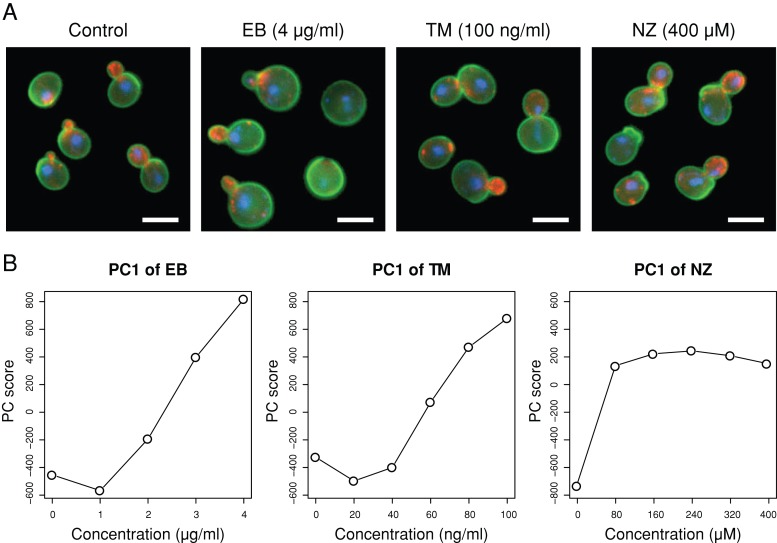
To better define how cell wall structure affects morphogenesis, the morphology of yeast cells was analyzed quantitatively after treatment with the three drugs that inhibit different aspects of cell wall synthesis. These drugs induced both similar effects, including broader necks and increased morphological variation, and distinct effects.
Abstract
The cell wall of budding yeast is a rigid structure composed of multiple components. To thoroughly understand its involvement in morphogenesis, we used the image analysis software CalMorph to quantitatively analyze cell morphology after treatment with drugs that inhibit different processes during cell wall synthesis. Cells treated with cell wall–affecting drugs exhibited broader necks and increased morphological variation. Tunicamycin, which inhibits the initial step of N-glycosylation of cell wall mannoproteins, induced morphologies similar to those of strains defective in α-mannosylation. The chitin synthase inhibitor nikkomycin Z induced morphological changes similar to those of mutants defective in chitin transglycosylase, possibly due to the critical role of chitin in anchoring the β-glucan network. To define the mode of action of echinocandin B, a 1,3-β-glucan synthase inhibitor, we compared the morphology it induced with mutants of Fks1 that contains the catalytic domain for 1,3-β-glucan synthesis. Echinocandin B exerted morphological effects similar to those observed in some fks1 mutants, with defects in cell polarity and reduced glucan synthesis activity, suggesting that echinocandin B affects not only 1,3-β-glucan synthesis, but also another functional domain. Thus our multivariate analyses reveal discrete functions of cell wall components and increase our understanding of the pharmacology of antifungal drugs.
INTRODUCTION
The fungal cell wall is a rigid structure that plays important roles in the establishment and maintenance of cell shape (Klis et al., 2006). Without the cell wall, fungal cells could not form their unique morphology. Cells of the budding yeast Saccharomyces cerevisiae have an oval shape, surrounded by the cell wall formed during vegetative cell growth. Dynamic remodeling of the yeast cell wall occurs during the cell cycle and is coordinated during cell morphogenetic events, including bud emergence, apical bud growth, isotropic bud growth, and cell division (Latgé, 2007).
Based on detailed analyses of yeast cell wall composition, it was proposed that it exhibits a highly organized dynamic network structure (Kollár et al., 1997). The cell wall is composed of interconnected filamentous polysaccharides and nonfilamentous glycoproteins. A major filamentous component is 1,3-β-glucan, whose nonreducing ends function as sites for the covalent attachment of other polysaccharides (Klis et al., 2006). 1,6-β-Glucan is another filamentous component, but it is relatively short and a minor component of the cell wall. Highly branched 1,6-β-glucan chains are found at the external face of the 1,3-β-glucan network. Chitin is an additional minor component normally deposited in a ring in the neck between a mother cell and its emerging bud, in the primary septum during division, and in the lateral walls of newly separated daughter cells, where it serves as an anchor for the glucan network (Orlean, 2012). The major nonfilamentous cell wall components are mannoproteins, which are mannose-containing glycoproteins. These heavily glycosylated proteins are typically found on the exterior of the cell wall. Most mannoproteins are glycosylphosphatidylinositol–anchored proteins and are connected to 1,6-β-glucan chains. Thus, covalent linkages between the various components of the cell wall give rise to its elastic and plastic properties, providing a strong, continuous fabric.
The fact that the different types of yeast cell wall components are interconnected suggests that the disruption of individual cell wall biosynthetic pathways would result in similar morphological phenotypes. Alternatively, the contribution of different components to cell morphology may differ because the physical strength and flexibility of the filamentous and nonfilamentous components vary. Several studies examined the effects of perturbing the synthesis or the cross-linking of cell wall components (de Groot et al., 2001; Nishiyama et al., 2002; Schmidt et al., 2003, 2005; Blanco et al., 2012). However, qualitative observation by eye has limited power to resolve the morphological differences caused by the mutation of genes encoding the enzymes involved in cell wall synthesis. To date, no quantitative morphological analyses of cell wall mutant strains have been performed.
To examine yeast cell morphology quantitatively, we developed an automatic image processing system called CalMorph (Ohtani et al., 2004). This system is a high-throughput, high-resolution image processing software package that enables the quantification of 501 cell morphology parameters from fluorescence images of the cell wall, actin, and nuclear DNA of yeast cells (Ohya et al., 2005). On the basis of a morphological analysis of the 4718 haploid nonessential gene deletion mutants, we created a morphological database named the Saccharomyces cerevisiae Morphological Database (SCMD; http://yeast.gi.k.u-tokyo.ac.jp; Saito et al., 2004). Analysis of the data for this large set of mutants revealed significant new relationships between morphological phenotypes and gene functions (Ohya et al., 2005). Further detailed phenotypic analysis of clusters of mutants with similar phenotypes revealed new insights into complex cellular pathways (Ohnuki et al., 2007; Okada et al., 2010; Yoshida et al., 2013). In addition, we were able to predict the intracellular targets of specific drugs by comparing their morphological effects to the database (Ohnuki et al., 2010; Iwaki et al., 2013).
To gain further insight into the involvement of the cell wall in morphogenesis, we quantitatively analyzed the morphological changes induced by echinocandin B (EB), tunicamycin (TM), and nikkomycin Z (NZ), drugs that block cell wall biosynthesis pathways. Specifically, EB is a noncompetitive inhibitor of 1,3-β-glucan synthase (GS; Sawistowska-Schröder et al., 1984; Douglas, 2001), TM targets N-acetylglucosamine (GlcNAc) phosphotransferase, which plays a role in the biosynthesis of mannoproteins (Barnes et al., 1984), and NZ is a competitive inhibitor of chitin synthase (Cabib, 1991). Multivariate analyses of the morphological changes induced by these drugs, as well as a comparison of the cell wall mutants, revealed discrete functions of the cell wall components in determining cell morphology.
RESULTS
Morphological effects of cell wall–affecting drugs
The morphological effects of treating haploid yeast with drugs that perturb the cell wall (EB, TM, and NZ) were examined by using fluorescence microscopy to observe the cell wall, actin, and nuclear DNA. Visual inspection revealed that cells treated with 4 μg/ml EB, 100 ng/ml TM, and 400 μM NZ exhibited abnormal morphologies (Figure 1A). To explore the dose-dependent changes, we quantified 501 morphometric parameters using CalMorph (Ohtani et al., 2004) after treatment with various concentrations of the drugs and performed a principal component analysis (PCA). PCA is an exploratory multivariate statistical technique for simplifying complex data sets, and it has been used for analyzing the effects of dose-dependent changes on morphology (Ohnuki et al., 2012). Figure 1B shows that the first principal component (PC1) scores increased in a dose-dependent manner for each of the cell wall–affecting drugs, suggesting that similar morphological changes were progressively more apparent in the data sets. Next we performed a second PCA to identify dose-dependent parameters that are independent of each other (see Materials and Methods). Accordingly, we identified seven, eight, and two representative parameters for EB, TM, and NZ, respectively (Supplemental Figures S1 and S2 and Supplemental Tables S1, a–c, and S2, a–c).
FIGURE 1:
Morphological changes induced by cell wall–affecting drugs. (A) Wild-type strains were cultured until the early log phase at 25°C in YPD with EB or TM, and in SD with NZ at the indicated concentrations. Cells were stained with fluorescein isothiocyanate–concanavalin A (green), rhodamine–phalloidin (red), or 4′,6-diamidino-2-phenylindole (blue) to visualize the cell wall, actin, or nucleus, respectively. Bar, 5 μm. (B) Dose-dependent changes in PC1 scores. Morphological data on cells treated with various concentrations of the drugs (n = 5) were subjected to PCA.
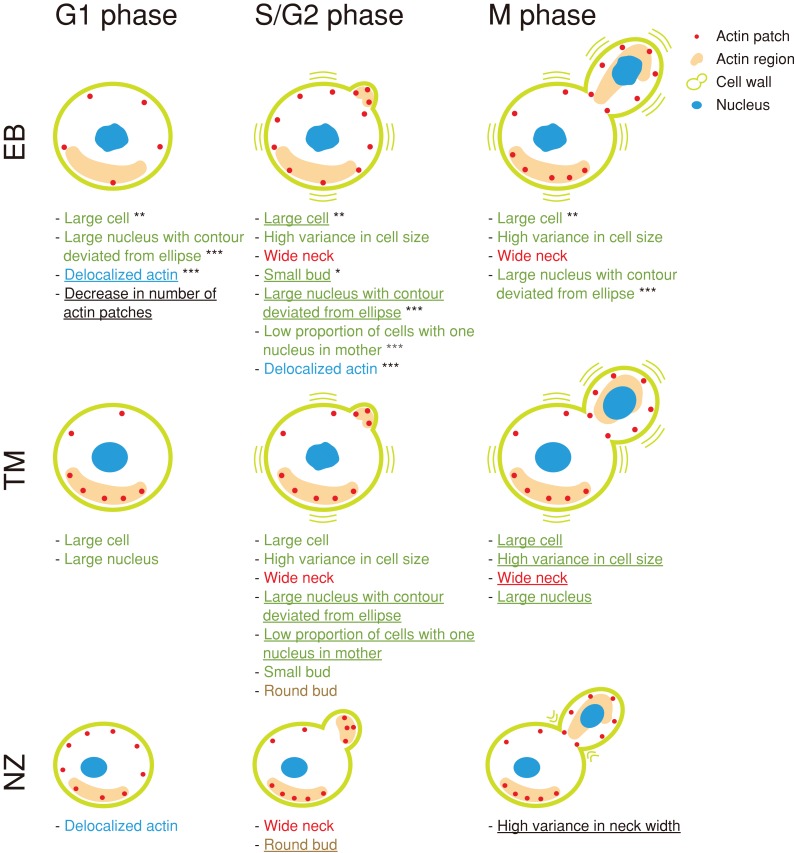
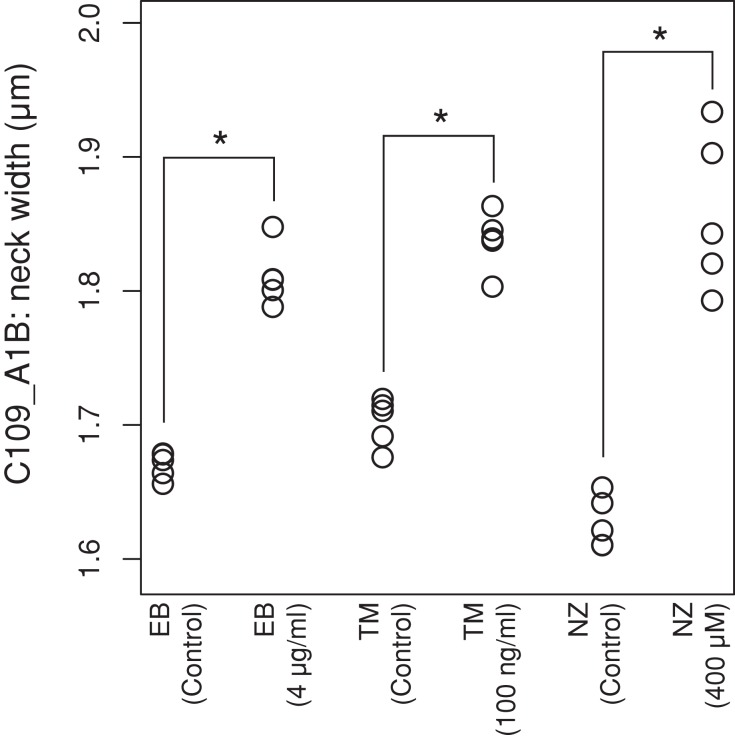
Figure 2 illustrates the morphological changes induced by EB, TM, and NZ, in which all representative parameters significantly affected by the drugs are shown (p < 0.0001 after Bonferroni correction using the t test), as well as the progression of the cell cycle stages, including unbudded cells (G1), budded cells with a single nucleus (S/G2), and budded cells with two nuclei (M). Of note, treatment with all three drugs resulted in an increased neck width (red). Neck width increased at 0.1–0.3 μm with the EB, TM, and NZ treatments (Figures 1A and 3), suggesting that preservation of the neck structure is a major role of the yeast cell wall. In addition to the unique features for each cell wall–affecting drug (black), we identified features common to EB and TM (green), EB and NZ (blue), and TM and NZ (brown). The morphological features induced by TM overlapped with those induced by EB. As reported previously, the proportion of small budded cells increased after EB treatment (Drgonová et al., 1999). Cells were enlarged after TM treatment, as described previously (Arnold and Tanner, 1982). In addition to the known cell morphological changes, we found that actin and the nucleus were also affected. The nuclear size increased after EB and TM treatment, and actin was delocalized in EB- and NZ-treated cells. These quantitative analyses revealed that each cell wall component had different and distinct functions in cell, actin, and nuclear morphology.
FIGURE 2:
Summary of the morphological changes induced by cell wall–affecting drugs. Morphological changes induced by the drugs were analyzed using two-step PCA to identify representative parameters for each drug (see Materials and Methods). Representative parameters for each drug (underlined) and those identified for another drug but significantly affected by the indicated drugs are shown with illustrations of the cells (Jonckheere–Terpstra test; <1 false positive was detected by chance at p = 0.0013, 0.0052, and 0.0033 for EB, TM, and NZ, respectively; Supplemental Table S3). Colored text indicates shared morphological features among drugs; red, green, blue, and brown represent features shared by EB–TM–NZ, EB–TM, EB–NZ, and TM–NZ, respectively. Asterisks denote features shared by EB and fks1-ts mutants; single, double, and triple asterisks indicate features shared by EB and class I and III, class II, and class II and III, respectively (Supplemental Table S5).
FIGURE 3:
Effect of cell wall–affecting drugs on neck width (C109_A1B). Morphological changes induced by treatment with (indicated concentrations) or without (control) the highest drug concentrations are plotted. Asterisk indicates significant difference (p < 0.05 by Mann–Whitney U test after Bonferroni correction).
Phenotypic variation after treatment with cell wall–affecting drugs
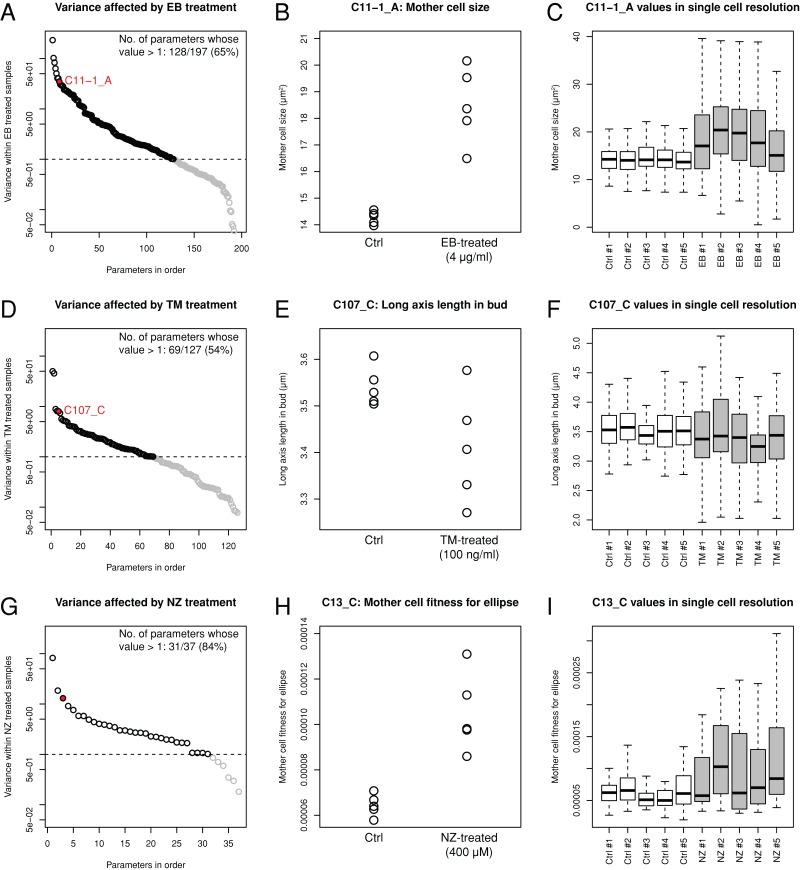
To investigate phenotypic variations, we compared the distribution of variance with and without cell wall drugs. Among the parameters with notable drug effects (Jonckheere–Terpstra test, p < 0.05), the variance was greater in EB-, TM-, and NZ-treated cells (Figure 4, A, D, and G). We found that 65% (128/197), 54% (69/127), and 84% (31/37) of the parameters showed a broad distribution after treatment with EB, TM, and NZ, respectively. Mother cell size (parameter C11-1_A) exhibited marked variance among the five replicates after the EB treatment (Figure 4B). Similarly, the long axis in bud (parameter C107_C) and mother cell fitness for ellipse (parameter C13_C) exhibited greater variance after the TM and NZ treatments, respectively (Figure 4, E and H). Phenotypic variation in each trait can be partitioned into the contribution of variations among the cell population and measurement errors. Significantly greater variations among the cell populations were detected after drug treatments (Figure 4, C, F, and I; p < 0.05 after Bonferroni correction, Mann–Whitney U test), which suggested that phenotypic variation could be explained in part by variation in the cell population.
FIGURE 4:
Marked morphological variation among drug-treated yeast populations. (A, D, G) Variance in the morphological parameters affected by the drugs (A, EB; D, TM; and G, NZ) was plotted in the highest to lowest order. Black and gray circles indicate parameters of higher and lower variance compared with the control (=1, dashed line), respectively. Red circles denote morphological parameters exemplified in B, E, and H. (B, E, H) Examples of morphological parameters with increased variance upon EB, TM, and NZ treatment. (C, F, I) Distributions of morphological parameters exemplified in B, E, and H visualized using a box plot with single-cell resolution. Gray and white boxes denote single-cell distribution with and without drug treatment, respectively.
Effects of EB, TM, and NZ on cell morphology
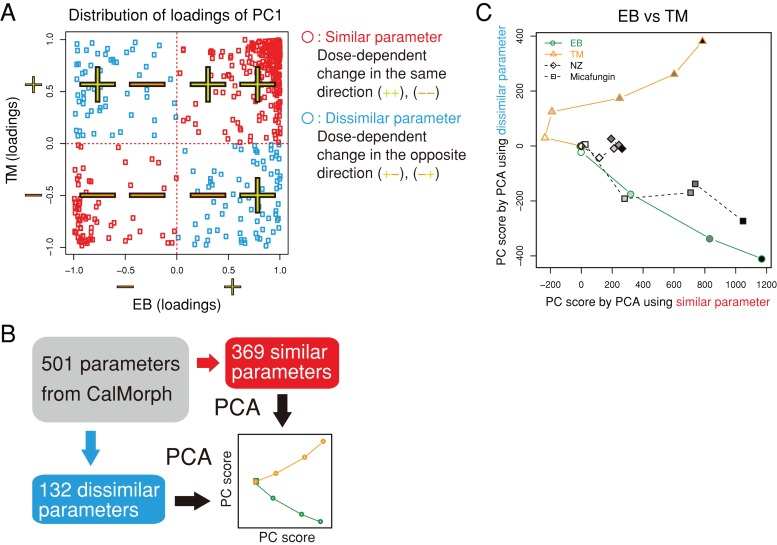
To compare the effects of the cell wall–affecting drugs, we plotted dose-dependent morphological changes in a two-dimensional (2D) space of “similarity” and “dissimilarity.” Similar parameters were defined as those with the same direction of changes after treatment with the two drugs, whereas dissimilar parameters exhibited changes in the opposite direction (see Materials and Methods). As shown in Figure 5A, similar and dissimilar parameters were located in the first/third quadrants and second/fourth quadrants, respectively. Representative similar and dissimilar features were then extracted by reducing the dimensions by PCA (Figure 5B and Supplemental Figure S3).
FIGURE 5:
Similarity and dissimilarity of the effects of the drugs on cell morphology. (A) Distribution of loadings for PC1 after PCA of the EB and TM data. Of the 501 parameters, 369 were defined as similar (red symbols) due to their distribution in the first/third quadrants; otherwise, 132 were dissimilar parameters that were distributed in the second/fourth quadrants (blue symbols). (B) Schematic representation of the extraction of similar/dissimilar effects on drug treatment. Morphological parameters were assigned to similar or dissimilar parameters based on the loading sign and independently subjected to the second PCA. Dose-dependent effects of the drugs were then visualized in a similar/dissimilar 2D space. (C) Similar and dissimilar effects of EB and TM on cell morphology. Morphological changes induced by NZ and micafungin were mapped onto the 2D space. The grayscale indicates the doses of drugs used (from dilute to concentrated).
The EB and TM treatments are compared in Figure 5C. The EB treatment decreased from the point of origin, but the TM treatment increased with increasing concentrations, separating the effects of the two drugs. Treatment with micafungin, another GS inhibitor, also decreased, similar to EB. Given that micafungin had similar effects to EB, drugs with the same activity showed similar behavior in the similar/dissimilar 2D space.
We also compared EB and NZ treatments and TM and NZ treatments (Supplemental Figure S4). Supplemental Figure S4A shows that EB, NZ, and TM behaved differently but that micafungin acted similarly to EB. TM, NZ, and EB also worked differently in the similar/dissimilar 2D space (Supplemental Figure S4B). Therefore EB, TM, and NZ had distinct effects on cell morphology, as expected from their rather different cellular effects.
Nonessential deletion mutants with morphologies similar to those induced by drug treatments
To validate the analytical system, we compared the morphological profile of cells treated with the cell wall–affecting drugs to those of the nonessential deletion mutants with defects in cell wall–related genes (Lesage and Bussey, 2006; Orlean, 2012). The deletion mutants of the cell wall targets should have similar morphological profiles as cells treated with the cell wall–affecting drugs.
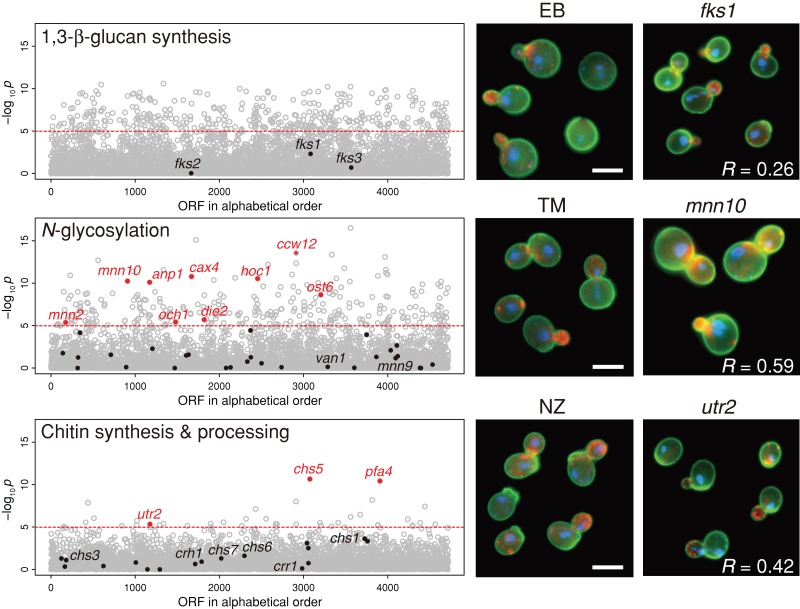
The morphological profiles induced by the TM treatment showed significant similarities to those induced by individual deletions of five genes (MNN10, ANP1, OCH1, MNN2, and HOC1) that encode mannosyltransferase in the Golgi (Figure 6; p < 0.05 after Bonferroni correction, t test). The correlation coefficient for mnn10 was 0.59. TM-treated cells were also similar to mutants of cax4, a dolichyl pyrophosphate phosphatase that supplies dolichol-linked oligosaccharide substrates for N-glycosylation in the endoplasmic reticulum (ER; van Berkel et al., 1999). The Gene Ontology term analysis with the significantly similar 85 gene deletion mutants (p < 0.0001 after Bonferroni correction, t test) showed enrichment of α-1,6-mannosyltransferase activity (Supplemental Table S4). These results are consistent with the mode of action of TM, which blocks N-glycosylation (Ballou, 1990; Jigami, 2008).
FIGURE 6:
Morphological profiling of the cell wall–affecting drugs. The drug-induced morphological profiles were compared with 4718 mutants of nonessential genes, as described previously (Ohnuki et al., 2010). The vertical axis indicates the similarity to each gene deletion mutant as a –log10 p value of a correlation coefficient (t test). The horizontal dashed line shows the threshold of p = 1.06 × 10−5 corresponding to a one-sided p = 0.05 after Bonferroni correction. Red dots denote cell wall–associated mutants (Lesage and Bussey, 2006; Orlean, 2012), with morphological profiles similar to those of drug-treated cells. Right, representative photographs of wild-type cells after drug treatment and representative deletion mutants (fks1, mnn10, and utr2). R is the correlation coefficient between drug-treated and mutant cells. Bar, 5 μm.
Of the three chitin synthase mutants, only the chs1 and chs3 mutants were viable and therefore testable in our assay. Neither of these mutants was morphologically similar to NZ-treated cells, probably because NZ targets more than one chitin synthase in vivo. NZ-treated cells were morphologically similar to the utr2 (crh2) mutant (Figure 6; r = 0.42), which was defective for the major chitin transglycosylase that functions in the transfer of chitin to 1,3-β- and 1,6-β-glucans (Cabib et al., 2007). In contrast, the morphologies of the crh1 mutant (defective in minor chitin transglycosylase) and the crr1 mutant (defective in cross-linking during sporulation) were different from NZ-treated cells (Rodríguez-Peña et al., 2000; Gómez-Esquer et al., 2004; Cabib et al., 2007). NZ-treated cells were also morphologically similar to mutants lacking CHS5 (Figure 6), which is involved in the trafficking of Chs3 and Utr2 (Rodríguez-Peña et al., 2002). These results highlight the critical role that chitin plays in the anchoring of the β-glucan network.
However, EB-treated cells had no resemblance to the deletion mutants of the major (Fks1) and minor (Fks2) subunits of GS (Figure 6). This is consistent with previous observations (Ohnuki et al., 2010; Okada et al., 2010) and is probably due to functional redundancy between Fks1 and Fks2 and/or the multifunctional properties of Fks1 (Ohnuki et al., 2010; Okada et al., 2010). Mutants subsets related to α-1,6-mannosyltransferase were enriched in 84 significantly similar mutants to EB-treated cells (p < 0.0001 after Bonferroni correction, t test; Supplemental Table S4), perhaps because EB- and TM-treated cells were somewhat similar.
Morphological similarities with temperature-sensitive fks1 mutants
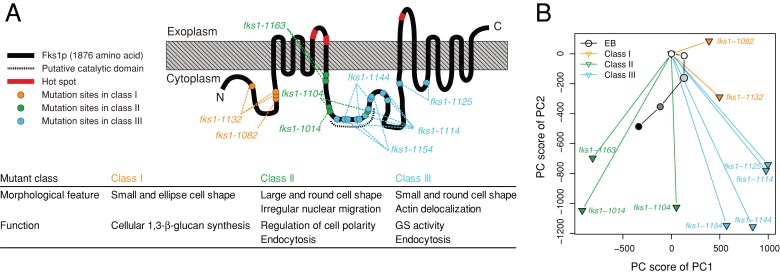
Because EB-treated cells were not similar to fks1Δ or fks2Δ deletion mutants, we compared them with the morphological profiles of temperature-sensitive GS mutants. Because FKS1 and FKS2 are redundant genes, we previously created 10 temperature-sensitive fks1 alleles in an fks2Δ-mutant strain background (Figure 7A; Okada et al., 2010). These mutants were classified into three groups (Classes I–III) according to their different phenotypes. After PCA analysis, the morphological profiles of the fks1 mutants were depicted in a 2D space that described the variety of mutant phenotypes (Figure 7B and Supplemental Figure S5). EB affects GS activity (Sawistowska-Schröder et al., 1984; Douglas, 2001). Because class III mutants showed decreased GS activity, we predicted that EB-treated cells would resemble class III mutants. However, the mapping of PC scores of EB-treated cells indicated similarity to the class II mutants (Figure 7B). Class II mutants showed normal GS activity and exhibited loss of Spa2 localization (Okada et al., 2010). Because Spa2 is a component of the polarisome (Snyder, 1989; van Drogen and Peter, 2002), loss of Spa2 localization is indicative of a cell polarity defect in class II mutants.
FIGURE 7:
Morphological phenotypes of EB-treated and fks1 cells. (A) Functional map of Fks1p (modified from Okada et al., 2010). The gray-hatched box denotes the plasma membrane. Red symbols on the black line indicate hot spots (the echinocandin resistance–conferring mutations). The amino acids mutated and the hot spots are listed in Supplemental Table S6. (B) PCA of the data sets of EB-treated and fks1 cells. Morphological data on EB-treated cells were mapped onto the 2D space obtained by PCA of the fks1-ts mutant (Okada et al., 2010). The grayscale within the circular symbols indicates the dose of EB used (from dilute to concentrated). PC1 and PC2 explained ∼60% of the variance.
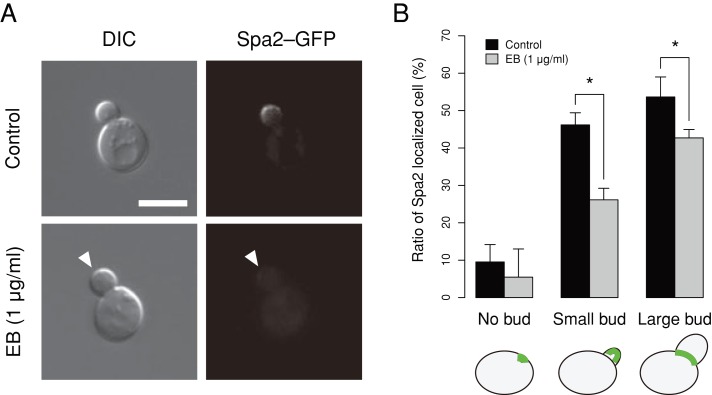
To determine whether EB-treated cells had a cell polarity defect, we assessed the localization of Spa2–green fluorescent protein (GFP) in living cells. We confirmed that Spa2 localization was lost in class II but not in class III mutants (Supplemental Figure S6), consistent with previous observations (Okada et al., 2010). We also found that Spa2–GFP localization decreased significantly after EB treatment (Figure 8), suggesting that EB influenced the regulation of cell polarity by inhibiting Spa2 localization, similar to class II mutants.
FIGURE 8:
Effect of EB on Spa2–GFP localization. (A) YOC5002 (spa2 [Spa2–GFP]) cells were incubated at 25ºC in SD–U medium with EB (1 μg/ml) or DMSO (control solvent) until the early log phase. Cells were then harvested and observed without fixation. The arrowhead indicates the absence of Spa2–GFP localization in the bud. Bar, 5 μm. (B) Spa2–GFP localized cells were enumerated in three independent experiments. The mean of the triplicates is plotted. Error bars denote 1 standard deviation. Asterisk indicates significant difference (p < 0.05 by t test after Bonferroni correction).
We next compared the morphological phenotypes of EB-treated cells with those of the class II and class III mutants. Among the several morphological features of EB-treated cells (Figure 2), either increased cell size or increased ratio of cells with small buds was observed in class II or class III mutants, respectively (Supplemental Table S5). The increased ratio of cells with delocalized actin was common to both classes (Supplemental Table S5). Thus some of the morphological parameters influenced by EB are also affected in class II and III mutants.
EB affects both class II and III domains
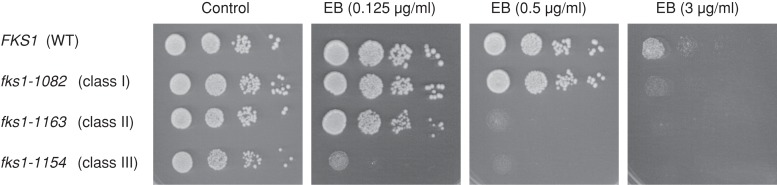
Recent topological analysis revealed that EB-resistance mutations accumulated as affected clusters of residues called hot spots at the extracellular surface of Fks1 (Johnson and Edlind, 2012). Hot spots 1–3 are located at amino acid residues of 635–649, 1354–1361, and 690–700, respectively, of Fks1 (Supplemental Table S6). Because these hot spots are considered to be the sites for direct interaction with EB, it is significant that class II and III mutations affected residues near these hot spots in the primary structure of Fks1p (Figure 7A and Supplemental Table S6). To determine whether EB inhibited the function of both class II and class III mutants, we examined the EB sensitivity of each class of GS mutants. Both class II (fks1-1163) and class III (fks1-1154) mutants failed to grow in the presence of 0.5 μg/ml EB, indicating an EB-sensitive phenotype (Figure 9). In contrast, wild-type and class I mutants (fks1-1082) were capable of growth in the presence of 0.5 μg/ml EB. These results suggested that EB affected the functions of the regions of Fks1 that are altered by the class II and class III mutations.
FIGURE 9:
EB sensitivity of the fks1-ts mutants. Cells were cultured in YPD medium until the log phase and diluted serially to 1 × 106, 1 × 105, 1 × 104, and 1 × 103 cells/ml. Suspensions of 5 μl were dropped on the plates contained EB or DMSO as a control solvent. Plates were incubated at 25ºC for 3 d. The experiments were replicated at least twice, and representative results are shown. FKS1, fks1-1082, fks1-1154, and fks1-1163 indicate YOC4318, YOC4320, YOC4327, and YOC4328, respectively.
We next determined whether the fks1 mutants with both class II and III defects were lethal. For this purpose, we constructed strains harboring single-domain (fks1-1163 and fks1-1154) and double-domain (fks1-11631154) mutations after a galactose-inducible FKS1 gene was introduced on a plasmid. We found that the double-domain mutants failed to grow even at 25°C, suggesting that class II and III domains had synergistic effects on the function of Fks1 (Supplemental Figure S7). Thus EB affected two related and important functions of Fks1: 1,3-β-glucan synthesis and cell polarity.
DISCUSSION
We used three drugs (EB, TM, and NZ) that cause distinct types of cell wall damage by inhibiting known targets to characterize the morphological changes induced by inhibiting the biosynthesis of the three major cell wall components, 1,3-β-glucan, mannoproteins, and chitin, in S. cerevisiae. Treatment with the three drugs induced several common morphological effects, including increased neck width and morphological variation. However, they also exhibited different effects on cell morphology, indicative of the distinct roles played by the cell wall components in cell morphogenesis. Results of chemical–genetic approaches using TM and NZ were consistent with the classical genetic approach, demonstrating that N-linked glycosylation and glucan/chitin cross-linking were important for morphogenesis. Moreover, EB had pleiotropic effects on cell morphology and affected multiple functions of Fks1, including maintenance of cell polarity and 1,3-β-glucan synthesis.
N-glycosylation pathway and cell morphogenesis
Statistical analysis revealed that the TM treatment induced several morphological features. Eight representative parameters were affected in a dose-dependent manner after treatment with TM (Supplemental Table S2b). For example, mother and daughter cells enlarged, nuclear size increased in unbudded cells, and the neck width increased. The increase in cell size was reported previously (Arnold and Tanner, 1982).
TM blocks the initial step of N-glycosylation by inhibiting Alg7 in the ER (Barnes et al., 1984). The morphological profile induced by the TM treatment showed significant similarity to individual deletions of the genes encoding mannosyltransferase in the Golgi, as well as cax4, die2, and ost6, which mediate N-glycosylation in the ER. TM-treated cells were similar to an och1 mutant (a mutant of cis-Golgi α-1,6-mannosyltransferase), deletion mutants of Golgi α-1,6-mannosyltransferase and the M-pol II complex (mnn10, anp1, and hoc1), and a mnn2 mutant (a mutant of α-1,2-mannosyltransferase), which are defective in the first, third, and fourth steps, respectively, of outer-chain elongation of N-linked oligosaccharides in the Golgi (Jigami, 2008). Mnn9 is also a component of M-pol II (A complex), but forms another mannosyltransferase complex known as M-pol I (V complex), which functions in the second step with Van1 (Hashimoto and Yoda, 1997; Jungmann and Munro, 1998; Jungmann et al., 1999). Given that the morphological phenotypes of mnn9 and van1 differ from that of TM-treated cells, M-pol I may play additional roles in cell morphogenesis.
Loss of mannosyltransferase activity reduces protein mannosylation (Lagunas et al., 1986). Mutants defective in early glycosylation reactions exhibit aberrant cell wall assembly (Orłowski et al., 2007). These results suggest that N-glycosylation is necessary for the activity or stability of the proteins responsible for cell wall assembly. Unexpectedly, we found that the effects of TM treatment were similar to the effects caused by the single-gene deletion of Ccw12 (Figure 6; r = 0.66), highlighting the role of this mannoprotein in cell wall function (Mrsa et al., 1999; Ragni et al., 2011).
Chitin synthesis and cell morphogenesis
NZ had no severe effects on S. cerevisiae growth, and therefore its effect on cell morphology was expectedly minor. We found that neck width and bud roundness increased, whereas the ratio of unbudded cells with localized actin decreased. The wide-neck phenotype was reported previously (Schmidt et al., 2003). Note that no changes in nuclear morphology were detected.
Chitin is a polymer of 1,4-β-linked GlcNAc, contributing only 1–2% of the dry weight of the cell wall (Lesage and Bussey, 2006), which serves as a scaffold for anchoring 1,3-β- and 1,6-β-glucans. Chitin occurs in three different and polydispersed forms in the wall. In addition to free chitin, some is bound to 1,3-β-glucan and is present predominantly in the neck region, whereas a lesser amount is found in lateral walls bound to 1,6-β-glucan (Cabib and Durán, 2005; Cabib, 2009). We found that NZ-treated cells were morphologically similar to utr2, which was defective in glucan/chitin cross-linking (Cabib et al., 2007; Cabib, 2009). In addition, chs5 and pfa4 mutants, but not chs3, chs6, or chs7, all of which contain reduced levels of chitin, also showed a significant correlation with the NZ treatment cells. Of interest, only chs5 and pfa4 have additional defects on the intracellular traffic of other proteins, including the mislocalization of Utr2 in the chs5 mutant (Rodríguez-Peña et al., 2002). These results highlight the critical role of the chitin–glucan linkage in morphogenesis, in clear agreement with a recent proposal (Cabib and Arroyo, 2013), but also uncover a redundant role of the different chitin synthases in forming the chitin–glucan network.
EB affects multiple functions of Fks1
Echinocandin-family drugs (e.g., EB and micafungin) noncompetitively inhibit incorporation of UDP-glucose into 1,3-β-glucan (Douglas, 2001). This has not been demonstrated directly, but several lines of evidence suggest that Fks1 is a direct target of echinocandins. For example, Fks1 is a putative catalytic subunit of GS (Inoue et al., 1995; Drgonová et al., 1996; Qadota et al., 1996); hot spots—regions of the mutations conferring strong echinocandin resistance—exist exclusively in Fks1 (Douglas, 2001; Perlin, 2007; Johnson et al., 2011); and specific mutations within these hot spots result in differential echinocandin resistance (Johnson et al., 2011; Healey et al., 2012).
Here we demonstrated that EB more strongly affected class II and III fks1 mutants, which carry substitutions in the cytosolic region. Johnson and Edlind (2012) reported that the three hot spots for echinocandin-resistance mutations affect residues that are located externally and proposed that all hot spot residues are juxtaposed in the three-dimensional structure of Fks1 to form a single echinocandin-binding pocket. Of interest, the residues affected by the class II and III mutations are adjacent to the hot spots, which suggests that binding of EB affects the central cytosolic domains of Fks1.
Preservation of the neck structure of the cell wall
We demonstrated that yeast cells had a broadened neck width after treatment with all cell wall–affecting drugs, suggesting that maintaining a narrow neck is a common function performed by cell wall components. Phenotypic analysis of several cell wall mutants supported this hypothesis. Mutation of genes involved in the outer-chain elongation of N-linked oligosaccharides in the Golgi (OCH1, MNN10, ANP1, and MNN2), CAX4 (N-glycosylation in the ER), and CCW12 (mannoprotein) resulted in significantly wider necks (data available at SCMD). Chs5, which is involved in the export of chitin synthase (Santos and Snyder, 1997; Santos et al., 1997), Kre6, which is required for 1,6-β-glucan biosynthesis (Roemer and Bussey, 1991), and 1,3-β-glucanosyltransferase (Gas1), which is required for cell wall assembly; Ram et al., 1998; Popolo and Vai, 1999), were also all necessary to maintain a narrow neck (data available at SCMD). Thus classical genetic and chemical–genetic approaches both resulted in common morphological phenotypes induced by the lesion of cell wall components.
All components of the yeast cell wall are localized at or near the bud neck. The filamentous components 1,3-β-glucan and 1,6-β-glucan are found on the interior of the cell wall around a constriction between the mother and daughter cells. Nonfilamentous component mannoproteins are also localized to this region on the exterior surface. In contrast, chitin is localized at the bud/birth scar and the primary septum. Several explanations of the wide-neck phenotypes caused by cell wall–affecting drugs are possible. Physical strength is presumably required to maintain the curvature structure of the neck. If the network structure of the cell wall is weakened, it may lead to morphological changes of the neck. Alternatively, neck morphology may be determined by many genes, among which those encoding enzymes responsible for synthesis of cell wall components may play important roles. Further studies are required to clarify the molecular mechanism underlying neck morphogenesis and demonstrate how it is related to cell wall functions.
Contribution of the cell wall to phenotypic robustness
We demonstrated that yeast cells were highly morphologically variable after perturbation of the cell wall components. We found significantly greater variations between the cell populations after treatment with cell wall–affecting drugs. Levy and Siegal (2008) identified yeast deletion mutants with highly variable phenotypes. In their study, the anp1 mutant exhibited the second-highest variability in overall morphology and mnn10 the 14th highest among all nonessential deletion mutants. Mutants (och1, hoc1, mnn2, cax4, and ccw12) defective in the mannoproteins, gas1, and kre6 were also identified to be highly variable. Although the molecular mechanism is unclear, the cell wall structure likely provides robustness to the cell by protecting the intracellular functional network from environmental conditions.
Multivariate analysis to explore complex phenotypes
We used several multivariate approaches to investigate the complex phenotypes induced by cell wall–affecting drugs. To identify the morphological features after obtaining high-dimensional morphology data, we performed two-step PCA (Ohnuki et al., 2012). This approach allowed us to demonstrate that all cell wall components had roles in the preservation of the neck structure and phenotypic robustness. To predict the gene deletion mutant whose morphological profile would be similar to that of the drug-treated cells, we used a high-content, image-based profiling method (Ohnuki et al., 2010), which validated our analysis and indicated a similar function of chitin synthesis for chitin transglycosylase in morphogenesis. A combination of genetic and statistical analyses facilitated examination of the complex effects of cell wall–affecting drugs. Using these approaches, we determined the mode of action of EB using temperature-sensitive fks1 mutants, which showed specific loss of the individual functions of Fks1.
Cell wall biosynthesis is a tightly coordinated dynamic biological process. Our results suggest mechanistic roles of the cell wall components in cell morphogenesis and increase our understanding of novel functions of the cell wall and the pharmacology of antifungal drugs. A similar approach could be used to investigate the functions of other cellular components in cell morphology.
MATERIALS AND METHODS
Strains and plasmids
The yeast strains and plasmids used in this study are listed in Supplemental Tables S7 and S8 and are deposited with the National BioResource Project Japan, Yeast Genetic Resource Center (Osaka, Japan). The fks1 mutants were constructed as described previously (Okada et al., 2010). Y02458 (as BY4741 his3::KanMX) was used as the wild-type strain. Standard procedures were used for all DNA manipulations and Escherichia coli transformations (Sambrook et al., 1989).
Media and reagents
Yeast-rich medium (YPD) contained 1% Bacto yeast extract (BD Biosciences, San Jose, CA), 2% Bacto peptone (BD Biosciences), and 2% glucose. Yeast synthetic medium (SD or SGS) contained 0.66% Bacto yeast nitrogen base lacking amino acids (BD Biosciences), 2% glucose (SD) or 2% galactose, plus 0.2% sucrose (SGS), and was supplemented with the appropriate nutrients. Luria–Bertani medium for E. coli contained 1% Bacto tryptone (BD Biosciences), 0.5% Bacto yeast extract, and 1% NaCl. Stock solutions of 2 mg/ml EB (a gift from O. Kondo, Chugai Pharmaceutical, Tokyo, Japan) or 10 mg/ml TM (Sigma-Aldrich, St. Louis, MO) were prepared in dimethyl sulfoxide (DMSO; Wako Pure Chemical Industries, Osaka, Japan). Stocks of 10 mM NZ (Sigma-Aldrich) and 10 mg/ml micafungin (Astellas Pharma, Tokyo, Japan) were prepared in distilled water.
Quantification of the effects of drug treatment on cell morphology
Wild-type yeast strains were cultured at 25°C in YPD until the early log phase (∼16 h) with 0, 1, 2, 3, or 4 μg/ml EB; 0, 20, 40, 60, 80, or 100 ng/ml TM; or 0, 10, 15, 20, 25, or 30 ng/ml micafungin. Strains treated with NZ (0, 80, 160, 240, 320, or 400 μM) were cultured in SD. The concentrations of cell wall–affecting drugs were determined based on the growth inhibition rates (∼10%, excluding NZ, which did not significantly inhibit the growth of S. cerevisiae). Cell fixation, staining, and observation were performed as described previously (Ohnuki et al., 2012). Images were analyzed using the image processing software CalMorph (version 1.2), which extracted a total of 501 morphological quantitative values from at least 200 individual cells in each experiment. Images were processed using Photoshop CS2 (Adobe Systems, San Jose, CA) for illustrative purposes.
Statistical analysis
Morphological profiling of the drugs using the Jonckheere–Terpstra test was performed to assess the morphological similarity between the cells treated with the cell wall–affecting drugs and nonessential deletion mutants (Jonckheere, 1954; Ohnuki et al., 2010). Other statistical analyses, including PCA, were performed using R, version 2.14.0 (www.r-project.org/).
Two-step PCA
Before applying PCA to morphological parameters, we transformed five replicated parameters to rank-sum values to unify the distribution of each parameter, as described previously (Ohnuki et al., 2012). PCA was performed using the prcomp of the R function without scaling.
To identify independent morphological features induced by the drugs, we performed a two-step PCA, as described previously (Ohnuki et al., 2012). In the first PCA, morphological data on cells treated with the drugs were subjected to PCA (Supplemental Figure S1). The dose-dependent parameters were correlated with the first principal component (PC1) significantly (Supplemental Table S1, a–c) and had high absolute loading values to PC1. Of the 501 parameters, significant PC loadings in PC1 were observed for 265, 168, and eight parameters in the EB, TM, and NZ data sets, respectively (false discovery rate = 0.1 by t test). In the second PCA, data obtained from 122 replicated experiments of wild-type cells (null-distributed data) in the correlated parameters were subjected to PCA after Box–Cox power transformation. The parameter-correlating PCs in the second PCA, explaining 60% of the variance, had significant loadings (>0.5) at p < 0.0001 based on the t test after Bonferroni correction (Supplemental Figure S2, A and B). Several independent PCs, named in alphabetical order (e.g., PC1, PC2, and PC3 were named PC1a, PC1b, and PC1c, respectively), were extracted for each drug (Supplemental Table S2, a–c). One or two dose-dependent parameters of the various morphological criteria were selected as representative parameters after taking the absolute loading values to each PC (>0.5) and the meaning of the parameters (colored in yellow in Supplemental Table S2, a–c) into account.
Estimation of the variation in cell morphology
Parameters showing notable dose-dependent drug effects were detected using the Jonckheere–Terpstra test (p < 0.05). Of the detected parameters (354 parameters in EB, 241 in TM, and 87 in NZ), the “mean” parameters (197, 127, and 37 parameters, respectively) representing the population average were analyzed further. The data on cells treated with a high concentration of drugs (4 μg/ml EB, 100 ng/ml TM, and 400 μM NZ; n = 5) were centered by subtracting the mean and scaled by dividing by the standard deviation of the control cells without drug treatment (n = 5) and then used to assess the effects of the drugs on cell morphology.
Extraction of similar/dissimilar effects of drug treatments
After PCA on the morphological data for the drug-treated cells, the loadings of 501 traits for PC1 were plotted on the 2D space of two or three arbitrary drugs. Because PC1 scores of all three drugs increased in a dose-dependent manner, we assumed that the traits correlating with PC1 in the same direction (a positive or negative correlation) between the drugs reflected similar effects. Conversely, traits in different directions reflected dissimilar effects. For pairwise comparisons among the three drugs (EB–TM, EB–NZ, and TM–NZ), we defined similar parameters (369, 289, and 293 traits) and dissimilar parameters (132, 212, and 208 traits, respectively) from 501 traits based on the loadings. To identify the primary effects of the drugs on each set of similar/dissimilar parameters, we applied PCA independently (Supplemental Figure S3) and visualized the distribution of PC1 scores.
Spa2 localization
To assess Spa2 localization, we constructed a plasmid expressing a Spa2–GFP fusion protein with GFP conjugated immediately before the stop codon. A DNA fragment of the SPA2 open reading frame, with its 0.5-kb-upstream region, was amplified from the S. cerevisiae genome by PCR using primers that generated a SmaI restriction site at each end. The PCR product was cloned into a low-copy plasmid harboring the URA3 marker and GFP-coding sequence with an FKS1 terminator to make pYO2642. pYO2642 was then used to transform Y01509, YOC1001, YOC1087, and YOC1089 on SD-without-uracil (SD–U) plates to obtain YOC5002 and YOC5041–5043, respectively. YOC5002 was cultured at 25ºC in liquid SD–U with EB (1 μg/ml) or DMSO (control solvent) until the early log phase and harvested. YOC5041–5043 were cultured with SD–U at 25ºC until early log phase, followed by additional incubation at 37ºC for 2 h. After a brief sonication of cells, Spa2–GFP localization was observed using fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank Osamu Kondo for providing EB, Satoru Nogami for initiating analysis of the morphology data, and members of the Laboratory of Signal Transduction for helpful discussions. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (21310127 and 24370002 to Y.O.), a grant from the Comisión Interministerial de Ciencia y Tecnología, Spain (BFU2010-18632 to C.R.), and a Public Health Service grant from the National Institute of Allergy and Infectious Diseases (AI-47837 to J.B.K.). H.O. and S.O. were Research Fellows of the Japan Society for the Promotion of Science.
Abbreviations used:
- 2D
two-dimensional
- DMSO
dimethyl sulfoxide
- EB
echinocandin B
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GlcNAc
N-acetylglucosamine
- GS
1,3-β-glucan synthase
- NZ
nikkomycin Z
- PCA
principal component analysis
- SCMD
Saccharomyces cerevisiae Morphological Database
- SD
synthetic growth medium
- SGS
synthetic growth medium containing 2% galactose and 0.2% sucrose as the carbon source
- TM
tunicamycin
- YPD
yeast-rich medium
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0396) on November 20, 2013.
The authors have no conflict of interest to declare.
REFERENCES
- Arnold E, Tanner W. An obligatory role of protein glycosylation in the life cycle of yeast cells. FEBS Lett. 1982;148:49–53. doi: 10.1016/0014-5793(82)81240-4. [DOI] [PubMed] [Google Scholar]
- Ballou CE. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- Barnes G, Hansen WJ, Holcomb CL, Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol. 1984;4:2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco N, Reidy M, Arroyo J, Cabib E. Crosslinks in the cell wall of budding yeast control morphogenesis at the mother-bud neck. J Cell Sci. 2012;125:5781–5789. doi: 10.1242/jcs.110460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. Differential inhibition of chitin synthetases 1 and 2 from Saccharomyces cerevisiae by polyoxin D and nikkomycins. Antimicrob Agents Chemother. 1991;35:170–173. doi: 10.1128/aac.35.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both beta(1-6)- and beta(1-3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot Cell. 2009;8:1626–1636. doi: 10.1128/EC.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Arroyo J. How carbohydrates sculpt cells: chemical control of morphogenesis in the yeast cell wall. Nat Rev Microbiol. 2013;11:648–655. doi: 10.1038/nrmicro3090. [DOI] [PubMed] [Google Scholar]
- Cabib E, Blanco N, Grau C, Rodríguez-Peña JM, Arroyo J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol Microbiol. 2007;63:921–935. doi: 10.1111/j.1365-2958.2006.05565.x. [DOI] [PubMed] [Google Scholar]
- Cabib E, Durán A. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J Biol Chem. 2005;280:9170–9179. doi: 10.1074/jbc.M414005200. [DOI] [PubMed] [Google Scholar]
- de Groot PW, et al. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp Funct Genomics. 2001;2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM. Fungal beta(1,3)-D-glucan synthesis. Med Mycol. 2001;39(Suppl 1):55–66. doi: 10.1080/mmy.39.1.55.66. [DOI] [PubMed] [Google Scholar]
- Drgonová J, Drgon T, Roh DH, Cabib E. The GTP-binding protein Rho1p is required for cell cycle progression and polarization of the yeast cell. J Cell Biol. 1999;146:373–387. doi: 10.1083/jcb.146.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonová J, Drgon T, Tanaka K, Kollár R, Chen GC, Ford RA, Chan CS, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- Gómez-Esquer F, Rodríguez-Peña JM, Díaz G, Rodriguez E, Briza P, Nombela C, Arroyo J. CRR1, a gene encoding a putative transglycosidase, is required for proper spore wall assembly in Saccharomyces cerevisiae. Microbiology. 2004;150:3269–3280. doi: 10.1099/mic.0.27314-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Yoda K. Novel membrane protein complexes for protein glycosylation in the yeast Golgi apparatus. Biochem Biophys Res Commun. 1997;241:682–686. doi: 10.1006/bbrc.1997.7888. [DOI] [PubMed] [Google Scholar]
- Healey KR, Katiyar SK, Raj S, Edlind TD. CRS-MIS in Candida glabrata: sphingolipids modulate echinocandin-Fks interaction. Mol Microbiol. 2012;86:303–313. doi: 10.1111/j.1365-2958.2012.08194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SB, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- Iwaki A, Ohnuki S, Suga Y, Izawa S, Ohya Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in Saccharomyces cerevisiae: application and validation of high-content, image-based profiling. PLoS One. 2013;8:e61748. doi: 10.1371/journal.pone.0061748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jigami Y. Yeast glycobiology and its application. Biosci Biotechnol Biochem. 2008;72:637–648. doi: 10.1271/bbb.70725. [DOI] [PubMed] [Google Scholar]
- Johnson ME, Edlind TD. Topological and mutational analysis of Saccharomyces cerevisiae Fks1. Eukaryot Cell. 2012;11:952–960. doi: 10.1128/EC.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Katiyar SK, Edlind TD. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob Agents Chemother. 2011;55:3774–3781. doi: 10.1128/AAC.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere AR. A test of significance for the relation between M rankings and K ranked categories. Br J Stat Psychol. 1954;7:93–100. [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Klis FM, Boorsma A, De Groot PWJ. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Kollár R, Reinhold BB, Petráková E, Yeh HJ, Ashwell G, Drgonová J, Kapteyn JC, Klis FM, Cabib E. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→)3-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Lagunas R, DeJuan C, Benito B. Inhibition of biosynthesis of Saccharomyces cerevisiae sugar transport system by tunicamycin. J Bacteriol. 1986;168:1484–1486. doi: 10.1128/jb.168.3.1484-1486.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J-P. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J Bacteriol. 1999;181:3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Uchida K, Yamaguchi H. Morphological changes of Candida albicans induced by micafungin (FK463), a water-soluble echinocandin-like lipopeptide. J Electron Microsc (Tokyo) 2002;51:247–255. doi: 10.1093/jmicro/51.4.247. [DOI] [PubMed] [Google Scholar]
- Ohnuki S, Kobayashi T, Ogawa H, Kozone I, Ueda J-Y, Takagi M, Shin-Ya K, Hirata D, Nogami S, Ohya Y. Analysis of the biological activity of a novel 24-membered macrolide JBIR-19 in Saccharomyces cerevisiae by the morphological imaging program CalMorph. FEMS Yeast Res. 2012;12:293–304. doi: 10.1111/j.1567-1364.2011.00770.x. [DOI] [PubMed] [Google Scholar]
- Ohnuki S, Nogami S, Kanai H, Hirata D, Nakatani Y, Morishita S, Ohya Y. Diversity of Ca2+-induced morphology revealed by morphological phenotyping of Ca2+-sensitive mutants of Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:817–830. doi: 10.1128/EC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki S, Oka S, Nogami S, Ohya Y. High-content, image-based screening for drug targets in yeast. PLoS One. 2010;5:e10177. doi: 10.1371/journal.pone.0010177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M, Saka A, Sano F, Ohya Y, Morishita S. Development of image processing program for yeast cell morphology. J Bioinform Comput Biol. 2004;1:695–709. doi: 10.1142/s0219720004000363. [DOI] [PubMed] [Google Scholar]
- Ohya Y, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. 2005;102:19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Abe M, Asakawa-Minemura M, Hirata A, Qadota H, Morishita K, Ohnuki S, Nogami S, Ohya Y. Multiple functional domains of the yeast l,3-β-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–1024. doi: 10.1534/genetics.109.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics. 2012;192:775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orłowski J, Machula K, Janik A, Zdebska E, Palamarczyk G. Dissecting the role of dolichol in cell wall assembly in the yeast mutants impaired in early glycosylation reactions. Yeast. 2007;24:239–252. doi: 10.1002/yea.1479. [DOI] [PubMed] [Google Scholar]
- Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- Ragni E, Piberger H, Neupert C, García-Cantalejo J, Popolo L, Arroyo J, Aebi M, Strahl S. The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC Genomics. 2011;12:107. doi: 10.1186/1471-2164-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AF, Kapteyn JC, Montijn RC, Caro LH, Douwes JE, Baginsky W, Mazur P, van den Ende H, Klis FM. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, Cid VJ, Arroyo J, Nombela C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol Cell Biol. 2000;20:3245–3255. doi: 10.1128/mcb.20.9.3245-3255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, Rodriguez C, Alvarez A, Nombela C, Arroyo J. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J Cell Sci. 2002;115:2549–2558. doi: 10.1242/jcs.115.12.2549. [DOI] [PubMed] [Google Scholar]
- Roemer T, Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci USA. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TL, Ohtani M, Sawai H, Sano F, Saka A, Watanabe D, Yukawa M, Ohya Y, Morishita S. SCMD: Saccharomyces cerevisiae Morphological Database. Nucleic Acids Res. 2004;32:D319–22. doi: 10.1093/nar/gkh113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santos B, Duran A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawistowska-Schröder ET, Kerridge D, Perry H. Echinocandin inhibition of 1,3-β-D-glucan synthase from Candida albicans. FEBS Lett. 1984;173:134–138. doi: 10.1016/0014-5793(84)81032-7. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Strenk ME, Boyer MP, Fritsch BJ. Importance of cell wall mannoproteins for septum formation in Saccharomyces cerevisiae. Yeast. 2005;22:715–723. doi: 10.1002/yea.1242. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Varma A, Drgon T, Bowers B, Cabib E. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol Biol Cell. 2003;14:2128–2141. doi: 10.1091/mbc.E02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel MA, Rieger M, te Heesen S, Ram AF, van den Ende H, Aebi M, Klis FM. The Saccharomyces cerevisiae CWH8 gene is required for full levels of dolichol-linked oligosaccharides in the endoplasmic reticulum and for efficient N-glycosylation. Glycobiology. 1999;9:243–253. doi: 10.1093/glycob/9.3.243. [DOI] [PubMed] [Google Scholar]
- van Drogen F, Peter M. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr Biol. 2002;12:1698–1703. doi: 10.1016/s0960-9822(02)01186-7. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ohnuki S, Yashiroda Y, Ohya Y. Profilin is required for Ca2+ homeostasis and Ca2+-modulated bud formation in yeast. Mol Genet Genomics. 2013;288:317–328. doi: 10.1007/s00438-013-0752-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.