Acetylation of α-tubulin is uniquely located on the microtubule lumen and away from most MAPs. It does not affect microtubule structure and must affect only proteins that bind directly to the lumen. αTAT1 can interact with the tubulin C-termini, which could facilitate access to its luminal site and make it responsive to other modifications.
Abstract
Tubulin undergoes posttranslational modifications proposed to specify microtubule subpopulations for particular functions. Most of these modifications occur on the C-termini of tubulin and may directly affect the binding of microtubule-associated proteins (MAPs) or motors. Acetylation of Lys-40 on α-tubulin is unique in that it is located on the luminal surface of microtubules, away from the interaction sites of most MAPs and motors. We investigate whether acetylation alters the architecture of microtubules or the conformation of tubulin, using cryo–electron microscopy (cryo-EM). No significant changes are observed based on protofilament distributions or microtubule helical lattice parameters. Furthermore, no clear differences in tubulin structure are detected between cryo-EM reconstructions of maximally deacetylated or acetylated microtubules. Our results indicate that the effect of acetylation must be highly localized and affect interaction with proteins that bind directly to the lumen of the microtubule. We also investigate the interaction of the tubulin acetyltransferase, αTAT1, with microtubules and find that αTAT1 is able to interact with the outside of the microtubule, at least partly through the tubulin C-termini. Binding to the outside surface of the microtubule could facilitate access of αTAT1 to its luminal site of action if microtubules undergo lateral opening between protofilaments.
INTRODUCTION
Microtubules perform a variety of vital functions within the cell as part of the eukaryotic cytoskeleton. These diverse functions often require microtubule specialization, which arises from the accumulation of posttranslational modifications (PTMs) on the tubulin subunits, which in turn likely affect binding of microtubule-associated proteins (MAPs) and/or dynamic instability parameters. Tubulin undergoes several types of PTMs, including detyrosination (removal of the C-terminal tyrosine), Δ2-tubulin generation (a nonreversible modification that results from the removal of the terminal tyrosine and glutamate residues), polyglutamylation, polyglycylation, and acetylation (Janke and Bulinski, 2011). Most of the tubulin PTMs occur within its unstructured C-terminal tails (or E-hooks), which extend outward on the surface of microtubules. The C-termini of tubulin are involved in interaction with most MAPs, either by themselves or in addition to the globular domain of tubulin, and specific PTMs influence tubulin–MAP interactions. Reversible acetylation of α-tubulin at Lys-40 (αK40) is unique in that it occurs on the luminal surface of the microtubule (Nogales et al., 1999). Acetylation of α-tubulin was first reported by L'Hernault and Rosenbaum (1983, 1985), who observed that the α-tubulin in flagella was posttranslationally modified and mapped it to the ε-amino group of a single lysine residue. A monoclonal antibody that recognizes acetylation of α-tubulin was initially generated by Piperno and colleagues (Piperno and Fuller, 1985) and later mapped to αK40 (LeDizet and Piperno, 1987; Piperno et al., 1987). This reagent is the only specific antibody that binds to acetylated tubulin. Other acetylation sites have been identified by enriching cell isolates using pan anti–acetylated lysine antibodies and mass spectrometry (Choudhary et al., 2009). However, acetylation of those sites is unlikely to be conserved (L'Hernault and Rosenbaum, 1985; Akella et al., 2010). Many of these acetylation sites have not been confirmed in vivo, and their physiological relevance is in question. An acetylation site on β-tubulin, which occurs at the tubulin dimer interface, has also been identified and shown to affect the polymerization kinetics of tubulin (Chu et al., 2011).
Determining the roles of the different modifications in vivo has been challenging because PTMs rarely occur in isolation and the level of each modification is distinctively regulated by the cell (Quinones et al., 2011). In fact, no clear role for αK40 acetylation has emerged despite the long time since its initial discovery. Acetylated microtubules in the cytoplasm are generally more stable than unmodified microtubules, in that they can withstand mild treatment with microtubule-depolymerizing drugs such as nocodazole (De Brabander et al., 1976) and colchicine (LeDizet and Piperno, 1986; Piperno et al., 1987). However, this modification does not directly influence tubulin polymerization or depolymerization kinetics in vitro (Maruta et al., 1986).
Studies that tracked the appearance of acetylated microtubules after release from nocodazole in interphase cells (Bulinski et al., 1988) showed that acetylation was restored before detyrosination, another major PTM that regulates interaction of microtubules with motors (Dunn et al., 2008; Konishi and Setou, 2009; Peris et al., 2009) and plus end–binding proteins (+TIPS) that track the polymerizing end of a microtubule, usually through interactions with end-binding protein 1 (EB1; Perez et al., 1999; Mimori-Kiyosue et al., 2000; Kumar and Wittmann, 2012). This result was confirmed for microtubules that specifically associate with the Golgi complex (Thyberg and Moskalewski, 1993). In the recovery of cilia of sea urchins after hypertonic salt treatment, microtubules were highly acetylated, both within the axoneme and in the matrix/membrane-associated tubulin fraction, before they reached mature levels of detyrosination (Stephens, 1992). However, acetylation is a late-stage event in myogenesis (Di Fulvio et al., 2011). These studies suggest that acetylation is adapted to specific processes and that cells carefully control the acetylation levels, depending on the microtubule structures being built. No direct link between αK40 acetylation and the binding of other proteins that are also predominantly located in stable, highly acetylated microtubule structures, such as cilia and axons, has been identified. Acetylated microtubules have also been identified in less stable structures, such as the microtubule network that assembles at the cleavage furrow to deposit new membrane during cytokinesis (Danilchik et al., 1998). Many other examples of diverse processes that correlate with acetylated microtubules were reported (Serrador et al., 2004; Reed et al., 2006; Cai et al., 2009; Creppe et al., 2009; Friedman et al., 2010; Sudo and Baas, 2010), but the direct role of this tubulin modification remains unclear.
The enzymes responsible for the removal of the acetyl group from αK40 were identified first and found to be homologues of histone deacetylases. Histone deacetylase 6 (HDAC6) and sirtuin 2 (SIRT2) both have deacetylase activity for αK40 (Hubbert et al., 2002; North et al., 2003) and localize to the cytoplasm rather than the nucleus. Both proteins deacetylate multiple substrates and operate on the free tubulin dimer. More recently it was found that acetylation of αK40 is performed by αTAT1 (Akella et al., 2010; Shida et al., 2010), which also shows homology to histone acetyltransferases (Friedmann et al., 2012; Kormendi et al., 2012; Taschner et al., 2012). Sequence alignments show that αTAT1 belongs to the GCN5 superfamily of acetyltransferases but may have a unique active site (Taschner et al., 2012). Despite the lack of a clear role for acetylated microtubules, αTAT1 is well conserved across many phyla and correlates perfectly with the presence of cilia, hinting at an essential role in this structure (Shida et al., 2010). Depletion of this enzyme impairs primary cilium assembly and reduces touch response in Caenorhabditis elegans (Shida et al., 2010). Consistent with these results, in Danio rerio, reduced touch response was also reported in knockdown animals along with abnormal development (Akella et al., 2010). Change in protofilament number of microtubules was also reported as a result of αTAT1 knockout (Cueva et al., 2012; Topalidou et al., 2012).
In the present study we sought to determine whether the acetylation state of tubulin directly influences the structure of tubulin and microtubules (MTs) in vitro by making use of the fact that preparations of fully acetylated or deacetylated tubulin are now possible with the identification of the tubulin acetyltransferase. Our studies show that acetylation per se has no significant effect on MT architecture or tubulin conformation. Thus it is likely that acetylation affects MT function indirectly, through the recognition of the modified αK40 by proteins that gain access to the luminal surface of MTs. Of interest, we found that the tubulin acetyltransferase, αTAT1, is itself not affected by acetylation state in its binding to MTs. Furthermore, we observe that αTAT1 can bind to the outside surface of the MT, which may serve as an initial step before gaining access to its luminal site of action.
RESULTS AND DISCUSSION
Cryo–electron microscope visualization shows no significant effect of tubulin acetylation on microtubule structural parameters
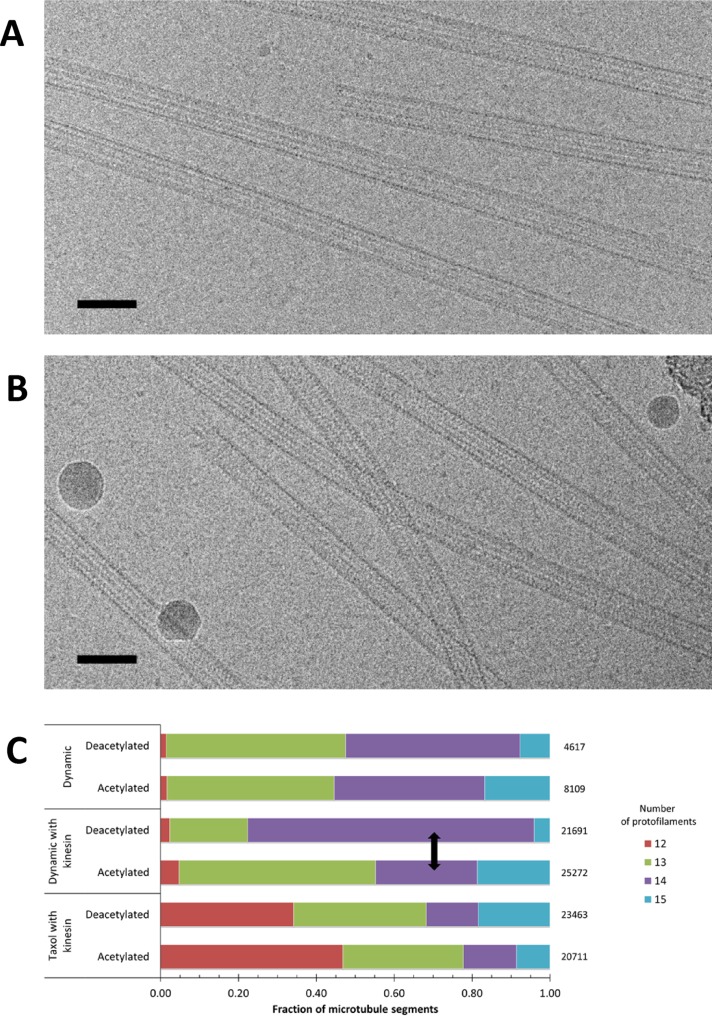
The recent identification of the major tubulin acetyltransferase, αTAT1 (Akella et al., 2010; Shida et al., 2010), now allows for in vitro preparation of both maximally acetylated and deacetylated microtubules and their subsequent characterization. Given the likely difficulty faced by proteins trying to gain access to the lumen of the microtubule and the generality in the binding of MAPs and microtubule motors to the external surface of microtubules, our starting hypothesis was that changes conferred by acetylation could either alter tubulin–tubulin contacts and microtubule lattice parameters or otherwise propagate to the microtubule external surface. Such changes would provide the means for proteins that bind to the outside of microtubules to detect this modification. To determine what kind of effect tubulin acetylation has on microtubule structure, mammalian brain tubulin was modified in vitro to generate either maximal or minimal levels of acetylation (D. Portran, T. Shida, and M. Nachury, unpublished data; Supplemental Figure S1). The modified tubulin was then assembled into either dynamic or Taxol-stabilized microtubules for analysis by cryo-electron microscopy (cryo-EM). Similar amounts of tubulin polymerized for acetylated and deacetylated tubulin samples, based on the apparent abundance of microtubules in micrographs, and both types of samples showed a similar increase in the amount of assembled microtubules upon addition of Taxol (unpublished data). In addition, no obvious differences were seen in the gross morphology or curvature of the microtubules (Figure 1, A and B).
FIGURE 1:
Tubulin acetylation does not affect the amount of polymerization or gross morphology of microtubules. Representative cryo-EM images of (A) acetylated and (B) deacetylated dynamic microtubules. Scale bar, 50 nm. (C) Protofilament number distribution of microtubules polymerized after acetylation or deacetylation into dynamic or Taxol-stabilized microtubules. Arrow shows largest observed change.
We visualized acetylated and deacetylated microtubules with the ultimate goal of generating high-resolution structures of the two types of polymers. As a first indication of whether acetylation alters the packing of tubulin dimers within the microtubule, we examined the distribution of protofilament numbers in acetylated and deacetylated microtubules. The percentage of microtubule segments with different protofilament numbers is shown in Figure 1C. Dynamic microtubules show a close to 50:50% mixture of microtubules with predominantly 13 and 14 protofilaments, irrespective of the acetylation state of tubulin. The binding of kinesin affects protofilament number in acetylated microtubules very marginally by slightly increasing the proportion of 13-protofilament microtubules. Of interest, the binding of kinesin to deacetylated tubulin significantly reduces the number of 13-protofilament microtubules while increasing the number of 14-protofilament microtubules. Addition of both Taxol and kinesin had the most severe effect in widening the range of protofilament number, dramatically increasing the fraction of 12-protofilament microtubules, a previously characterized effect of Taxol (Andreu et al., 1992). Of importance, this effect was very similar for acetylated and deacetylated microtubules. These results indicate that Taxol and kinesin have a much more significant effect on protofilament number than the acetylation state of tubulin. The abundance of 14 protofilaments in dynamic, deacetylated microtubules in the presence of kinesin is a curious result for which we do not have a clear interpretation. However, it seems to indicate that kinesin has an apparent destabilizing effect on 13-protofilament (but not 14-protofilament) microtubules that are devoid of acetylation, perhaps by having lower affinity for the 13-protofilament lattice of the deacetylated tubulin. Considering that the 13-protofilament structure is likely the predominant microtubule architecture in the cell, our in vitro assay may reflect a higher affinity of kinesin for the 13-protofilament microtubule made of acetylated versus deacetylated tubulin, which would have biological implications for the effect of this modification on intracellular motility.
Previous experiments showed that whereas the sensory dendrites of wild-type C. elegans touch receptor neurons contain microtubules of almost exclusively 15 protofilaments, mutants lacking the acetyltransferase have fewer microtubules, and protofilament number ranges between 11 and 16 (Cueva et al., 2012; Topalidou et al., 2012). Given our in vitro results, it is likely that the change in protofilament number range with acetylation state is due not to the direct effect of this modification on microtubule structure, but to its effect on the binding of specific MAPs, sensitive to the tubulin acetylation state. Given the proximity of αK40 to the lateral contacts between tubulin dimers, binding of a MAP in this region could have a constraining effect on the angle of the interaction, thus fixing the number of protofilaments.
To further investigate whether acetylation alters the packing of tubulin dimers within the microtubule, we next examined the helical lattice parameters for each of the microtubule conditions. Such parameters are obtained during the process of image analysis. Our three-dimensional (3D) cryo-EM reconstruction of microtubules follows an iterative refinement process (see Materials and Methods), by which estimates for the longitudinal (rise) and lateral (twist) spacing of tubulin subunits are optimized by improving the alignment of individual microtubule segments with respect to an evolving model (Egelman, 2007). Estimates for the rise and twist of a 14-protofilament microtubule at each iteration of the reconstruction are shown in Supplemental Figure S2. Final estimates for 13- and 14-protofilament microtubules are given in Table 1. We observed no influence of acetylation on the longitudinal or lateral spacing of the tubulin dimers. When we examined acetylated and deacetylated microtubules bound to kinesin, which serves as a marker of the tubulin dimer repeat (allowing one to discriminate α- and β-tubulin during the reconstruction; see later discussion), we saw a small but detectable increase in the longitudinal spacing of tubulin dimers of ∼1–2%, irrespective of whether tubulin was acetylated or deacetylated. Our data show remarkable structural similarity between the acetylated and deacetylated microtubule lattices, demonstrating that helical packing of tubulin dimers within the microtubules is not altered by the acetylation state of αK40. Previous computational models suggested the formation of a salt bridge between neighboring α-tubulin subunits across protofilaments upon αK40 acetylation (Cueva et al., 2012). Our data show that if this salt bridge forms, it must not alter the packing of tubulin dimers within the microtubule.
TABLE 1:
Tubulin acetylation does not change microtubule lattice parameters.
| Untreated | Deacetylated | Acetylated | Untreated + kinesin | Deacetylated + kinesin | Acetylated + kinesin | |
|---|---|---|---|---|---|---|
| 13-protofilament microtubule | ||||||
| Rise | 9.44 | 9.36 | 9.36 | 9.23 | 9.49 | 9.50 |
| Twist | −27.68 | −27.69 | −27.69 | −27.70 | −27.70 | −27.69 |
| 14-protofilament microtubule | ||||||
| Rise | 8.70 | 8.72 | 8.88 | 8.82 | 8.80 | 8.79 |
| Twist | −25.75 | −25.76 | −25.76 | −25.77 | −25.76 | −25.77 |
Final estimations for rise and twist of tubulin dimers in 13- and 14-protofilament microtubules from reconstructions of dynamic microtubules assembled from untreated, acetylated, or deacetylated tubulin, in the presence or absence of kinesin.
The subnanometer structures of tubulin in acetylated and deacetylated microtubules are indistinguishable
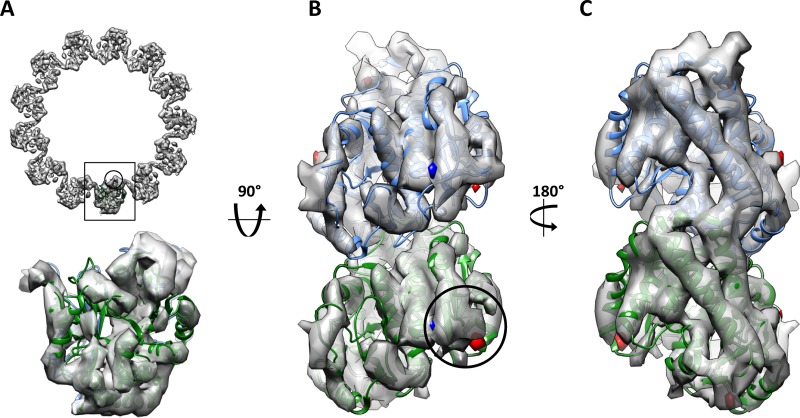
To investigate whether acetylation locally altered the structure of tubulin without modifying the lattice parameters, perhaps having an allosteric effect that would be propagated to the outside surface of the microtubule, we generated subnanometer-resolution 3D cryo-EM structures of acetylated and deacetylated microtubules (Figures 2 and 3). We carried out parallel reconstructions for coexisting 13- and 14-protofilament microtubules in each sample. The conclusions were the same for both types of microtubules, so we describe in detail the results for just one of them. We found that the reconstructions of 13-protofilament microtubules made of maximally acetylated tubulin (8.5 Å resolution by 0.5 Fourier shell correlation [FSC]) and completely deacetylated tubulin (8.6 Å resolution; Figure 2) look very similar to each other, as well as to previous reconstructions of mammalian brain tubulin (Li et al., 2002; Sui and Downing, 2010), which was reported to be acetylated by ∼5% (Eddé et al., 1990) but we find to correspond to 25–30% of the total (Portran et al., unpublished data). We isolated a single tubulin dimer from the acetylated and deacetylated microtubule structures, aligned them to each other, and calculated a difference map using Fourier space subtraction and amplitude correction (see Materials and Methods). Figure 2, B and C, displays the positive and negative difference densities in red and blue, respectively, with isosurfaces contoured at three standard deviations. No statistically significant changes in density are observed at the present resolution.
FIGURE 2:
Acetylation state does not alter the structure of dynamic microtubules. (A) End-on view of the deacetylated, dynamic microtubule reconstruction with the docked electron crystallographic structure of tubulin (1JFF). (B) Inside view (from the microtubule lumen) of the difference maps between acetylated and deacetylated reconstructions contoured at 3σ, with extra density in acetylated microtubules shown in blue and extra density in deacetylated microtubules shown in red. The differences are superimposed on the cryo-EM reconstruction of deacetylated microtubules, shown as gray isosurface. The location of the loop containing the modified residue is indicated by the circle. (C) Outside view, colors as in B. α-Tubulin is shown in green and β-tubulin in blue.
FIGURE 3:
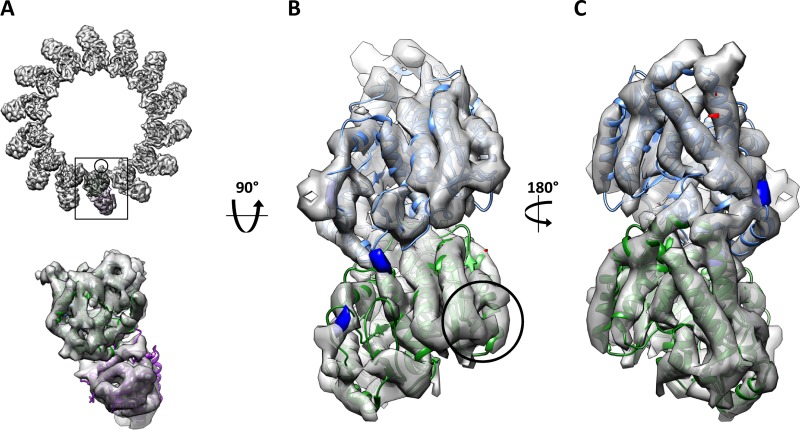
Preventing averaging of α- and β-tubulin by kinesin decoration does not reveal any difference between acetylation states. (A) End-on view of the deacetylated, dynamic microtubule reconstruction with kinesin. (B) Inside and (C) outside views of an isolated tubulin dimer, with differences superimposed as in Figure 2. No significant differences were observed within the tubulin dimer. The location of the loop containing the modified residue is indicated by the circle. α-Tubulin is shown in green, β-tubulin in blue, and kinesin (1BG2) in purple.
Because of the similarity between α- and β-tubulin, microtubules in cryo-EM images cannot be aligned correctly with respect to the two subunits, resulting in their averaging. Thus, if subtle differences upon acetylation were to affect the two subunits differently (e.g., causing changes only in α-tubulin), these differences would be “blurred out” during this unintentional averaging, making their detection harder. To prevent averaging of α- and β-tubulin, we decorated microtubules with a monomeric mutant kinesin (K349-E236A) that has high affinity for the microtubule lattice (Rice et al., 1999). The large kinesin density on the outside of the microtubule provides a strong signal for the tubulin dimer, ensuring that α and β subunits are not averaged together. More segments were available for the reconstruction of the 14-protofilament deacetylated condition, leading to higher resolution than for the 13-protofilament reconstruction. The reconstructions for acetylated and deacetylated 14-protofilament microtubules went to 8.8 and 9.2 Å, respectively. The 14-protofilament acetylated microtubule is shown in Figure 3A. Difference maps were again calculated as described, and no significant differences were seen in the tubulin structure (Figure 3, B and C). Taxol-stabilized microtubules were also prepared, imaged, and reconstructed. The reconstructions obtained were of marginally lower resolution than those from dynamic microtubules (Supplemental Table S1) but also showed no differences between acetylated and deacetylated states (unpublished data).
The work presented here indicates that acetylation of α-tubulin at Lys-40 has no appreciable structural effect on overall microtubule assembly and architecture or on tubulin structure as visualized at 8 to 9 Å resolution. Although we cannot discard the possibility that acetylation has a small effect on the structure/stability of the loop where Lys-40 resides, detectable only in structures closer to atomic resolution, our analyses strongly suggest that the effect of acetylation must be small and local. We also cannot completely exclude the possibility that the modification might alter the local packing of the Lys-40 loop in a manner that would not persist if the microtubule was depolymerized and reformed. On the other hand, the remarkable structural similarity we observe between the acetylated and deacetylated microtubule samples could mean that this modification is invisible to the majority of proteins that bind the outside of the microtubule. We propose that the “biological readout” of this modification must require protein factors that directly recognize the acetyl group of the modified lysine or a small local structural rearrangement by the site. Thus it is likely that acetylation affects microtubule function by the binding of proteins that gain access to the luminal surface of microtubules where acetylation resides. Of interest, the microtubule lumen, where tubulin acetylation occurs, appears not to be empty in the cell, as a number of electron tomographic studies demonstrated the presence of luminal particles in neurons, astrocytes, and stem cells (Burton, 1984; Garvalov et al., 2006; Bouchet-Marquis et al., 2007). The identity of these particles is unknown, but it is intriguing that the greater abundance of luminal particles in neurons correlates with an increased degree of acetylation in this microtubule population. Furthermore, mutants lacking the acetyltransferase have much fewer luminal particles (Topalidou et al., 2012).
αTAT1 binding to microtubules is not influenced by accessibility to the lumen
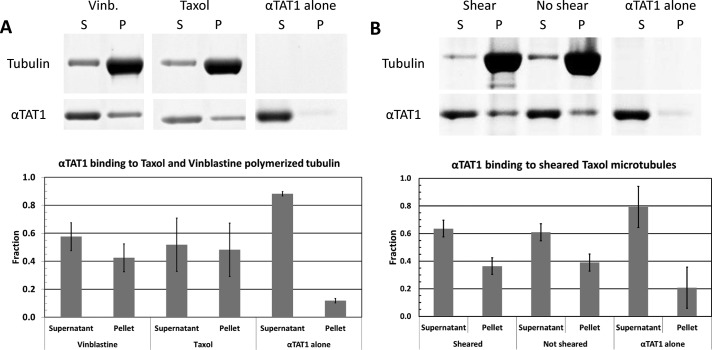
Given that αTAT1 acts on the luminal surface of the microtubule and that its enzymatic activity is higher for assembled microtubules than for free tubulin dimers, it must be able to gain access to the inside of the microtubule to carry out its enzymatic activity. Thus αTAT1 may represent the first protein strongly predicted to bind to the inside of microtubules. However, this raises the question of how it accesses the luminal surface. To test whether αTAT1 binding to microtubules is limited by accessibility to the acetylation site, we compared the results of cosedimentation assays for two alternative tubulin polymer forms: Taxol-stabilized microtubules and vinblastine-induced coils. Vinblastine creates open, spiral polymers of tubulin where the luminal surface of the microtubule is fully exposed (Nogales et al., 1995; Gigant et al., 2005). Surprisingly, no increase in αTAT1 binding was observed for this type of polymer with respect to binding to Taxol-stabilized microtubules (Figure 4A). Because the structure of tubulin is different in the vinblastine-induced polymers (curved vs. the straight conformation within microtubules), a decrease in affinity could have compensated for the better accessibility. Thus we carried out a second test in which Taxol microtubules were sheared by passing them through a small-diameter needle This creates more free ends, which should allow the enzyme to access the microtubule lumen more readily if access is gained through the microtubule ends. However, we did not observe any significant increase of αTAT1 binding to sheared compared with unsheared microtubules (Figure 4B). These results indicate that αTAT1 binding to Taxol microtubules is not limited by accessibility to the microtubule lumen. All of our pelleting assays show that a significant fraction of αTAT1 remains unbound even when mixed with an excess of tubulin, indicating that the binding affinity of αTAT1 for microtubules is modest.
FIGURE 4:
αTAT1 accessibility to its binding sites on tubulin polymers. (A) SDS gel (top) and quantitation (bottom) of pelleting assays showing that αTAT1 binds similarly to tubulin polymers assembled with vinblastine (coils) or Taxol (microtubules). (B) Shearing microtubules to create more microtubule ends does not increase binding of αTAT1 to microtubules. Data from three independent cosedimentation assays. Error bars, SD.
αTAT1 binds the exterior surface and interacts with the C-terminal tails of tubulin
In an attempt to directly observe how αTAT1 binds microtubules, we polymerized dynamic microtubules in the presence of stoichiometric amounts of αTAT1. The structures obtained are shown in Figure 5A. We observed extensive microtubule bundling, mediated by a complex coat of protein around the microtubules. The size and granularity of that “coat” indicate that it likely involves both αTAT1 and tubulin assembled into curved structures, a type of arrangement previously observed for other microtubule-binding proteins (Tan et al., 2006). This result can only be explained if, irrespective of its binding to the lumen, αTAT1 binds also to the outside surface of the microtubule, where it can interact with unassembled tubulin and promote microtubule bundling. Such a “sandwich” of αTAT1 between tubulin structures could be obtained if the enzyme either had more than one tubulin binding site per molecule (e.g., one site could bind the globular part of the tubulin structure, and the other could interact with the acidic C-tails of tubulin) or by oligomerization of the enzyme (e.g., dimers for which each monomer binds identical regions in tubulin). Our gel filtration indicates that, while full-length αTAT1 can dimerize, the truncated construct used in this study does not (Supplemental Figure S5). Further study will be required to determine the molecular origin of the bundling and whether there is any biological relevance for this in vitro observation.
FIGURE 5:
Effect of αTAT1 binding on microtubule assembly and bundling. (A) Cryo-EM image showing that the presence of αTAT1 during the assembly of dynamic microtubules gives rise to large protein structures around the microtubule lattice (black arrow) and promotes bundling (white arrow). (B) Negative-stain EM image of microtubules copolymerized with αTAT1 in the presence of Taxol at a 1:5 αTAT1:tubulin molar ratio, showing incomplete microtubule closure and numerous defects. Scale bar, 50 nm.
We also studied the effect of adding Taxol to the acetylation reactions. Note that in the presence of Taxol there is little unassembled tubulin (as demonstrated in the cosedimentation assays; e.g., Figure 4A). Microtubules formed with αTAT1:tubulin molar ratios from 1:3 to 1:10 had numerous defects and were often opened (Figure 5B). Taxol microtubules open laterally (between and along the length of protofilaments) more often than dynamic microtubules (or perhaps just as often, but whereas dynamic microtubules may depolymerize upon opening in the absence of other stabilizing proteins, Taxol microtubules do not) and contain more discontinuities in the helical packing of tubulin dimers (Arnal and Wade, 1995). This effect is dramatically enhanced in the presence of the acetyltransferase, suggesting that its binding may sterically preclude reclosure. At the high excess ratio of 10:1 αTAT1:tubulin, microtubule assembly is inhibited entirely. Thus an excess of αTAT1 has a deleterious effect on microtubule assembly, perhaps by steric hindrance while bound to its luminal site. The destabilizing effect of αTAT1 that we see in vitro agrees with the decrease in microtubule stability observed upon overexpression of the enzyme in mammalian cells, even if the overexpressed enzyme is a catalytically dead mutant (Kalebic et al., 2012), again hinting that it is the presence of the enzyme rather than the tubulin modification that has an effect on microtubule stability.
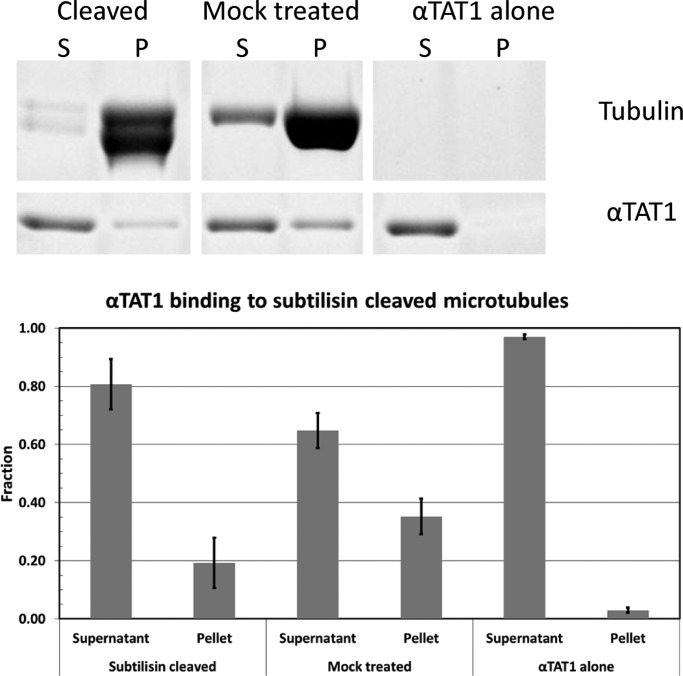
Neither the dynamic nor the Taxol-stabilized microtubule samples containing αTAT1 were amenable to 3D reconstruction, preventing us from directly visualizing the binding site(s) of αTAT1 on microtubules. To further probe the possible interaction of αTAT1 with the outside surface of the microtubule, hinted at by the bundling effect of the enzyme, we used cosedimentation assays with microtubules treated with subtilisin, a protease that removes the acidic C-terminal tails of tubulin that extend from the microtubule surface. Indeed, we observed a clear decrease in the amount of αTAT1 that cosediments with subtilisin-treated microtubules (Figure 6). Consistent with αTAT1 interacting with microtubules through this charged region, we also observed that the binding of αTAT1 decreases with increasing salt concentration (Supplemental Figure S4.) This result lends further support to the idea that αTAT1 binds tubulin at additional site(s) from its site of action on the microtubule luminal surface.
FIGURE 6:
αTAT1 binding to microtubules is affected by the absence of the C-terminal tails of tubulin. SDS gel (top) and quantitation (bottom) of pelleting assays showing that αTAT1 cosediments less with subtilisin-treated microtubules than with untreated tubulin. Data from three independent cosedimentation assays. Error bars, SD.
Interaction of αTAT1 with the external surface of the microtubule may help to target the enzyme. This binding event may serve to concentrate the enzyme, which could then be used to funnel it to the ends and/or make use of lattice fluctuations/defects to gain access to its luminal site. The pores between protofilaments obtained in this study and previous work (Meurer-Grob et al., 2001; Li et al., 2002) are ∼20 Å in diameter. This is smaller than the smallest dimension of the αTAT1 monomer, which can be approximated by an ellipsoid with major and minor axes of 60 and 30 Å, respectively (Kormendi et al., 2012; Taschner et al., 2012). Concepts like defects in the microtubule wall and/or temporary openings of the lattice (perhaps at the seam) have been used to explain the fast binding of Taxol (Díaz et al., 2003) and the ability of the anti–acetylated tubulin antibody to gain access to the lumen (Odde, 1998). On the basis of the importance of the tubulin E-hook for αTAT1 association with microtubules, we speculate that αTAT1 could be responsive to other PTMs that also occur on the tails. This may be a mechanism for coordinating acetylation with other tubulin PTMs. Future studies could test the possibility that the N-terminal catalytic domain of αTAT1 binds to the luminal surface of the microtubule, whereas other regions of the protein mediate binding to the outside.
The affinity of αTAT1 for microtubules is not significantly affected by the tubulin acetylation state
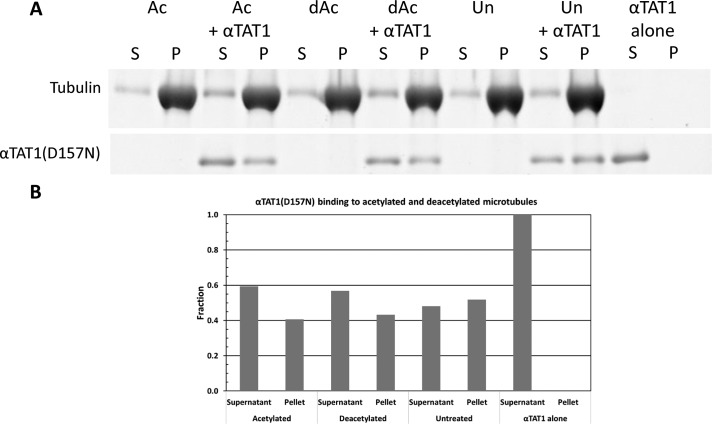
Histone acetyltransferases associate with transcriptional cofactors containing bromodomains that bind acetylated histones, thus allowing for epigenetic spreading of this modification in chromatin (Filippakopoulos and Knapp, 2012). Although no such cofactors have been discovered that would “spread” tubulin acetylation inside microtubules, and although no classical bromodomain exists within αTAT1, we decided to test whether αTAT1 would bind preferentially to modified αK40. Tubulin was acetylated using 100-fold less αTAT1 compared with the treatments performed for the cryo-EM studies, in order to minimize any possible residual binding to undetectable levels. Acetylation levels were ∼80% of the level obtained using previous concentrations of αTAT1 (Supplemental Figure S1B). Deacetylase levels were kept the same, as residual enzyme is not observed after the microtubule pellet is resuspended. Catalytically inactive αTAT1 (D157N mutation) was then added after microtubules were assembled in the presence of Taxol. Our pelleting results show that αTAT1 does not bind differently to microtubules with different tubulin acetylation states (Figure 7). This result suggests that TAT1 is not sufficient to spread the Ac-αK40 mark.
FIGURE 7:
αTAT1 binding to microtubules is not affected by the acetylation state of tubulin. αTAT1(D157N) cosedimented similarly with microtubules polymerized from tubulin that was previously acetylated or deacetylated. Ac, acetylated; dAc, deacetylated; Un, untreated.
Several groups reported the structure of αTAT1 (Friedmann et al., 2012; Kormendi et al., 2012; Li et al., 2012; Taschner et al., 2012), but none of these structures was solved with a bound tubulin peptide. Of interest, whereas HATs are fully active on histone tail peptides, αTAT1 has no activity on a 19-residue peptide corresponding to the loop where Lys40 resides (Li et al., 2012). Contrary to histone tails, the H1-S2 loop in α-tubulin is structured, and recognition of the site by αTAT1 may require the proper secondary structure. Furthermore, αTAT1 is more active on microtubules than on soluble tubulin dimers due to a higher Kcat rather than increased affinity (Shida et al., 2010), suggesting that the loop containing αK40 requires the context of the tubulin dimer or microtubule to be an optimal substrate for αTAT1.
Conclusions
Our studies demonstrate that the acetylation of Lys-40 in α-tubulin has no significant effect on microtubule structure or tubulin conformation. Obviously, the biological role of tubulin acetylation does not require that this modification produce a direct effect on microtubule structure, stability or other intrinsic properties. Instead, it could operate to alter the binding of proteins that access the luminal surface of the microtubule. Although the identity of these proteins is unknown, the presence of luminal particles has been confirmed for many cell types (Burton, 1984; Garvalov et al., 2006; Bouchet-Marquis et al., 2007). Furthermore, the abundance of luminal particles appears to positively correlate with the degree of acetylation reported for these cell types (Garvalov et al., 2006). Of interest, our in vitro studies indicate that the enzyme carrying out this tubulin modification, αTAT1, itself has the potential to affect microtubule structure and stability. Our attempts to decorate microtubules with αTAT1 to directly visualize its binding were unsuccessful. This was due to αTAT1 interfering with the assembly of microtubules and/or having multiple tubulin-binding sites. Our in vitro observation of αTAT1 effect on microtubule stability is in good agreement with what was seen in in vivo overexpression experiments, which showed that the effect is not dependent on the enzymatic activity of αTAT1. Instead, our study shows that the effect involves either the incorporation of the enzyme into the microtubule lumen or its interaction with the microtubule surface. We believe that the latter precedes luminal access and may serve as a funneling mechanism to take advantage of protofilament breathing and/or lattice defects.
MATERIALS AND METHODS
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Recombinant αTAT1 (amino acids 2–236) and SIRT2 (full length) were provided by the Nachury lab and purified as previously described (Shida et al., 2010). Antibodies for αK40 acetylated tubulin (T7451, clone 6-11B-1) and goat anti-mouse immunoglobulin G (PA42002; GE Healthcare, Piscataway, NJ) were used in Western blots to quantify acetylation levels.
Protein and plasmid for monomeric kinesin (Kif5b), defective in ATP hydrolysis (E236A), was supplied by the Vale lab (Rice et al., 1999). Kinesin purification from Escherichia coli was performed by growing cells to OD600 nm ≈ 0.6 at 37°C, inducing with 0.4 mM isopropyl-β-d-thiogalactoside, and cooling to 22°C for overnight expression. Cells were harvested and resuspended in 50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 2 mM MgCl2, 10% (vol/vol) glycerol, 10 mM imidazole, 0.001% (vol/vol) 1-thiolglycerol, and protease inhibitors (11836153001; Roche Applied Science, Indianapolis, IN). Cells were lysed by sonication and the lysate clarified at 104,000 × g for 30 min. Nickel beads (1 ml of packed beads volume/l culture; P6611) were equilibrated in lysis buffer and incubated with the supernatant from the clarified lysate for 2 h with gentle mixing. Beads were washed with twice with lysis buffer containing 200 μM ATP and no protease inhibitors and then washed twice with 50 mM phosphate buffer. pH 8.0, 150 mM NaCl, 2 mM MgCl2, 10% (vol/vol) glycerol, 0.001% (vol/vol) 1-thiolglycerol, and 30 mM imidazole. Protein was eluted with 2× beads volume using 50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 2 mM MgCl2, 10% (vol/vol) glycerol, 0.001% (vol/vol) 1-thiolglycerol, and 250 mM imidazole. Kinesin was concentrated using a 10-kDa cutoff membrane and loaded onto a S200 Superdex column equilibrated in 25 mM Tris, pH 7.5, 150 mM KCl, 10% (vol/vol) glycerol, 2 mM MgCl2, and 1 mM dithiothreitol (DTT). Fractions were pooled and the kinesin concentrated to 9.6 mg/ml and flash frozen.
Tubulin modification in vitro
Porcine tubulin was purchased from Cytoskeleton (Denver, CO; T240), reconstituted to 10 mg/ml in BRB80 buffer (80 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.9, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM MgCl2) with 10% (vol/vol) glycerol, 1 mM GTP, and 1 mM DTT, and flash frozen in 10-μl aliquots until needed. Acetylation and deacetylation reactions were carried out in ADE buffer (40 mM PIPES, pH 6.9, 0.8 mM EGTA, 0.5 mM MgSO4) with 30% glycerol, 0.5 mM GTP, and 1 mM DTT. Tubulin concentrations were 40–50 μM for all acetylation and deacetylation reactions. αTAT1 and acetyl-CoA were used at final concentrations of 6–20 and 50 μM, respectively. SIRT2 and NADH were used at final concentrations of 5 μM and 1 mM, respectively. Acetylation levels were estimated using Western blots probed with an anti–acetylated tubulin antibody (T7451) and a fluorescent secondary antibody (PA43009; GE Healthcare). Quantification of the in vitro acetylation levels was estimated by normalizing the Western blot signal to the amount of tubulin in the replicated lane and using Tetrahymena ciliary tubulin as a standard for completely acetylated tubulin (Supplemental Figure S1A). For cosedimentation assays where the catalytically inactive mutant was used, the acetylation reaction was performed with 100× lower αTAT1 to minimize residual αTAT1 that remained bound after pelleting the microtubule. All reactions were allowed to proceed for 2 h at 22°C. Modified tubulin was then cooled to 4°C and incubated with fresh GTP for 20 min before polymerization into dynamic or Taxol-stabilized microtubules.
Cosedimentation assays
Taxol-stabilized microtubules were prepared by warming reconstituted tubulin to 37°C for 10 min and adding Taxol (TXD01; Cytoskeleton) in two additions, spaced 10 min apart, to a final concentration of 180 μM. Microtubules were spun down in a tabletop centrifuge at 17,000 × g for 20 min and resuspended in binding buffer BRB80 with 5% (vol/vol) sucrose, 50 mM KCl, and 1 mM DTT supplemented with Taxol. Sheared microtubules were formed by passing the microtubules through a 10-μl syringe (60373-946; VWR, Radnor, PA) ∼100 times. Vinblastine polymers were generated by adding vinblastine (V1377) to 1 mM and polymerizing for 3 h at room temperature. Polymers were spun down and the concentration assayed using A280 absorbance after depolymerization with 50 mM CaCl2. Subtilisin-cleaved microtubules were prepared by incubating preformed microtubules (∼2 mg/ml) with 0.05 mg/ml subtilisin (572909; EMD Millipore, Billerica, MA) in BRB80 at 37°C for 20 min. Digestion was halted by the addition of phenylmethanesulfonyl fluoride to 2 mM final. Attempts to digest tubulin further to completely remove the C-terminal tails of both monomers resulted in the cleavage of the N-terminal loop of α-tubulin, as judged by the signal obtained in a Western blot of the tubulin preparations using the anti–acetylated tubulin antibody (Supplemental Figure S3). This suggests that subtilisin can access the lumen of microtubules, where it can then cleave the flexible loop containing αK40. Tubulin polymers were mixed with αTAT1 that had been desalted into the binding buffer and allowed to equilibrate at room temperature for 20 min. Binding buffer was supplemented with KCl to investigate the effect of different salt concentrations. Mixtures were layered on top of a 50% glycerol cushion and spun at 352,000 × g for 10 min to sediment polymers. Supernatant and pellets were analyzed by SDS–PAGE stained with SYPRO (170-3125; BioRad, Hercules, CA) or Flamingo (161-0491; BioRad) fluorescent stains. Gels were analyzed using ImageJ (National Institutes of Health, Bethesda, MD). The amount of αTAT1 that pelleted in the absence of microtubules (self-pelleting) was subtracted when calculating the bound fraction of αTAT1.
Analytical gel filtration
αTAT1 buffer was exchanged into BRB80 with 1 mM DTT and KCl at either 10 or 200 mM using Zeba spin desalting columns (89882; Thermo Scientific, Rockford, IL). A Superdex 200 PC 3.2/30 column (17-1089-01; GE Healthcare) equilibrated in the appropriate buffer was loaded with 50 μl of αTAT1 and run at 4°C with a flow rate of 50 μl/min.
Cryo-EM grid preparation and imaging
Kinesin was desalted into EM buffer BRB80 with 0.05% (vol/vol) Nonidet P-40 (11332473001; Roche) and 2 mM DTT, supplemented with 0.5 mM ATP, using a Zeba spin desalting column and diluted to 1 mg/ml in the foregoing buffer supplemented with GTP to 1 mM (to maintain microtubule dynamics while making grids). After dilution, kinesin was spun at 80,000 × g at 4°C to remove any aggregates. Microtubules were diluted to 0.5 mg/ml in EM buffer supplemented with 1 mM GTP and applied to a glow-discharged C-flat grid (Protochips, Raleigh, NC) in the chamber of a Virtrobot (Mark II; Maastricht Instruments, Maastricht, Netherlands). Microtubules were allowed to adhere to the grid for 30 s. Kinesin was then applied to the grid twice, 4 μl each addition, briefly manually blotting after the first addition, to ensure complete decoration of the microtubules. The grid was then blotted for 4 s and plunged into ethane slush. Micrographs were collected on a Tecnai F20 transmission electron microscope (FEI, Hillsboro, OR) operating at 120 kV and equipped with a Gatan 4k Ultrascan charge-coupled device. Micrographs were collected using Leginon (Suloway et al., 2005) with a dose of 20 e−/Å2 and nominal magnification of 80,000×, giving a final size of 1.37 Å/pixel.
Image analysis and data processing
Images were processed using the Appion EM image processing suite (Lander et al., 2009). Overlapping square boxes of 700 Å, spaced 80 Å apart, were masked from the raw images and grouped into two-dimensional reference-free class averages. Particles belonging to class averages that appeared to contain high-resolution information were further processed using EMAN2 multimodel refinement (Tang et al., 2007) against initial models of 12-, 13-, 14-, and 15-protofilament microtubules (Sui and Downing, 2010) low-pass filtered to 20-Å resolution and iterative helical real-space reconstruction (Egelman, 2007) to obtain the 3D reconstruction. The number of particles that matched each model was used to estimate the total number of microtubules of each protofilament number. Resolutions of the reconstructions were estimated using FSC cutoff of 0.5 between volumes using half the particles. Difference maps were obtained by applying a negative B-factor to the reconstructions, isolating a dimer, and aligning the isolated dimers using the Fit in Map command within UCSF Chimera (Pettersen et al., 2004). The rotation and translations were then applied to the isolated dimers from the unsharpened (no B-factor applied) reconstructions. Differences between the aligned dimers were then calculated using the DIFFMAP program (Grigorieff lab, Brandeis University, Waltham, MA) after padding the dimer volumes by a factor of 2. Differences at the edges due to the alignment after the isolations were ignored. All figures were prepared using UCSF Chimera.
Supplementary Material
Acknowledgments
We thank Erik Jonsson and Ronald Vale (Department of Cellular and Molecular Pharmacology and Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, CA) for the generous gift of monomeric kinesin protein and expression plasmids. We also thank Gabriel Lander and Tom Houweling for computer support and Patricia Grob and Gigi Kemalyan for maintenance of the electron microscope facility. This work was supported by National Institute of General Medical Sciences Grants P01GM051487 to E.N. and R01GM089933 to M.V.N. E.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations used:
- αTAT1
alpha-tubulin acetyltransferase 1
- EM
electron microscopy
- FSC
Fourier shell correlation
- HDAC
histone deacetylase
- MAP
microtubule-associated protein
- PTM
posttranslational modification
- SIRT2
sirtuin 2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0387) on November 13, 2013.
Present addresses: *Cell Biology and Physiology Center, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892;
†Laboratory for Protein Conformation Diseases, RIKEN Brain Science Institute, Wako, Saitama 351-0198, Japan.
REFERENCES
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Gaertig J. MEC-17 is an α-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu JM, Bordas J, Diaz JF, García de Ancos J, Gil R, Medrano FJ, Towns-Andrews E. Low resolution structure of microtubules in solution. Synchrotron X-ray scattering and electron microscopy of Taxol-induced microtubules assembled from purified tubulin in comparison with glycerol and MAP-induced microtubules. J Mol Biol. 1992;226:169–184. doi: 10.1016/0022-2836(92)90132-4. [DOI] [PubMed] [Google Scholar]
- Arnal I, Wade RH. How does Taxol stabilize microtubules. Curr Biol. 1995;5:900–908. doi: 10.1016/s0960-9822(95)00180-1. [DOI] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Zuber B, Glynn A-M, Eltsov M, Grabenbauer M, Goldie KN, Chrétien D. Visualization of cell microtubules in their native state. Biol Cell. 2007;99:45–53. doi: 10.1042/BC20060081. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, Richards JE, Piperno G. Posttranslational modifications of alpha tubulin: detyrosination and acetylation differentiate populations of interphase microtubules in cultured cells. J Cell Biol. 1988;106:1213–1220. doi: 10.1083/jcb.106.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR. Luminal material in microtubules of frog olfactory axons: structure and distribution. J Cell Biol. 1984;99:520–528. doi: 10.1083/jcb.99.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chu CW, Hou F, Zhang J, Phu L, Loktev AV, Kirkpatrick DS, Zou H. A novel acetylation of β-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol Biol Cell. 2011;22:448–456. doi: 10.1091/mbc.E10-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert M-L, Gillard M, Close P, Malaise O, Nguyen L. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Cueva JG, Hsin J, Huang KC, Goodman MB. Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr Biol. 2012;22:1066–1074. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchik MV, Funk WC, Brown EE, Larkin K. Requirement for microtubules in new membrane formation during cytokinesis of Xenopus embryos. Dev Biol. 1998;194:47–60. doi: 10.1006/dbio.1997.8815. [DOI] [PubMed] [Google Scholar]
- De Brabander MJ, Van de Veire RM, Aerts FE, Borgers M, Janssen PA. The effects of methyl (5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl) carbamate, (R 17934; NSC 238159), a new synthetic antitumoral drug interfering with microtubules, on mammalian cells cultured in vitro. Cancer Res. 1976;36:905–916. [PubMed] [Google Scholar]
- Díaz JF, Barasoain I, Andreu JM. Fast kinetics of Taxol binding to microtubules. Effects of solution variables and microtubule-associated proteins. J Biol Chem. 2003;278:8407–8419. doi: 10.1074/jbc.M211163200. [DOI] [PubMed] [Google Scholar]
- Di Fulvio S, Azakir BA, Therrien C, Sinnreich M. Dysferlin interacts with histone deacetylase 6 and increases alpha-tubulin acetylation. PloS One. 2011;6:e28563. doi: 10.1371/journal.pone.0028563. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunn S, Morrison EE, Liverpool TB, Molina-París C, Cross RA, Alonso MC, Peckham M. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Aguilar A, Fan J, Nachury MV, Marmorstein R. Structure of the α-tubulin acetyltransferase, αTAT1, and implications for tubulin-specific acetylation. Proc Natl Acad Sci USA. 2012;109:19655–19660. doi: 10.1073/pnas.1209357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Zuber B, Bouchet-Marquis C, Kudryashev M, Gruska M, Beck M, Cyrklaff M. Luminal particles within cellular microtubules. J Cell Biol. 2006;174:759–765. doi: 10.1083/jcb.200606074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang C, Ravelli RBG, Roussi F, Steinmetz MO, Curmi PA, Knossow M. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- Hubbert CC, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yao T-P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Kalebic N, Martinez C, Perlas E, Hublitz P, Bilbao-Cortes D, Fiedorczuk K, Heppenstall PA. The tubulin acetyltransferase αTAT1 destabilizes microtubules independently of its acetylation activity. Mol Cell Biol. 2012;33:1114–1123. doi: 10.1128/MCB.01044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Kormendi V, Szyk A, Piszczek G, Roll-Mecak A. Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus. J Biol Chem. 2012;287:41569–41575. doi: 10.1074/jbc.C112.421222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wittmann T. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 2012;22:418–428. doi: 10.1016/j.tcb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J Cell Biol. 1986;103:13–22. doi: 10.1083/jcb.103.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci USA. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified in the flagella during flagellar assembly. J Cell Biol. 1983;97:258–263. doi: 10.1083/jcb.97.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 A resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- Li W, Zhong C, Li L, Sun B, Wang W, Xu S, Zhang T, Wang C, Bao L, Ding J. Molecular basis of the acetyltransferase activity of MEC-17 towards α-tubulin. Cell Res. 2012;22:1707–1711. doi: 10.1038/cr.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol. 1986;103:571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer-Grob P, Kasparian J, Wade RH. Microtubule structure at improved resolution. Biochemistry. 2001;40:8000–8008. doi: 10.1021/bi010343p. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Nogales E, Medrano FJ, Diakun GP, Mant GR, Towns-Andrews E, Bordas J. The effect of temperature on the structure of vinblastine-induced polymers of purified tubulin: detection of a reversible conformational change. J Mol Biol. 1995;254:416–430. doi: 10.1006/jmbi.1995.0628. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Odde D. Diffusion inside microtubules. Eur Biophys J. 1998;27:514–520. doi: 10.1007/s002490050161. [DOI] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones GB, Danowski BA, Devaraj A, Singh V, Ligon LA. The posttranslational modification of tubulin undergoes a switch from detyrosination to acetylation as epithelial cells become polarized. Mol Biol Cell. 2011;22:1045–1057. doi: 10.1091/mbc.E10-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Serrador JM, Cabrero JR, Sancho D, Mittelbrunn M, Urzainqui A, Sánchez-Madrid F. HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity. 2004;20:417–428. doi: 10.1016/s1074-7613(04)00078-0. [DOI] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RE. Tubulin in sea urchin embryonic cilia: post-translational modifications during regeneration. J Cell Sci. 1992;101:837–45. doi: 10.1242/jcs.101.4.837. [DOI] [PubMed] [Google Scholar]
- Sudo H, Baas PW. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci. 2010;30:7215–7226. doi: 10.1523/JNEUROSCI.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H. Kinesin-13s form rings around microtubules. J Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Taschner M, Vetter M, Lorentzen E. Atomic resolution structure of human α-tubulin acetyltransferase bound to acetyl-CoA. Proc Natl Acad Sci USA. 2012;109:19649–19654. doi: 10.1073/pnas.1209343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Relationship between the Golgi complex and microtubules enriched in detyrosinated or acetylated alpha-tubulin: studies on cells recovering from nocodazole and cells in the terminal phase of cytokinesis. Cell Tissue Res. 1993;273:457–466. doi: 10.1007/BF00333700. [DOI] [PubMed] [Google Scholar]
- Topalidou I, Keller C, Kalebic N, Nguyen KCQ, Somhegyi H, Politi KA, Chalfie M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22:1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.