W, Engelke K, Xia L, McEver RP, Koni PA, Silberstein LE, von Andrian UH. Bone marrow is a major reservoir and site of recruitment for central memory CD8+ T cells. Immunity. 2005 Feb;vol. 22:259–270. doi: 10.1016/j.immuni.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Woodland DL, Blackman MA. Immunity: it's in our bones. Immunity. 2005 Feb;vol. 22:143–144. doi: 10.1016/j.immuni.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Lawler A. Remaking NASA: How much space for science? Science. 2004 Jan 30;vol. 303:610–612. doi: 10.1126/science.303.5658.610. 2004. [DOI] [PubMed] [Google Scholar]
- 43.Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998 Apr;vol. 63:405–417. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]
- 44.Bjornsson JM, Larsson N, Brun ACM, Magnusson M, Andersson E, Lundstrom P, Larsson J, Repetowska E, Ehinger M, Humphries RK, Karlsson S. Reduced Proliferative Capacity of Hematopoietic Stem Cells Deficient in Hoxb3 and Hoxb4. Mol. Cell. Biol. 2003 Jun 1;vol. 23:3872–3883. doi: 10.1128/MCB.23.11.3872-3883.2003. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauvageau G, Thorsteinsdottir U, Hough MR, Hugo P, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997 Jan;vol. 6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 46.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997 May 15;vol. 89:3636–3643. [PubMed] [Google Scholar]
- 47.LeCouter JE, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 Gene. Mol. Cell. Biol. 1998 Dec 1;vol. 18:7455–7465. doi: 10.1128/mcb.18.12.7455. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attwooll C, Denchi EL, Helin K. The E2F family: specific functions and overlapping interests. Embo J. 2004 Dec 8;vol. 23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enos ME, Bancos SA, Bushnell T, Crispe IN. E2F4 Mmdulates differentiation and gene expression in hematopoietic progenitor cells during commitment to the lymphoid lineage. J. Immunol. 2008 2008 Mar 15;vol. 180:3699–3707. doi: 10.4049/jimmunol.180.6.3699. [DOI] [PubMed] [Google Scholar]
- 50.Lawler A. Scientists Hope to Adjust the President's Vision for Space. Science. 2008 Feb 1;vol. 319:564–566. doi: 10.1126/science.319.5863.564. 2008. [DOI] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Statistical Soc. Series B. 1995;vol. 57:289–300. 1995. [Google Scholar]