The midbody ring (MR) is asymmetrically segregated during asymmetric divisions of germline stem cells (GSCs) in Drosophila. Male GSCs, which inherit the mother centrosome, exclude the MR, whereas female GSCs, which inherit the daughter centrosome, inherit the MR. Moreover, stem cell identity correlates with the mode of MR inheritance.
Abstract
Many stem cells, including Drosophila germline stem cells (GSCs), divide asymmetrically, producing one stem cell and one differentiating daughter. Cytokinesis is often asymmetric, in that only one daughter cell inherits the midbody ring (MR) upon completion of abscission even in apparently symmetrically dividing cells. However, whether the asymmetry in cytokinesis correlates with cell fate or has functional relevance has been poorly explored. Here we show that the MR is asymmetrically segregated during GSC divisions in a centrosome age–dependent manner: male GSCs, which inherit the mother centrosome, exclude the MR, whereas female GSCs, which we here show inherit the daughter centrosome, inherit the MR. We further show that stem cell identity correlates with the mode of MR inheritance. Together our data suggest that the MR does not inherently dictate stem cell identity, although its stereotypical inheritance is under the control of stemness and potentially provides a platform for asymmetric segregation of certain factors.
INTRODUCTION
Asymmetric stem cell division is critical for tissue homeostasis by balancing the production of stem cells and differentiating daughters (Morrison and Kimble, 2006). The centrosome has become increasingly recognized as playing key roles in asymmetric stem cell division (Yamashita et al., 2003, 2007; Rebollo et al., 2007; Rusan and Peifer, 2007; Cheng et al., 2008; Wang et al., 2009). As a microtubule-organizing center (MTOC), the centrosome position within the cell can dictate the orientation of cell division, often leading to asymmetric stem cell division. The centrosome also plays critical roles in cytokinesis (Piel et al., 2000, 2001; Doxsey et al., 2005; Gromley et al., 2005; Goss and Toomre, 2008; Pohl and Jentsch, 2008; Prekeris and Gould, 2008). In some cell types, cytokinesis has been shown to be asymmetric, in that abscission occurs on only one side of the midbody and the other side inherits the midbody ring (MR; Gromley et al., 2005). In other cases, the MR was reported to be released into the extracellular space (Dubreuil et al., 2007). Kuo et al. (2011) reported that cells containing the mother centrosome inherit the MR upon abscission. They also showed that pluripotent stem cells (embryonic stem cells and induced pluripotent stem cells) and cancer cells tend to accumulate the MRs, probably correlating with their inheritance of the mother centrosome, leading them to propose that MR inheritance may play a role in stem cell identity (Kuo et al., 2011). In contrast, Ettinger et al. (2011) reported that stem cells are characterized by a high capacity of MR release into the extracellular space. Therefore it is unclear whether the MR carries any information relevant to stem cell behavior and how MR fate is determined, possibly depending on cell type.
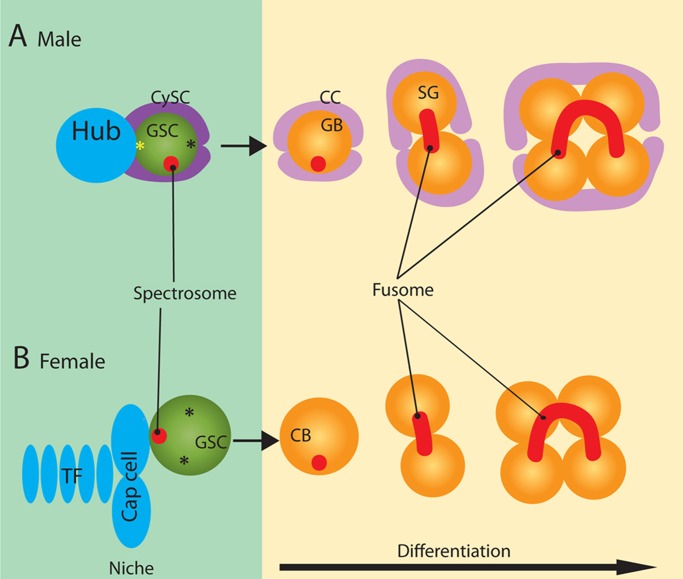
Drosophila male and female germline stem cells (GSCs) divide asymmetrically to produce one stem cell and one differentiating cell. In the Drosophila testis, GSCs attach to somatic hub cells, which, together with cyst stem cells (CySCs), create a signaling microenvironment—the niche—to specify GSC identity (Figure 1A; Fuller and Spradling, 2007; Yamashita et al., 2010). Similarly, in the Drosophila ovary, GSCs attach to cap cells, which form the niche together with the terminal filaments and escort cells (Figure 1B; Decotto and Spradling, 2005; Morris and Spradling, 2011). Germline cells that remain within these niches maintain stem cell identity, whereas those that are displaced away from the niches initiate differentiation. The asymmetric outcome of GSC division is mainly governed by spindle orientation, which is achieved by the stereotypical movement of centrosomes during interphase in male GSCs (Figure 1A; Yamashita et al., 2003) or anchoring of one spindle pole to the spectrosome (a germline-specific organelle) in female GSCs (Figure 1B; Deng and Lin, 1997).
FIGURE 1:
Diagram of male and female germline stem cell niches. (A) At the apical tip in the Drosophila testis, GSCs attach to the hub cells, whereas their daughters, GBs, are displaced away from the hub. Centrosome orientation prepares for perpendicular spindle orientation; the mother centrosome (yellow asterisk) is consistently located near the hub, whereas the daughter centrosome (black asterisk) migrates toward the opposite side of the GSC. GSCs contain the spectrosome (red circle), which assumes a spherical morphology, whereas differentiating spermatogonia (SG) contain the fusome (red line), which is branched and runs through the ring canals. GSCs are encapsulated by a pair of CySCs. GBs and SG are encapsulated by a pair of CCs, progeny of CySCs. (B) In the germarium in the Drosophila ovary, GSCs attach to the cap cells, whereas their daughters, CBs, are displaced away from the cap cells. Although the centrosomes (asterisks) are not stereotypically oriented in female GSCs, the spectrosome (red circle) is located close to the cap cells, orienting the mitotic spindle. Cap cells and terminal filaments (TFs) provide niche signals to GSCs. Escort cells (not shown) exist in the germarium that closely associate with the GSCs and developing germ cells. Unlike CySCs, they do not normally proliferate or move along with the developing germ cells. However, they provide supportive signals for germ cell development, similar to CySCs and CCs in the testis.
Here we demonstrate that male and female GSCs segregate the MR asymmetrically with strikingly distinct processes. Our data show that the MR is inherited by the cell containing the daughter centrosome and that the MR is not always inherited by stem cells in the Drosophila germline. We propose that, whereas asymmetry in MR inheritance can potentially serve as a platform for carrying information to impose asymmetric behavior of cells, the MR does not inherently confer stem cell identity.
Results
The MR is inherited by the differentiating daughter during male GSC division
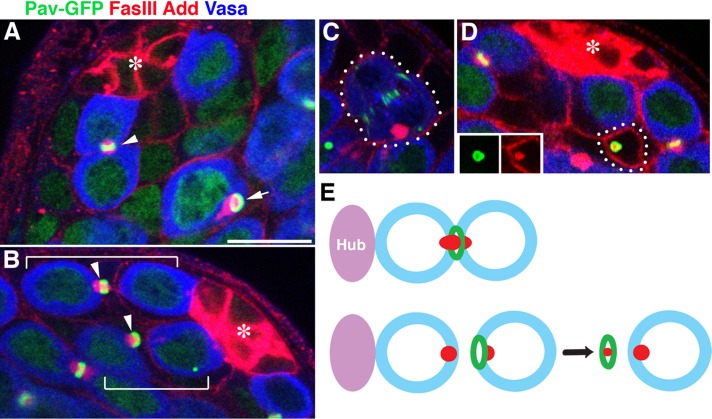
To examine MR inheritance during male GSC division, we used Pavarotti–green fluorescent protein (GFP; Minestrini et al., 2002) to visualize the MR. Pavarotti is a homologue of MKLP1, a kinesin-like protein that is required for cytokinesis (Adams et al., 1998; Minestrini et al., 2002, 2003). Consistent with the reported localization of Pavarotti in other cell types, Pavarotti-GFP was observed at the spindle midzone, the MR, and ring canals, the structures that connect differentiating spermatogonia and spermatocytes (Figure 2A). The spectrosome, a germline-specific membranous organelle marked by Hts/adducin-like (Add), was observed to run through the MR (Figure 2A), similar to previous observations in females (Deng and Lin, 1997; de Cuevas and Spradling, 1998). As in female GSCs, cytokinesis in male GSCs is prolonged, and final resolution of the cytokinesis takes place in the G2 phase of the next cell cycle (Sheng and Matunis, 2011). As a result, GSCs stay connected to the gonialblasts (GBs), the differentiating daughter of GSCs, for ∼50% of the cell cycle time. Of interest, when cytokinesis was complete, the MR was inherited by the GBs (>92%; n > 200 GSC-GB pairs; Figure 2B). We limited our analysis to cases in which the pairing of GSCs and GBs was evident by the presence of a thin thread of spectrosome material (positive for Add) connecting the GSCs and GBs. As a result of asymmetric cytokinesis, GBs containing the MR were frequently observed, even after clear separation of GSCs and GBs (Figure 2A, arrow). These observations are distinct from findings in mammalian cells, in which it was proposed that the stem cells inherit and accumulate MRs (Kuo et al., 2011). GBs in the subsequent mitosis (to become two-cell spermatogonia) never contained MR remnants (Figure 2C), suggesting that GBs somehow dispose of the MR before mitosis. Surprisingly, we found that the MR is released from GBs into somatic cells (CySCs or cyst cells [CCs]; Figure 2D). The MR found in CySCs/CCs is indeed derived from GBs rather than from somatic cells, based on the following observations: 1) the Pavarotti-GFP–marked MR was found in CySCs/CCs even when Pavarotti-GFP is expressed only in the germline (using nos-gal4 driver; Figure 2D), and 2) the MRs in CySCs/CCs are still associated with the spectrosome material, suggesting that they must have derived from the germline (Figure 2D, inset). In addition, by changing the focal plane of the microscopy, we confirmed that such MRs were not part of germ cells that exist outside of the focal plane. Because MRs were not internalized into the GB, it is likely that CySCs/CCs engulf the MR from the surface of GBs after the completion of abscission. In addition, we never observed CySCs/CCs containing multiple MR remnants, suggesting that the CySCs/CCs somehow remove MRs; indeed, we often observed that the MRs in CySCs/CCs were associated with lysosomes (Supplemental Figure S1). Of 126 testes observed, 109 had MR remnants in CySCs/CCs, 49% of which were associated with lysosomes. The MR was found in either CySCs (58%) or CCs (42%), suggesting that MR ingestion was not related to the identity of CySCs or CCs. This observation is reminiscent of mouse neural stem cells, in which the MR is released into the extracellular space (Dubreuil et al., 2007; Ettinger et al., 2011). Our observation also showed that even when the MR was eventually released into the extracellular space, the initial abscission was stereotypically asymmetric as to which cell inherits the MR. On the basis of these findings, we conclude that cytokinesis in male GSCs is asymmetric: the MR is almost always inherited by GBs and is eventually released into somatic CySCs/CCs, where it is degraded (Figure 2E).
FIGURE 2:
Midbody ring inheritance is asymmetric during male GSC cytokinesis, and the GSC excludes the MR in the GB. (A) An example of a testis apical tip containing a GSC connected to a GB via the MR (Pav-GFP; arrowhead) and a GB containing the MR after complete separation from the GSC (arrow). Ubi-Pav-GFP flies were used. Green, Pavarotti-GFP (Pav-GFP) marking MRs and ring canals; red, FasIII (fasciclin III) marking hub cells and adducin-like (Add) marking the spectrosome/fusome; Blue, Vasa marking germ cells. The asterisk indicates the hub. Bar, 10 μm. (B) An example of a testis apical tip containing two GSC-GB pairs with their MR (arrowheads) inherited by GBs. GSC-GB pairs are indicated by brackets. Ubi-Pav-GFP flies were used. (C) An example of a GB in mitosis, demonstrating that the GB does not contain the MR from the previous mitosis. nos-gal4>UAS-Pav-GFP flies were used. (D) MR observed in CySCs (the attachment of CySCs to the hub is not visible in this focal plane). The inset shows separate channels of Pav-GFP (green) and Add (red). (E) Schematic diagram of MR inheritance during male GSC cytokinesis. nos-gal4>UAS-Pav-GFP flies were used.
The MR is inherited by GSCs in the female germline
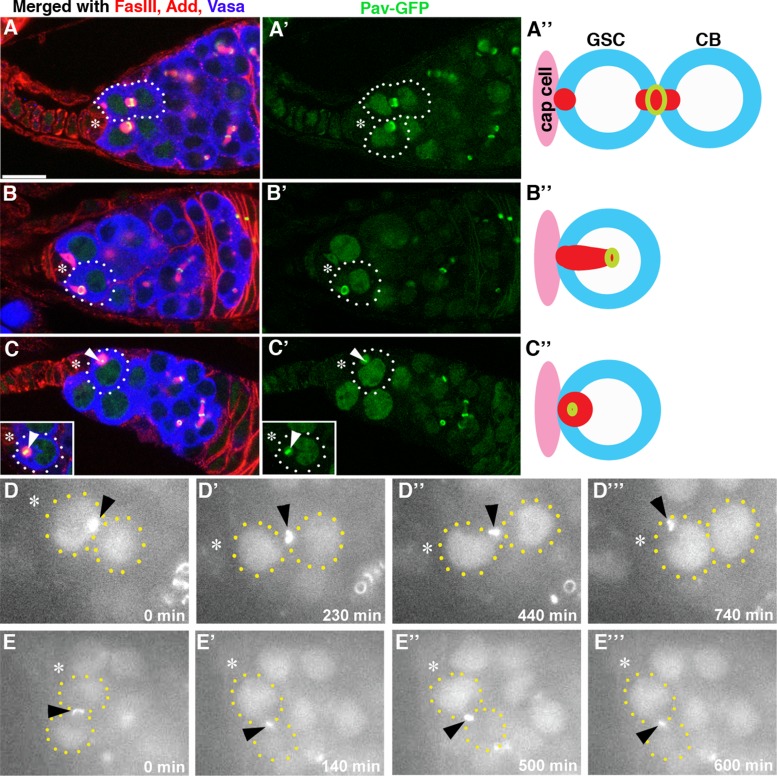
We next examined MR inheritance in female GSCs. Surprisingly, cytokinesis in female GSCs is also asymmetric but in the opposite manner: the MR is almost always inherited by GSCs (92%; n = 61 GSC-cystoblast [CB] pairs; Figure 3). Immediately after cytokinesis, the MR was observed between GSCs and CBs (Figure 3A). The female spectrosome is known to display dynamic morphological changes during the cell cycle (Deng and Lin, 1997; de Cuevas and Spradling, 1998; Hsu et al., 2008) and eventually splits between GSCs and CBs, when about two-thirds of the spectrosome material is inherited by GSCs. When the spectrosome is split between GSCs and CBs, the MR is almost always inherited by GSCs associated with the GSC-inherited spectrosome (Figure 3B). MRs appear to be degraded within the spectrosome of GSCs, as a small Pav-positive structure (often retaining MR morphology) was observed in the spectrosome of GSCs (Figure 3C).
FIGURE 3:
MR inheritance is asymmetric during female GSC cytokinesis, and GSCs inherit MR. (A–C) Examples of germaria containing distinct stages of MR inheritance and degradation. The arrowhead in C indicates an MR that lost its ring shape and became smaller. The arrowhead in the inset of C indicates an MR in the round-stage spectrosome before it becomes smaller. Note that the entire spectrosome becomes weakly positive for GFP at this stage. The arrowheads point to MRs, which maintain stronger a GFP signal and appear as yellow in C. Ubi-Pav-GFP flies were used. Green, Pav-GFP; red, FasIII and Add; blue, Vasa. The asterisk indicates cap cells. Bar, 10 μm. (D, E) Selected frames from time-lapse live observation of MR behavior in female GSCs from Pav-GFP flies (from Supplemental Movies S1 and S2). (D) An example of an MR being inherited by the GSC. (E) An example of an MR becoming small at the site of cytokinesis.
We conducted time-lapse live observation of ovaries from Pav-GFP flies to complement our observation using fixed samples. Whereas fixed samples stained for multiple cellular markers may correlate the spatial relationship between the MR and spectrosome, they do not unambiguously tell the temporal order of MR movement and inheritance. Live observation could add clarity to the temporal order of MR inheritance. Live observation of ovaries to track MR inheritance proved to be challenging: the MR typically stayed in the middle of GSCs and CBs for a long time (often >10 h), and maintaining the focal plane to visualize the MR throughout this period was often impossible. Tracking a small structure such as the MR requires flattening of the sample to some extent, limiting the duration of live culture. Yet we obtained four cases in which the MR was clearly inherited by the GSC or CB. In three of four cases, the MR was inherited by GSCs, consistent with our result obtained from fixed samples (Figure 3D and Supplemental Movie S1). In other cases (n = 15), the MR stayed between GSCs and CBs until the end of the imaging (typically 10–16 h). The cause may be that MR inheritance takes a long time and/or the culture condition compromised cell cycle progression. Yet, in four cases of such movies, we observed that the MR gradually became small without being inherited by GSCs or CBs (Figure 3E and Supplemental Movie S2). Because we observed small MRs between GSCs and CBs even in fixed samples, this likely reflects MR behavior in vivo. Observed variations in the timing of MR inheritance might indicate that MR inheritance is not synchronized with other cell cycle–dependent events, such as changes in spectrosome morphology. However, the MR is clearly degraded by the following mitosis, because we never observed MR remnants in mitotic cells. It should be noted that the scoring of MR inheritance during female GSC mitosis was limited to GSC-CB pairs in which the directionality of MR inheritance was clear. Therefore we conclude that MR is predominantly inherited by GSCs when the inheritance is asymmetric. However, from our data, it cannot be conclusively determined whether all MRs are eventually inherited by GSCs (or CBs) or some MRs may be resolved at the site of cytokinesis.
Previously, it was reported that the MR, detected by staining with anti-anillin antibody, was resolved at the site of cytokinesis instead of being incorporated into the GSC spectrosome (de Cuevas and Spradling, 1998). Although we observed cases where the MR becomes small at the site of cytokinesis, our results suggest that the MR is frequently inherited by GSCs. To reconcile these potentially conflicting observations, we stained the germaria that express Pav-GFP with anti-anillin antibody. We noted that the signal intensity ratio (signal on the MR/signal in the nucleus) was much higher with the Pav-GFP marker than with anti-anillin staining (Supplemental Figure S2), possibly because Pav-GFP was overexpressed and/or because of differences in their endogenous localization. As a result, the MR was more prominently visualized with Pav-GFP than with anti-anillin antibody. Particularly after abscission, when the MR was incorporated into the GSCs, the strong nuclear staining of the anillin often made it difficult to see the MR localization, although anillin was indeed still detectable on the MR (Supplemental Figure S2). Thus it is likely that the spectrosome-incorporated MR was missed in the previous study that used anti-anillin antibody staining. Some MRs became small before being inherited by GSCs or CBs as described, and this might correspond to the population of cells that showed resolution of anillin-positive MRs at the site of cytokinesis. It was not possible to determine whether such small MRs are eventually inherited by either side or resolved at the site of cytokinesis.
The MR inheritance pattern depends on a functional centrosome
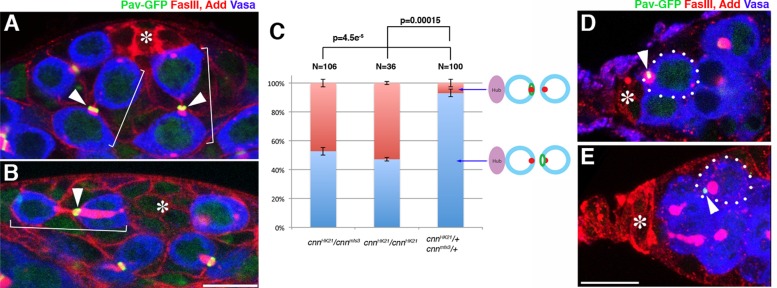
Because the centrosome plays a key role in abscission in mammalian cells (Piel et al., 2000, 2001; Gromley et al., 2005; Lee et al., 2008; Kuo et al., 2011), we tested whether a functional centrosome is required for stereotypical MR inheritance. Indeed, we found that MR inheritance was randomized in male GSCs mutant for centrosomin (cnn), a core component of pericentriolar material required for the majority of centrosome functions (Megraw et al., 1999, 2001; Vaizel-Ohayon and Schejter, 1999): in 47% of cytokineses (n = 106 GSC-GB pairs in which the directionality of MR inheritance is clear), the MR was inherited by GSCs instead of GBs in cnn-mutant animals (either in animals homozygous for the strong loss-of-function allele cnnHK21, which has a stop codon mutation after 105 amino acids out of a total of >1000 amino acids, or in animals transheterozygous for cnnHK21 and cnnmfs3, which has a 190–amino acid truncation at the C-terminus; Figure 4, A–C). Furthermore, the dsas-4 mutant (dsas-4S2214, a loss-of-function allele; Basto et al., 2006), which does not contain any centriole, also showed randomization of MR inheritance during male GSC division (Supplemental Figure S3), suggesting that the stereotypical inheritance of the MR indeed depends on the function of the centrosome.
FIGURE 4:
A functional centrosome is required for stereotypical MR inheritance in male and female GSCs. (A, B) Examples of a testis apical tip from control (cnnHK21/+) and cnnHK21/cnnmfs3 mutant flies stained for FasIII and Add (red), Vasa (blue), and Pav-GFP (green). In control flies (A), the MR was segregated to the GBs, whereas the MR was frequently segregated to GSCs in the cnnHK21/cnnmfs3 mutant (B). Brackets indicate GSC-GB pairs. The asterisk indicates hub cells. Bar, 10 μm. (C) Frequency of MR inheritance by GSCs or GBs in cnnHK21/cnnmfs3, cnnHK21/cnnHK21, or control flies. p value of Student's t test (two tailed). N, number of GSC-GB pairs scored. (D, E) Examples of germaria from control (cnnHK21/+) and cnnHK21/cnnmfs3 mutant flies stained for FasIII and Add (red), Vasa (blue), and Pav-GFP (green). In control flies (D), the MR was segregated to GSCs, whereas the MR was frequently segregated to CBs in the cnnHK21/cnnmfs3 mutant (E). The asterisk indicates cap cells.
In contrast to male GSCs, it has been reported that asymmetric division of female GSCs relies on the spectrosome but not on a functional centrosome (Deng and Lin, 1997; Stevens et al., 2007). A more recent study suggested that centrosomes are oriented toward the GSC–cap cell interface throughout the cell cycle, and such centrosome positioning plays a role in asymmetric division of female GSCs (Lu et al., 2012). We found that MR inheritance was randomized in cnn-mutant female GSCs and the MR was inherited by CBs in 42% of the cases observed (n = 64 GSC-CB pairs; Figure 4, D and E), whereas MR was inherited by CBs only in 8% of the wild type (n = 61) and in 22% of the heterozygous controls (cnnHK21/+, n = 37). These results demonstrate that a functional centrosome is required for asymmetric MR inheritance in both male and female GSCs.
Female GSCs inherit the daughter centrosome
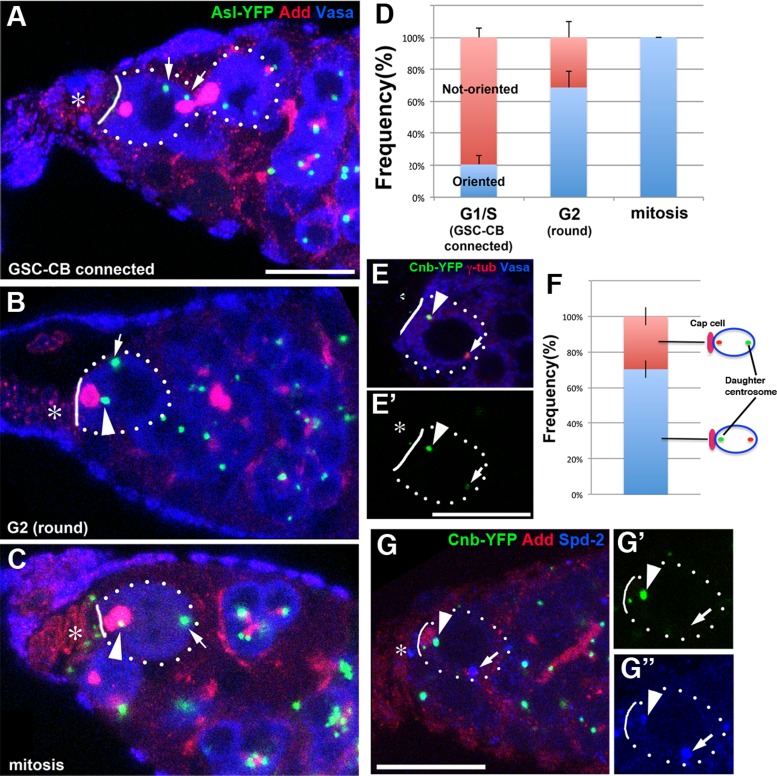
Intrigued by the opposite pattern of MR inheritance (by the differentiating cell in the testis and the GSCs in the ovary) and requirement of cnn in MR inheritance in female GSCs, we decided to characterize centrosome positioning during the female GSC cell cycle in more detail. Under our experimental and culture conditions, we found that centrosomes were not oriented in 57.5% of female GSCs (n = 332 GSCs). However, when we examined the centrosome orientation in relation to the spectrosomal morphology, which corresponds to cell cycle stage (Hsu et al., 2008), it became clear that one centrosome closely associates with the spectrosome (or GSC–cap cell interface) as cells approach mitosis (Figure 5). This nicely bridges two previous reports that one centrosome is associated close to the cap cells in mitosis (Deng and Lin, 1997) but not in interphase (Stevens et al., 2007). As shown in Figure 5, A–D, centrosomes were mostly unoriented during earlier phases of the cell cycle, but in the “round-fusome” stage, which corresponds to the G2 phase of the cell cycle (Hsu et al., 2008), one centrosome was closely associated with the apical side (with either the apically localized spectrosome or GSC–cap cell interface; 68.7 ± 12.7%, n = 141 “round-stage” GSCs; Figure 5, B and D). The centrosome–spectrosome association subsequently reached 100% in mitosis (Figure 5, C and D), as reported previously (Deng and Lin, 1997). These data demonstrate that female GSCs also orient their centrosomes with respect to the niche but during a limited period of the cell cycle (i.e., G2 to mitosis). We do not know where the discrepancy arises between the report by Stevens et al. (2007), which is similar to our observation, and the report by Lu et al. (2012). The cause may be differences in cell cycle distribution under different culture conditions. Lu et al. used a very protein-rich diet (Lu et al., 2012; C. Ferguson, personal communication), whereas we did not supplement our regular fly culture medium with additional dry yeast. We showed that, in male GSCs, the diet influences centrosome orientation, leading to regulation of GSC division frequency, depending on the availability of nutrients (Roth et al., 2012). A similar regulation might be operating in female GSCs.
FIGURE 5:
Female GSCs inherit the daughter centrosomes upon division. (A–C) Centrosome positioning during the cell cycle in female GSCs. (A) The stage when the GSC is still connected to the CB (corresponding to G1/S). (B) The “round-fusome” stage (corresponding to G2). (C) Mitosis. In mitosis (C), Vasa, which is excluded from the nucleus in interphase (A, B), is evenly distributed within the cell, except for the mitotic chromosomes on the metaphase plate. Condensed metaphase chromosomes are also visible in the 4′,6-diamidino-2-phenylindole channel (not shown). The GSC (and CB when still connected) is indicated by dotted circles. The GSC–cap cell interface is indicated by solid lines. The niche component (cap cell/terminal filament) is indicated by asterisks. Arrows indicate centrosomes that are not associated with the cap cells. Arrowheads indicate centrosomes that are associated with the spectrosome. Green, Asl-YFP (centrosome); red, Add; blue, Vasa. Bar, 10 μm. (D) Centrosome orientation during the cell cycle of female GSCs. A total of 332 GSCs were scored. (E) Cnb-YFP labels the apically localized centrosome. Green, Cnb-YFP (F′); red, γ-tubulin; blue, Vasa. (F) The frequency of Cnb-YFP labeling either the apical or proximal centrosomes in female GSCs with oriented centrosomes (N = 54 GSCs with oriented centrosomes). (G) Cnb-YFP labels the centrosome that is associated with the spectrosome at the “round stage.” Green, Cnb-YFP; red, Add; blue, Spd-2.
Januschke et al. (2011) reported Drosophila centrobin (Cnb) as the first molecular marker that distinguishes mother and daughter centrioles in Drosophila. Using this marker and live observation (Conduit and Raff, 2010; Januschke et al., 2011), it was shown that the larval neuroblasts inherit the daughter centrosome upon division. When we scored female GSCs with apparently oriented centrosomes in which only one of the two centrosomes was marked with Cnb-YFP, we found a strong bias toward the Cnb-yellow fluorescent protein (YFP)–labeled daughter centrosome being close to the cap cells/spectrosome (70%; n = 54, Figure 5, E and F). The frequency of daughter centrosomes associated with the cap cells/spectrosome is lower than that of mother centrosomes associated with the hub cells in male GSCs (>90%; Yamashita et al., 2007). This might be because of the contribution of female GSCs in early phases: at this time, ∼20% of GSCs have “apparently oriented” centrosomes (Figure 5D), but these cells might not actually be oriented yet, and either the mother or daughter centrosome may be randomly located near the cap cells. To test this idea, we combined Cnb-YFP and stained for the spectrosome and centrosome to score GSCs exclusively at the “round stage,” which corresponds to late G2 phase of the cell cycle (Hsu et al., 2008). This approach proved to be very challenging because at the “round stage,” many GSCs had two Cnb-YFP–positive centrosomes, presumably due to the maturation of both centrosomes (i.e., both centrosomes are composed of mother and daughter centrioles): of 585 GSCs at the round stage, only 66 cells had one Cnb-positive and one Cnb-negative centrosome, with the remainder containing two Cnb-positive centrosomes. Of these 66 cells, 31 had centrosomes that were not oriented. Among the remaining 35 cells, however, the Cnb-marked daughter centrosome was associated with the cap cells in 30 cells (86 ± 4.2%; Figure 5G). The strong tendency toward association of the daughter centrosome with the cap cells/spectrosome during late stages of the cell cycle suggests that the daughter centrosome is inherited by female GSCs. In the testes, almost all GSCs had two Cnb-YFP–positive centrosomes, likely because centrosome splitting occurs after centrosome duplication, and it was not possible to distinguish mother and daughter centrosomes using Cnb-YFP (Llamazares and Gonzalez, personal communication and unpublished data). We concluded that the daughter centrosome is preferentially inherited by the female GSCs, similar to the situation in Drosophila neuroblasts. These results also reveal a strong correlation between centrosome age and MR inheritance: male GSCs inherit the mother centrosome (Yamashita et al., 2007) and exclude the MR, whereas female GSCs inherit the daughter centrosome and inherit the MR. In other words, in both male and female GSCs, the cell that inherits the daughter centrosome inherits the MR as well. Of interest, this pattern is opposite to the previous observation of (Kuo et al., 2011), in which stem cells were shown to inherit the mother centrosome and MR. This finding may suggest that the MR inheritance pattern and its relationship to centrosome inheritance are cell type–specific (or species-specific) phenomena.
The MR inheritance pattern is influenced by stem cell factors
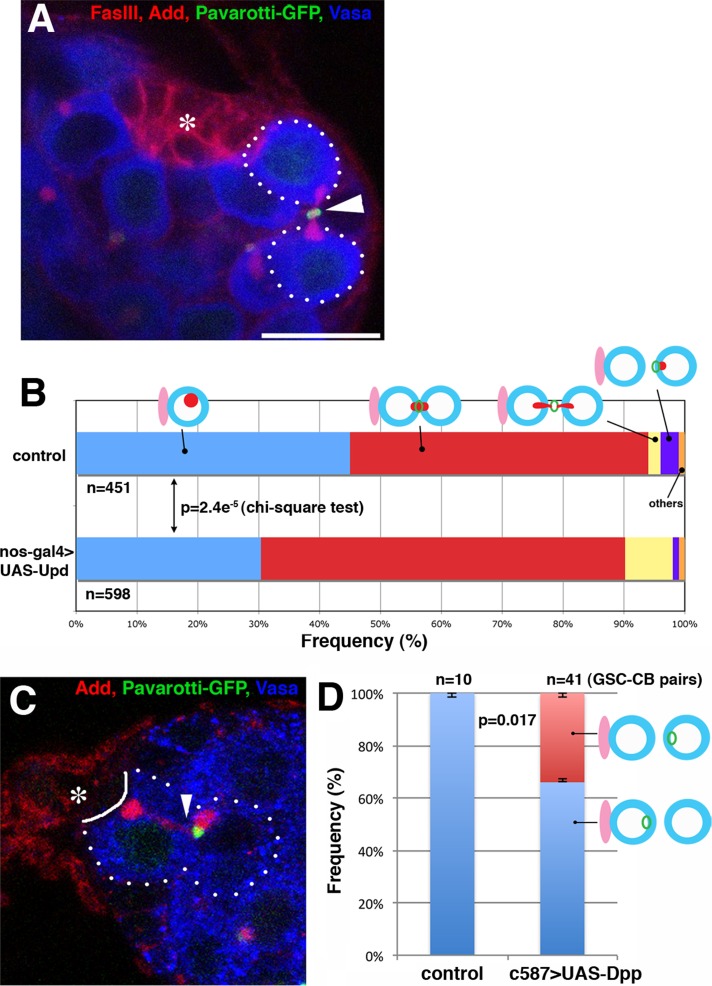
What is the biological relevance of asymmetric MR inheritance during stem cell division? To address whether asymmetric MR inheritance correlates with cell fate, we examined the effect of ectopic expression of niche factors, which leads to symmetric stem cell divisions, on the MR inheritance pattern. Unpaired (Upd) is secreted from hub cells to specify male GSC identity, and overexpression of Upd in the germline is known to lead to GSC tumors (Kiger et al., 2001; Tulina and Matunis, 2001). The MR inheritance pattern was scored upon overexpression of Upd in the germline (nos-gal4>UAS-Upd; Figure 6, A and B). We focused on GSCs that were attached to the hub because GSC tumors outside the normal niche do not have a reference point (landmark) to address the pattern of MR inheritance. On overexpression of Upd, the frequency of GSCs that had completed the abscission (and thus are not connected with the GBs) decreased mildly but significantly compared with controls (31% in Upd-expressing testes vs. 45% in control testes, p = 2.4e−5 by chi-squared test), concomitant with an increase in the frequency of GSCs that are still connected to GBs (61% in Upd-expressing testes vs. 49% in control testes, p = 2.4e−5 by chi-squared test). This finding may suggest that GSC cytokinesis is stalled, possibly due to a problem in deciding on which side of the MR abscission should occur. Consistent with this idea, we also frequently observed GSC-GB pairs in which the MR was apparently pinched from both sides or stuck in the middle upon expression of Upd (Figure 6A). Such instances were very rare in wild-type or control testes (2% of total GSCs, n = 451 GSCs) but increased to 8% (n = 598 GSCs) in Upd-expressing testes (Figure 6B). We speculate that in Upd-expressing testes, GSC division yields two GSCs, both of which activate the program to pinch off the MR. We also speculate that the similar (albeit low frequency) observation in wild-type/control testes might reflect a low frequency of symmetric GSC divisions, as reported recently (Sheng and Matunis, 2011). Together these results indicate that Upd, and presumably stem cell fate, regulates the abscission site during GSC divisions.
FIGURE 6:
MR inheritance correlates with stem cell fate. (A) An example of an MR that is apparently pinched off from both sides upon GSC division in a Upd-expressing testis (nos-gal4>UAS-Upd). Green, Pav-GFP; red, Fas III and Add; blue, Vasa. The asterisk indicates hub cells. Bar, 10 μm. (B) The MR inheritance pattern in control vs. Upd-expressing testes. (C) An example of MR inherited by the cell that is displaced away from the niche (i.e., the cell at the position of the cystoblast; c587-gal4>UAS-Dpp, raised at 18°C, shifted to 29°C for 7 d). Arrowheads indicate the site of abscission. Note that Dpp expression only caused a mild increase in the GSC number under our experimental conditions. (D) The MR inheritance pattern in control vs. Dpp-expressing ovary. Only GSC-CB pairs in which the direction of MR inheritance was clear were scored.
Next we addressed whether GSC fate correlates with the MR inheritance pattern in the female germline. Dpp is a major factor that regulates female GSC identity, and its overexpression leads to GSC tumors (Xie and Spradling, 1998). On overexpression of Dpp, we frequently observed that the cell located away from the cap cell (i.e., at the CB position) inherited the MR more frequently (34%; n = 41 GSC-CB pairs, Figure 6, C and D) than in controls (0%; n = 10 GSC-CB pairs), suggesting that overexpression of Dpp disrupts the stereotypical MR inheritance pattern. Together these results suggest that GSC fate conferred by Upd or Dpp governs the MR inheritance pattern.
DISCUSSION
Here we show that the MR is inherited asymmetrically during GSC divisions in the Drosophila germline and that this correlates with centrosome age and depends on a functional centrosome. Of interest, inheritance of the MR by the cell containing the daughter centrosome is opposite to a recent observation in mammalian cells (Kuo et al., 2011). Further studies are required to determine whether the asymmetrically inherited MR, or factors associated with it, regulates stem cell behavior, and whether this regulation occurs in a species- or cell type–dependent manner. Of importance, mutations that randomize MR inheritance (cnn and dsas-4) do not drastically modulate stem cell identity, and cnn and dsas-4 mutants show apparently normal progression of differentiation regarding the cell fate (Yamashita et al., 2003; Stevens et al., 2007). Furthermore, the MR is inherited by the differentiating daughter in the male germline, whereas it is inherited by the stem cell in the female germline. Therefore it is unlikely that the MR harbors an inherent fate determinant. However, it is tempting to speculate that certain fate determinants “hitchhike” the MR in certain cell types, taking advantage of its stereotypical inheritance. In addition, it is possible that the MR regulates an aspect of stem cell behavior rather than identity per se; for example, the MR could regulate the rate of stem cell division. The fact that we never see multiple MRs in a single cell (GSC or CySC) may indicate that removal of the MR is a prerequisite of cell cycle progression into the next cell cycle. Moreover, the MR that is transferred from the GB to the CySC/CC might function as a messenger to coordinate the division frequency between GSCs and CySCs (Inaba et al., 2011).
The reports by Kuo et al. (2011) and Ettinger et al. (2011) are seemingly contradictory in that the former reported that stem cells are characterized by the accumulation of MRs, whereas the latter reported that they are characterized by the high capacity for MR release into the extracellular space. Our study using male and female GSCs demonstrates that MR fates are highly stereotypical yet strikingly distinct, depending on the cell type. This finding indicates that each cell type handles MRs with its own elaborate cellular program. The reason why MR must be handled in such an elaborate manner awaits future investigation. Nonetheless, our study reveals that a basic cellular asymmetry such as MR inheritance correlates with asymmetry during stem cell division.
MATERIALS AND METHODS
Fly husbandry and strains
All fly stocks were raised in standard Bloomington medium at 25°C. The following fly stocks were used: nos-gal4 (Van Doren et al., 1998), cnnHK21 (Megraw et al., 2001), and UAS-dpp (obtained from the Bloomington Drosophila Stock Center, Bloomington, IN); Ubi-Pavarotti-GFP and UAS-Pavarotti-GFP (Minestrini et al., 2002; obtained from David Glover, University of Cambridge); cnnmfs3 (Megraw et al., 1999; obtained from Thom Kaufman, Indiana University); dsas-4S2214 (Basto et al., 2006; obtained from the Bloomington Stock Center); and UAS-Upd (Zeidler et al., 1999), Asl(asterless)-YFP (Varmark et al., 2007), and Ubi-Cnb-YFP (Januschke et al., 2011; obtained from Cayetano Gonzalez, IRB Barcelona). To assess MR inheritance in cnn mutants, a cnnHK21/CyO; Ubi-PavGFP/TM3 fly stock was generated, which was subsequently crossed with cnnmfs3/CyO to obtain transheterozygous mutant flies (cnnHK21/cnnmfs3; Ubi-Pav-GFP/+), as well as control siblings (cnn/CyO; Ubi-Pav-GFP/+).
Immunofluorescence microscopy
Samples were fixed for 30–60 min with 4% formaldehyde in phosphate-buffered saline (PBS) and permeabilized for 30 min in PBST (0.1% Triton X-100 in PBS). Samples were then incubated overnight at 4°C with primary antibodies, washed three times with PBST for 20 min, incubated overnight at 4°C with Alexa Fluor–conjugated secondary antibodies (1:200; Molecular Probes, Eugene, OR), and washed again with PBST (three times for 20 min). For lysosome staining, testes were dissected into PBS, incubated with LysoTracker (conjugated with Alexa 594; Invitrogen, Carlsbad, CA) for 30 min, and fixed with 4% formaldehyde for 30 min, followed by a standard immunofluorescence staining procedure as described. Stained samples were mounted in Vectashield (H-1200; Vector Laboratories, Burlingame, CA). The primary antibodies used were mouse anti-fasciclin III (1:20; developed by C. Goodman, University of California, San Francisco, and obtained from the Developmental Studies Hybridoma Bank [DSHB], University of Iowa, Iowa City, IA), mouse anti–adducin-like (1:20; developed by H. D. Lipshitz, University of Toronto, and obtained from the DSHB), goat anti-Vasa (1:100; dC-13; Santa Cruz Biotechnology), rabbit anti-Vasa (1:100; Santa Cruz Biotechnology), rat anti-Vasa (1:40; developed by Allan Spradling, Carnegie Institution, and obtained from the DSHB), rabbit anti-Spd-2 (1:100; a gift from Maurizio Gatti, Sapienza University of Rome; Giansanti et al., 2008), and rabbit anti-anillin antibody (1:1300; a gift from Christine Field, Harvard University; Field and Alberts, 1995). Images were taken using a Leica TCS SP5 confocal microscope with a 63× oil immersion objective (numerical aperture, 1.4) and processed using Photoshop software (Adobe).
Time-lapse live-imaging methods
Newly eclosed Pav-GFP flies were dissected inside Drosophila culture medium containing Schneider's Drosophila medium and 10% fetal bovine serum. The ovaries were placed inside a sterile glass-bottom chamber covered with a gas-permeable membrane and were mounted on a three-axis, computer-controlled piezoelectric stage and imaged using an inverted microscope equipped with an electron multiplier cooled charge-coupled device camera. Image sequences were acquired every 600 s. The supplemental movies were generated using ImageJ software (National Institutes of Health, Bethesda, MD).
Supplementary Material
Acknowledgments
We thank Adelaide Carpenter, David Glover, Salud Llamazares, Cayetano Gonzalez, Thom Kaufman, Maurizio Gatti, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for reagents and Steve Doxsey for bringing Pavarotti/MKLP1 to our attention. We also thank Salud Llamazares and Cayetano Gonzalez for sharing unpublished results, Chip Ferguson and the Yamashita lab members for discussion, and anonymous reviewers for their constructive criticism. This work was supported by National Institutes of Health Grants R21HD067692 (to Y.M.Y.) and R01GM07200606 (to M.M. and Y.M.Y.). Y.M.Y. is supported by the MacArthur Foundation.
Abbreviations used:
- CB
cystoblast
- CC
cyst cell
- CySC
cyst stem cell
- GB
gonialblast
- GFP
green fluorescent protein
- GSC
germline stem cell
- MR
midbody ring
- MTOC
microtubule-organizing center
- SG
spermatogonia
- UAS
upstream activation sequence
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0541) on November 13, 2013.
REFERENCES
- Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. Pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger AW, Wilsch-Brauninger M, Marzesco AM, Bickle M, Lohmann A, Maliga Z, Karbanova J, Corbeil D, Hyman AA, Huttner WB. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun. 2011;2:503. doi: 10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Goss JW, Toomre DK. Both daughter cells traffic and exocytose membrane at the cleavage furrow during mammalian cytokinesis. J Cell Biol. 2008;181:1047–1054. doi: 10.1083/jcb.200712137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Yamashita YM. String (Cdc25) regulates stem cell maintenance, proliferation and aging in Drosophila testis. Development. 2011;138:5079–5086. doi: 10.1242/dev.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Kuo TC, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Casanueva MO, Mahowald AP, Kato M, Lauterbach D, Ferguson EL. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 2012;10:e1001357. doi: 10.1371/journal.pbio.1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Minestrini G, Harley AS, Glover DM. Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol Biol Cell. 2003;14:4028–4038. doi: 10.1091/mbc.E03-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini G, Mathe E, Glover DM. Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J Cell Sci. 2002;115:725–736. doi: 10.1242/jcs.115.4.725. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Morris LX, Spradling AC. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138:2207–2215. doi: 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Gould GW. Breaking up is hard to do—membrane traffic in cytokinesis. J Cell Sci. 2008;121:1569–1576. doi: 10.1242/jcs.018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Roth TM, Chiang CY, Inaba M, Yuan H, Salzmann V, Roth CE, Yamashita YM. Centrosome misorientation mediates slowing of the cell cycle under limited nutrient conditions in Drosophila male germline stem cells. Mol Biol Cell. 2012;23:1524–1532. doi: 10.1091/mbc.E11-12-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng XR, Matunis E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development. 2011;138:3367–3376. doi: 10.1242/dev.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, Johnston D, St, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–898. doi: 10.1016/s0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC. Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Yuan H, Cheng J, Hunt AJ. Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a001313. doi: 10.1101/cshperspect.a001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Perrimon N, Strutt DI. Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling. Genes Dev. 1999;13:1342–1353. doi: 10.1101/gad.13.10.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.