Lipid droplet formation and degradation are pivotal processes in preventing lipotoxicity and providing energy sources and signaling molecules. This is the first demonstration of lipid droplet turnover in yeast by microautophagy. Lipophagy is distinct from ER-phagy, mitophagy, and pexophagy and contributes to neutral lipid homeostasis by vacuolar lipolysis.
Abstract
Cytosolic lipid droplets (LDs) are ubiquitous organelles in prokaryotes and eukaryotes that play a key role in cellular and organismal lipid homeostasis. Triacylglycerols (TAGs) and steryl esters, which are stored in LDs, are typically mobilized in growing cells or upon hormonal stimulation by LD-associated lipases and steryl ester hydrolases. Here we show that in the yeast Saccharomyces cerevisiae, LDs can also be turned over in vacuoles/lysosomes by a process that morphologically resembles microautophagy. A distinct set of proteins involved in LD autophagy is identified, which includes the core autophagic machinery but not Atg11 or Atg20. Thus LD autophagy is distinct from endoplasmic reticulum–autophagy, pexophagy, or mitophagy, despite the close association between these organelles. Atg15 is responsible for TAG breakdown in vacuoles and is required to support growth when de novo fatty acid synthesis is compromised. Furthermore, none of the core autophagy proteins, including Atg1 and Atg8, is required for LD formation in yeast.
INTRODUCTION
Lipids are essential to sustain life, as they are fundamental constituents of biological membranes and metabolic energy stores and important players in many signaling pathways. The metabolic demand for lipids differs greatly in growing, differentiating, or resting cells. Therefore rapid adaptation of lipid content and composition in response to fluctuating environmental conditions is crucial to support cellular function. A key role in these lipid metabolic fluxes is played by fatty acids, which are the building blocks for membrane phospholipids and storage lipids but are subject to multiple modifications, such as elongation and desaturation, and degradation (Tehlivets et al., 2007). On the other hand, high concentrations of fatty acids are harmful and the cause of lipotoxicity, with detrimental pathological consequences (Listenberger et al., 2003; Kohlwein, 2010a). The storage of fatty acids as triacylglycerols (TAGs), which are packaged into cytosolic lipid droplets (LDs), provides a highly efficient way of dealing with fluctuating nutritional supply and physiological demand for fatty acids: fatty acids are stored as TAGs in times of excess and mobilized by lipolytic breakdown to support membrane proliferation or signaling processes in growing cells or oxidized to generate cellular energy in times of starvation (Zechner et al., 2012). Thus molecular mechanisms regulating LD formation and turnover have gained extensive biomedical attention in view of prevalent lipid-associated metabolic diseases, such as obesity and type 2 diabetes (Greenberg et al., 2011; Cusi, 2012). The neutral lipid core of LDs, consisting of TAGs and steryl esters, is delimited by a phospholipid monolayer that is decorated by a unique set of lipogenic and lipolytic enzymes and their regulators that catalyze lipid storage and degradation and interaction with other organelles (Farese and Walther, 2009; Walther and Farese, 2012; Kohlwein et al., 2013). Evidence suggests that LDs derive from the endoplasmic reticulum (ER) and may remain largely associated with this membrane after maturation, which may be functionally relevant to facilitate lipid and protein exchange and cellular dynamics (Szymanski et al., 2007; Kohlwein, 2010b; Jacquier et al., 2011; Wolinski et al., 2011). Release of fatty acids from TAG stores is controlled by LD-resident lipases and hydrolases in the cytosol, such as adipose triglyceride lipase (ATGL) and hormone-sensitive lipase in mammals (Zechner et al., 2012), Brummer lipase in Drosophila (Grönke et al., 2005), and Tgl3 and Tgl4 lipases in Saccharomyces cerevisiae (Athenstaedt and Daum, 2005; Kurat et al., 2006). The yeast Tgl4 lipase is a functional orthologue of mammalian ATGL and, together with Tgl3, shares structural features of the patatin domain–containing family of phospholipases (Kienesberger et al., 2009), indicating that the lipolytic process is highly conserved from yeast to mammals.
In addition to the lipolytic enzymes acting on cytosolic LDs, mammalian cells also express lysosomal hydrolases that catabolize neutral lipids. This process provides the main source of cellular cholesterol, which is taken up as cholesteryl ester from the bloodstream by receptor-mediated endocytosis (Jerome, 2010). Degradation of lipids in the yeast vacuole (the functional equivalent to mammalian lysosomes) is less well defined. However, some evidence suggests that Atg15 may be responsible for lipid degradation in the course of autophagic internalization of membrane-bound organelles, such as mitochondria and peroxisomes, into the vacuole (Epple et al., 2001; Teter et al., 2001). Of note, evidence suggests that in mammalian organisms, autophagic uptake and degradation of LDs by lysosomes (“lipophagy”) plays an important role in lipid metabolism and contributes to reverse cholesterol transport, and as such opposes atherosclerotic plaque formation (Singh et al., 2009a; Ouimet et al., 2011; Dugail, 2014). Thus, besides a highly regulated cytosolic lipolysis, lipophagy provides an additional important pathway to maintain cellular and organismal lipid and fatty acid homeostasis (for review see Dugail, 2014). Controversy exists, however, on whether a key protein in autophagic degradation, LC-3, also affects neutral lipid storage and LD formation (Shibata et al., 2009, 2010). Whether the conserved yeast orthologue of LC-3, namely Atg8, plays a role in neutral lipid homeostasis has not been resolved.
Two main mechanisms of autophagy exist, namely microautophagy and macroautophagy, which can act either selectively or nonselectively. Selective autophagic processes have been reported for various cellular components, such as mitochondria, peroxisomes, ribosomes, and ER, and are referred to as mitophagy, pexophagy, ribophagy, and ER-phagy, respectively (Rabinowitz and White, 2010). During microautophagy, pieces of the cytoplasm are directly engulfed by the lysosomal or vacuolar membranes, internalized, and degraded by resident hydrolases (acid lipases, esterases, proteases). Macroautophagy initiates by the formation of a double membrane that sequesters part of the cytoplasm and, upon completion (termed the autophagosome), fuses with the lysosome/vacuole. The origin of the autophagosomal membrane is quite controversial and may be derived from the ER, mitochondria, or plasma membrane (Ravikumar et al., 2010; Hamasaki et al., 2013). The autophagy machinery is highly conserved, and some 36 autophagy (Atg) proteins have been identified (Meijer et al., 2007; Reggiori and Klionsky, 2013). Autophagy is constitutively active at a basal level but highly inducible by various stress and starvation conditions, such as nitrogen or carbon limitation.
Lipid metabolism and autophagy are highly conserved processes, which led us to examine the molecular mechanisms and physiological role of lipophagy in yeast. This study identifies a unique subset of components of the autophagy machinery required for microautophagic degradation of LDs, including the vacuolar lipase Atg15. No indications were obtained that any of the key Atg proteins, such as Atg1 or Atg8, are required for TAG formation and their storage into cytoplasmic LDs in yeast.
RESULTS
Lipid droplets are taken up by vacuoles in yeast by a process resembling microautophagy
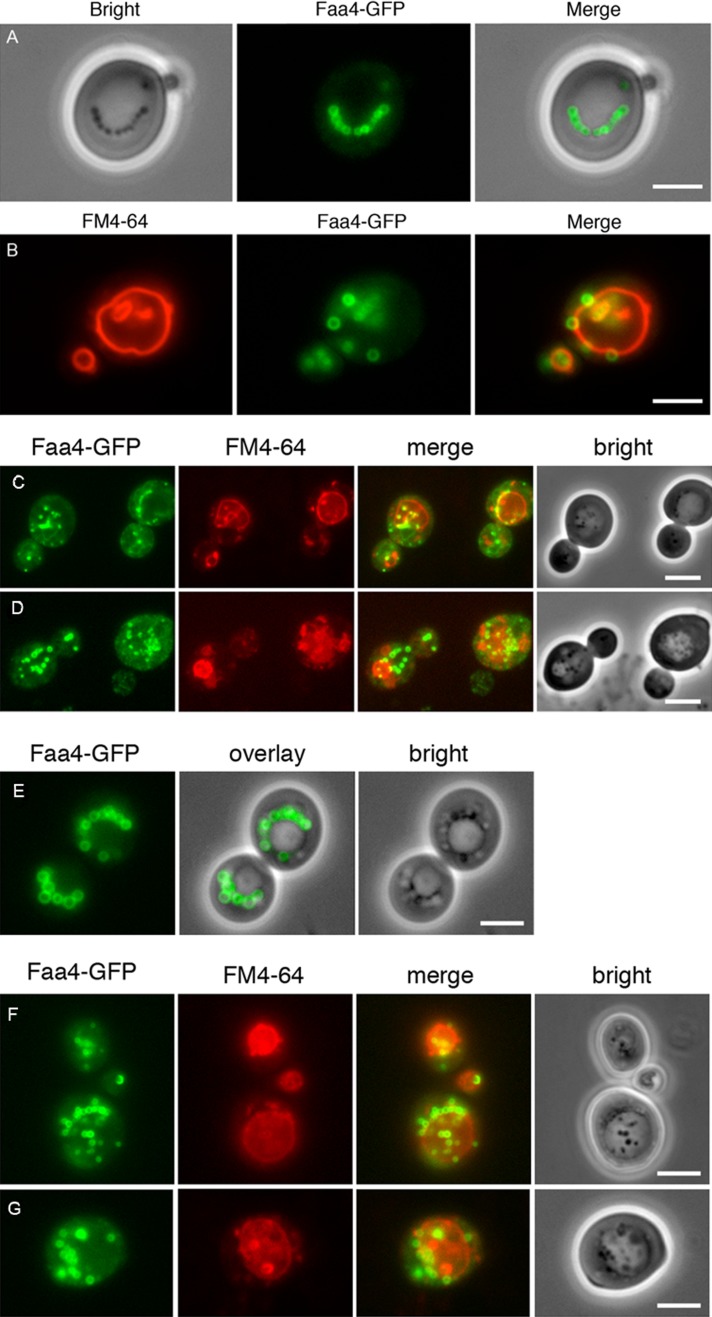
Although yeast LDs, like their mammalian counterparts, harbor a full set of lipases involved in TAG and steryl ester degradation (Kohlwein, 2010b; Kohlwein et al., 2013; Henry et al., 2012), internalization of LDs into the vacuole is frequently observed in growing cells. To characterize vacuolar LD uptake and the underlying molecular mechanisms in greater detail, we first used wild-type S. cerevisiae cells expressing from their chromosomal locus the lipid droplet–resident protein Faa4–green fluorescent protein (GFP; Kurat et al., 2006). Cells were grown in minimal media containing 0.5% glucose to the late stationary growth phase. Under this growth condition, numerous LDs are present in the cells, often in clusters, but frequently also localized in strings adjacent to the vacuole (Figure 1A). However, LDs were also frequently observed inside the vacuole and could easily be distinguished under the microscope from cytosolic LDs by their increased mobility (see later discussion). Internalization of the Faa4-GFP–labeled LDs into the vacuole was confirmed by staining the vacuolar membrane with FM4-64 (Figure 1B).
FIGURE 1:
Lipid droplet–vacuole interaction and uptake in glucose- and oleate-grown yeast cells. LDs are labeled with endogenously expressed Faa4-GFP in cells grown on 0.5% glucose for 21 h (A) and 46 h (B). LDs are typically localized in strings adjacent to the vacuole (A) or randomly distributed in the cytosol. They are also frequently observed inside the vacuole, especially in the stationary phase of growth (absence of glucose; B). Cells expressing Faa4-GFP were pregrown on glucose and subsequently shifted to oleate-containing media. After 6 (C) and 12 (D) h of incubation, LDs are massively induced in the cytosol and are also present inside the vacuoles. In stationary phase (28 h of incubation) distinct LDs are no longer detectable in the vacuole (E). After shift of these cells to fresh oleic acid–containing medium lacking a nitrogen source, LDs are rapidly incorporated into the vacuole: after 1 h (F) and 5 h (G). Vacuolar membranes are stained with FM4-64. Scale bar, 5 μm.
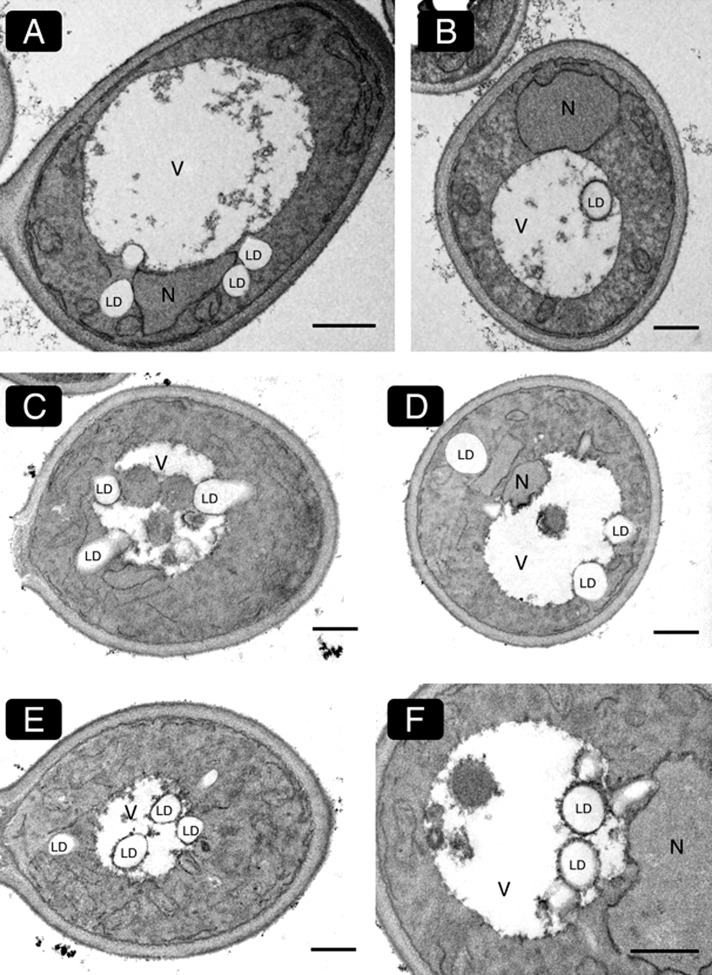
Because LD formation in growing cells is limited by the availability of fatty acids, which are preferentially channeled into membrane phospholipids (Kohlwein et al., 2013), we next grew cells in the presence of oleate, a condition that increases TAG synthesis and LD formation (Grillitsch et al. 2011). Indeed, after 6 h (Figure 1C) and 12 h (Figure 1D) of cultivation, massive LD proliferation was observed in the cytosol, and so was an increased appearance in the vacuole. LDs inside the vacuole were reduced in size compared with cytosolic LDs, and their Faa4-GFP fluorescence was attenuated (Figure 1, C and D). Live-cell phase contrast imaging again revealed a higher mobility of LDs inside the vacuole relative to those residing in the cytosol. In the late stationary growth phase, that is, after 28 h of incubation, LDs were no longer detectable inside the vacuole by fluorescence or phase contrast imaging (Figure 1E), indicating that vacuolar internalization of LDs leads to their subsequent degradation. Internalization of LDs into the vacuole was also confirmed at the electron microscopic level (Figure 2, A and B).
FIGURE 2:
Electron microscopy of vacuolar lipid droplet internalization. Cells were grown in the absence of a nitrogen source (A, B) or for 5 h in oleic acid–containing media (C–F) and processed for electron microscopy. Both conditions lead to a stimulated internalization of LDs into the vacuole. Various stages of LD internalization are shown. Lipid droplets that enter the vacuole are partially covered by an electron-dense vacuolar membrane (B, E; higher magnification in F). These morphological characteristics suggest that LD internalization into the vacuole occurs via microautophagy in yeast. Scale bar, 1 μm.
To further characterize the vacuolar incorporation of LDs, we next tested whether induction of autophagy stimulated their uptake. Cells were grown overnight in the presence of oleate and shifted to the same medium without a nitrogen source up to 8 h. Under these conditions, LDs were rapidly taken up by the vacuole (Figure 1, F and G). We also used coherent anti-Stokes Raman scattering (CARS; see later discussion) and electron microscopy to unequivocally confirm vacuolar localization of unlabeled LDs in living cells or in fixed and sectioned yeast cells, respectively. Data in Figure 2, C–F, show various stages of internalization of LDs into the vacuole after 5 h of incubation in the presence of oleate. From these electron microscopy images it is evident that LDs are typically associated with invaginations of the vacuolar membrane rather than any additional membranes such as autophagosomal membranes. These morphological data demonstrate that LD uptake into the vacuole occurs in a process resembling microautophagy. Similar observations were made under nitrogen starvation conditions that induce autophagy (see later discussion).
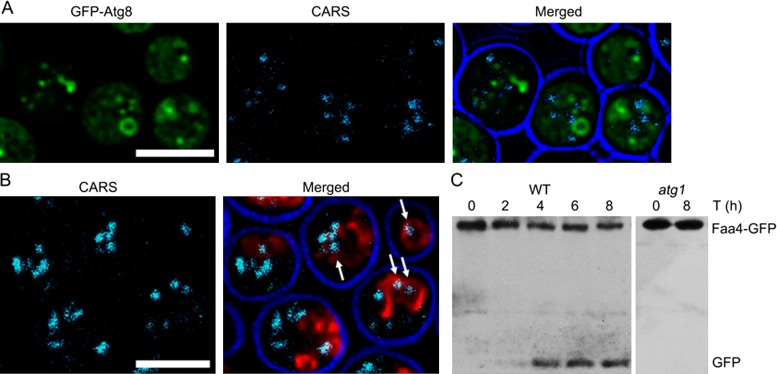
To further support the hypothesis that microautophagy is responsible for LD internalization into the vacuole, we expressed the autophagosomal marker GFP-Atg8 in ypt7 mutant cells. These mutants still can form autophagosomes, which are, however, unable to fuse with the vacuole (Kirisako et al., 1999). As expected, upon induction of autophagy, ample cup-shaped and ring-like GFP-Atg8–containing structures were present in the ypt7 cells. However, we never observed any of these structures surrounding LDs, consistent with the view that macroautophagy is not responsible for LD degradation (Figure 3A). As an alternative method to visualize LD uptake into the vacuole in living cells, we used label-free CARS microscopy, which yielded essentially identical results to Faa4-GFP– or BODIPY 493/503–labeled LDs (Figure 3B).
FIGURE 3:
Lipid droplets are degraded in the yeast vacuole upon induction of autophagy. (A) ypt7 cells expressing GFP-Atg8 show the accumulation of autophagosomes that lack LDs. (B) Detection of LDs inside the vacuole of wild-type cells with CARS imaging; vacuolar membranes are labeled with FM4-64. Cells were shifted to nitrogen starvation medium for 8 h in the presence of PMSF before microscopy to induce autophagy. Scale bar, 5 µm. (C) Western blot of cell extracts of wild-type cells expressing the LD marker Faa4-GFP, using an anti-GFP antibody. Late exponential cells grown in rich medium were shifted for 8 h to medium lacking a nitrogen source. The appearance of one or two bands at ∼27 kDa is indicative of vacuolar proteolytic processing of the Faa4-GFP fusion protein. This band is absent in atg1 cells.
Taken together, these data support the notion that LDs can be taken up and degraded by vacuoles by a process resembling microautophagy. Vacuolar internalization of LDs is observed in various stages of growth but is pronounced upon induction of autophagy under nitrogen-limiting conditions.
Core autophagic components are not required for LD formation in yeast
Some controversy exists as to the role of the Atg8 orthologue LC-3 in LD autophagy and/or LD biogenesis in mouse model systems (Shibata et al., 2009, 2010; Singh et al., 2009a). To address this issue, we investigated LD formation in mutants of the autophagy machinery, using Faa4-GFP as well as CARS microscopy. As shown in Supplemental Figure S1, atg1 and atg8, as well as atg15 mutants, are able to develop cytosolic LDs in growing cells that are morphologically indistinguishable from wild type. These observations exclude a significant role of Atg8 and other core components of autophagy in LD formation in yeast.
Identification of the molecular machinery of LD autophagy
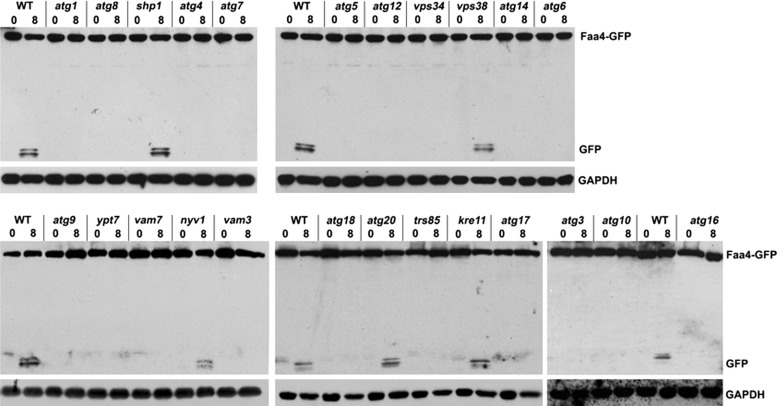
To identify the molecular components involved in LD autophagy, we used mutant strains expressing the LD markers Faa4-GFP (Figures 3C and 4; see later discussion) and Erg6-GFP (Supplemental Figure S2) and assessed their proteolytic processing in the vacuole. The remarkably stable β-barrel structure of GFP is more resistant to vacuolar proteolysis, and the appearance of one or two bands at ∼27 kDa is indicative of vacuolar internalization of the fusion protein (Cheong and Klionsky, 2008; Kraft et al., 2008; Manjithaya et al., 2010). The identity of these GFP-fusion protein–derived bands was confirmed by mass spectrometry (unpublished data). As expected, cleavage of Faa4-GFP was readily observed in wild-type cells under nitrogen-limiting conditions but was completely absent in mutants lacking the key autophagy regulator, Atg1 (Figure 3C). We next analyzed other atg mutants to determine the critical factors required for LD autophagy. We observed a block in Faa4-GFP and Erg6-GFP degradation in atg8 cells (Figure 4 and Supplemental Figure S2), as well as in mutants of the Atg8-activating machinery (atg3, atg4, atg5, atg7, atg10, atg12, and atg16). However, Shp1, an Atg8 cofactor that functions in macroautophagy and piecemeal autophagy of the nucleus (Krick et al., 2010), was not required. LD internalization was absent in cells lacking Atg9, which is required to deliver vesicles to the developing autophagosome (Mari et al., 2010), and was also blocked in mutants defective in the vacuole-specific phosphoinositide 3-kinase complex—mutants lacking the Vps34 kinase itself, the vacuole-specific factor Atg14, and the beclin homologue Atg6, but not Vps38, the Golgi-specific member of this complex. We also observed an essential function in LD autophagy for the vacuole fusion machinery that is involved in macroautophagy in yeast, except for Nyv1. The TRAPPIII-specific subunit Trs85, which recruits the GTPase Ypt1-containing complex to the vacuole and is implicated in autophagy, was also required. In contrast, the TRAPPII-specific subunit Kre11 (Lynch-Day et al., 2010) does not appear to be involved in LD autophagy.
FIGURE 4:
Lipid droplet autophagy requires the core autophagy machinery and additional factors. Western blots were prepared from crude extracts of the indicated mutant cells, which were grown to the late logarithmic growth phase in rich medium and shifted to synthetic minimal medium lacking nitrogen for 8 h. Blots were decorated with anti-GFP and anti-GAPDH antibodies.
Taken together, all members of the core machinery required for various types of autophagy are also involved in LD autophagy. We also identified several additional factors, such as Atg17 and Trs85, required for that process, whereas other organelle-specific autophagy proteins, such as Atg20, Nyv1, and Shp1, are not. Both LD marker proteins, Faa4-GFP and Erg6-GFP, yielded essentially identical results, confirming that the analysis indeed identified components relevant for LD autophagy. This analysis defines a unique subset of autophagy proteins that play an essential role in LD autophagy.
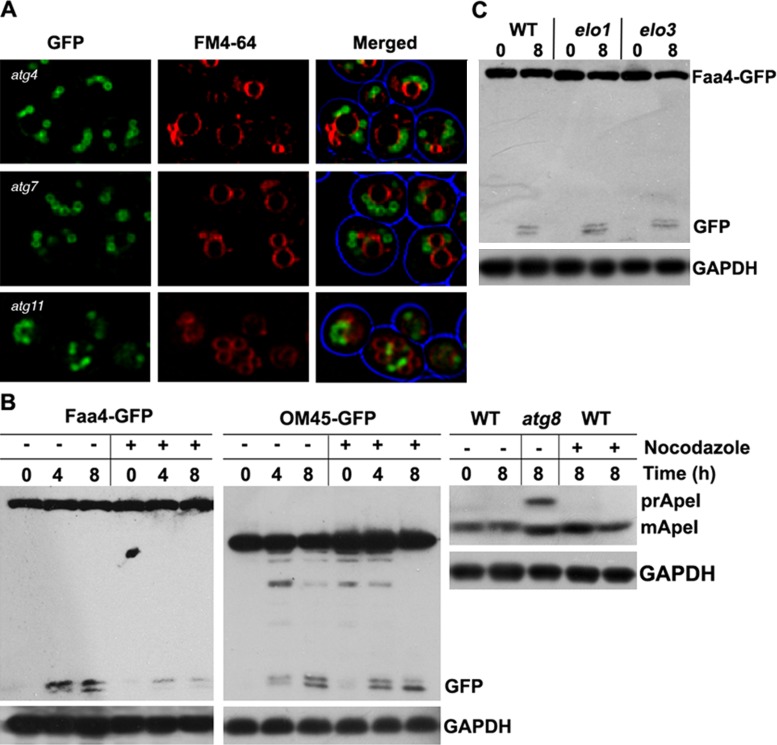
During macroautophagy, Atg11 is required to deliver cargo to the vacuole, as well as for assembly of the phagophore-assembly site, together with several other Atg proteins, such as Atg1 and Atg8 (Backues and Klionsky, 2012; Lipatova et al., 2012). Because we observed LDs frequently adjacent to the vacuole, we determined whether this localization depends on Atg proteins and phagophore assembly by analyzing LD localization in several autophagy mutants. Data summarized in Figure 5A show that autophagy is not required for LD recruitment to the vacuole.
FIGURE 5:
Lipid droplet autophagy requires tubulin. (A) atg4-, atg7-, and atg11-mutant cells expressing Faa4-GFP were shifted to synthetic minimal medium lacking nitrogen for 8 h. LDs are closely associated with the cytoplasmic site of the vacuolar membrane (labeled with FM4-64). Scale bar, 5 μm. (B) Western blots were prepared from crude extracts of wild-type cells expressing either Faa4-GFP or Om45-GFP or no marker, as indicated. Cells were incubated in synthetic minimal medium lacking nitrogen supplemented with 15 μg/ml nocodazole for 4 or 8 h. Blots were decorated with anti-GFP, anti–aminopeptidase I, or anti-GAPDH antibodies. Faa4-GFP degradation is strongly reduced, suggesting that nocodazole treatment inhibits LD internalization into the vacuole. In contrast, processing of Om45-GFP is not affected, consistent with previous results that tubulin is not required for mitophagy (Kanki et al., 2009). (C) Western blot of cell extracts prepared from Faa4-GFP–expressing elo1 and elo3 mutant cells, which display highly fragmented vacuoles (Kohlwein et al., 2001). Cells were grown to the late logarithmic growth phase in rich medium and shifted to synthetic minimal medium lacking nitrogen for 8 h. Both, elo1 and elo3 mutants show normal Faa4-GFP processing, indicating that vacuolar fragmentation does not affect LD autophagy. Blots were decorated with anti-GFP and anti-GAPDH antibodies.
LD autophagy depends on tubulin
We previously observed that actin is required for LD dynamics in growing cells, whereas tubulin destabilization did not affect this process (Wolinski et al., 2011). Thus we next analyzed whether tubulin is required for LD autophagy by treating cells with the tubulin-destabilizing drug nocodazole. As shown in Figure 5B, nocodazole caused a strong inhibition of LD autophagy. This is in marked contrast to mitophagy, as shown with the marker Om45-GFP, which was largely unaffected by the inhibitor. Processing of aminopeptidase I, which is delivered to the vacuole via the cytoplasm-to-vacuole pathway, was also not affected (Figure 5B). Because nocodazole induces vacuole fragmentation (Guthrie and Wickner, 1988), we also determined LD autophagy in elo1 and elo3 mutants, which display fragmented vacuoles (Kohlwein et al., 2001). No inhibition of Faa4-GFP cleavage was observed in these mutants (Figure 5C), indicating that tubulin is involved in targeting of LDs to the vacuole, independent of its function in maintaining vacuole morphology (Reggiori et al., 2005). This finding further corroborates the distinct difference between LD-phagy and ER/mito/pexophagy.
Vac8 is required for lipid droplet autophagy
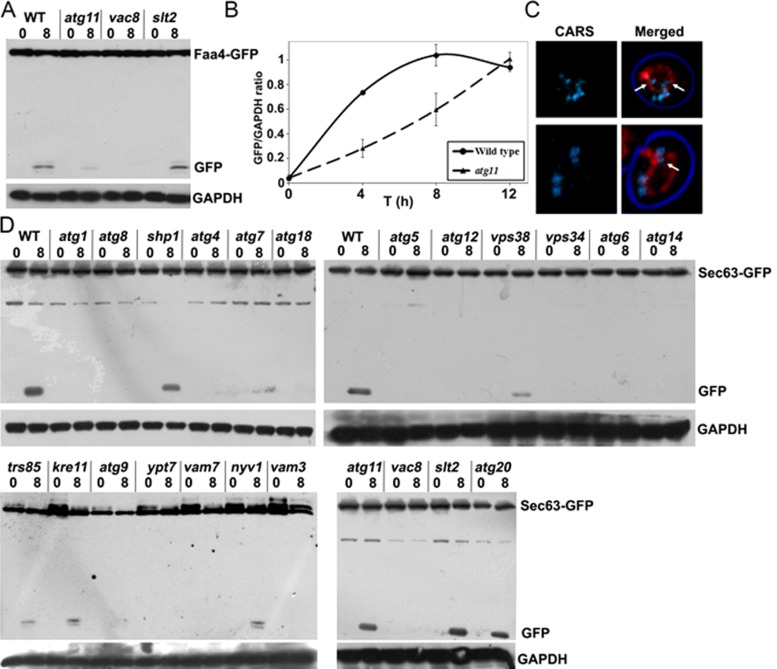
Selective autophagy requires specific adaptor proteins that often bind to receptors on the organelle or macromolecule and connect it to the core autophagy machinery (Suzuki, 2013). By testing these components for their role in LD autophagy, we found a requirement for the armadillo-repeat protein Vac8, which is involved in multiple vacuolar processes but not in general autophagy (Figure 6). In the atg11 mutant, Faa4-GFP processing was significantly delayed (Figure 6, A and B), indicating that the Atg11 protein may function as an efficiency factor rather than a key adaptor protein. To confirm the delayed uptake determined by vacuolar GFP cleavage of the LD marker, we also analyzed LD uptake by label-free CARS microscopy, which indeed showed LDs inside the vacuole (Figure 6C). On the other hand, the mitogen-activated protein kinase Slt2, a pathway recently implicated in several selective types of autophagy (Manjithaya et al., 2010; Mao et al., 2011), had no apparent function in LD autophagy.
FIGURE 6:
Lipid droplet autophagy requires selective adapters and differs from ER-phagy. (A) Protein extracts of various mutant cells expressing Faa4-GFP were grown to the late logarithmic growth phase in rich medium and shifted to synthetic minimal medium lacking nitrogen for the indicated time intervals. This analysis shows the requirement for Vac8 and a partial requirement for Atg11 for Faa4-GFP cleavage. Blots were decorated with anti-GFP and anti-GAPDH antibodies. (B) Quantification of cleaved Faa4-GFP at different time points after the shift to starvation medium in wild-type and atg11 mutant cells expressing Faa4-GFP relative to the GAPDH loading control. (C) CARS images of atg11-mutant cells shifted to nitrogen starvation medium for 8 h in the presence of PMSF. LDs are internalized into vacuoles of atg11 cells that are labeled with FM4-64. (D) Protein extracts from various mutant cells expressing the ER marker Sec63-GFP analyzed by Western blotting. Cells were grown to the late logarithmic growth phase in rich medium and shifted to synthetic minimal medium lacking nitrogen for indicated times. Blots were decorated with anti-GFP and anti-GAPDH antibodies. This analysis shows that LD autophagy is distinct from ER-phagy. See the text for details.
Lipid droplet autophagy is distinct from ER-phagy
Although the view of LDs as individual organelles has been largely accepted, extensive interactions with or even attachment to or a continuum with the ER membrane are frequently observed (Szymanski et al., 2007; Jacquier et al., 2011; Wolinski et al., 2011). Through this interaction, transmembrane proteins may even relocate from the ER to LDs (Jacquier et al., 2011). These close interactions raise the question of whether LDs are targeted by autophagy independently or degraded as part of the ER. To analyze the protein requirements for ER-phagy, we expressed Sec63-GFP in various atg mutants and examined the appearance of GFP fragments (Figure 6D). For the core autophagy machinery—the Atg8-activating machinery, Vps34, Atg6, Atg14, Atg9, and Atg18—we observed similar results as for LD autophagy. Of note, we also found defective Sec63-GFP processing in mutants lacking Atg5 and Atg12, which were reported to be dispensable for ER-phagy (Mijaljica et al., 2006). It should be noted, however, that studies on ER-phagy have also been conducted after induction of the unfolded protein response (UPR), which may be responsible for the observed differences in genes involved (Bernales et al., 2006). In contrast to LD-phagy, trs85-mutant cells were not fully blocked in ER turnover, although a substantial decrease in the appearance of vacuolar GFP was observed (Figure 6D). Deletion of Atg11 did not affect Sec63-GFP internalization into the vacuole, whereas deletion of Atg15 completely blocked its uptake (see discussion of Figure 7), in contrast to LD internalization. These data are in marked contrast to findings obtained for Faa4-GFP (and Erg6-GFP), arguing that LD autophagy requires a distinct set of proteins and is not merely a segment of ER-phagy.
FIGURE 7:
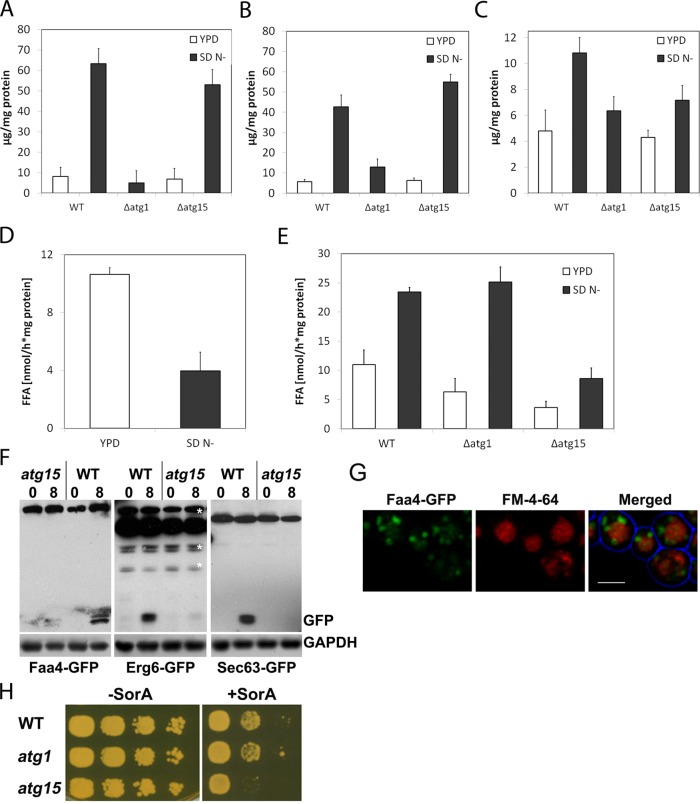
The yeast vacuole has lipase activity that depends on Atg15. Steryl ester (A), triacylglycerol (B), and free fatty acid (C) content of vacuolar fractions of wild-type, atg1, and atg15 cells grown on either rich (YPD) or autophagy-inducing (SD N−) media. Lipase activity in isolated lipid droplet (D) and vacuole fractions (E). Western blot (F) of proteins in crude extracts of wild-type and atg15 cells expressing either Faa4-GFP or Erg6-GFP to analyze lipid droplet autophagy or Sec63-GFP to determine ER-phagy. Cells were grown to the end of the logarithmic growth phase and shifted to SD N− medium for 8 h. Single optical sections (G) of atg15-mutant cells expressing Faa4-GFP (green) and labeled with FM4-64. Cells were cultivated in SD N− for 8 h, showing accumulation of GFP in the vacuole lumen. Scale bar, 5 µm. Lack of the vacuolar lipase Atg15 renders cells sensitive to the inhibitor soraphen A, which blocks de novo fatty acid synthesis (H).
LD autophagy is physiologically relevant and supports growth
Internalization of LD into the vacuole by autophagy requires the activity of lipases to make their lipid constituents available for the cell. Thus we first aimed at identifying lipase activities in vacuolar fractions that were purified according to Zinser and Daum (1995). External LD-resident lipases (Athenstaedt and Daum, 2005; Kurat et al., 2006) and other proteins were removed from purified vacuoles by trypsin treatment, thus leaving putative vacuolar lipases in the lumen intact; the vacuole membrane is known to be resistant against trypsin (Horst et al., 1999). In highly purified vacuoles from nitrogen-starved wild-type cells we observed ∼10-fold increase in vacuolar neutral lipid levels compared with logarithmically grown cells on yeast extract/peptone/glucose medium, further demonstrating the massive internalization of LDs under starvation conditions in wild-type cells (Figure 7, A–C). Similarly, increased neutral lipid levels were observed in vacuoles prepared from atg15 cells, consistent with a proposed role of Atg15 as a vacuolar TAG lipase in Fusarium graminearum (Nguyen et al., 2011; Figure 7, A–C). In contrast, hardly any neutral lipids were detectable in purified vacuoles from atg1-mutant cells, confirming the essential role of Atg1 in LD autophagy (Figure 7). To analyze this further, we next determined cellular lipase activities in these mutants. Lipase activities in cytosolic LD fractions under autophagy-inducing conditions were reduced in wild-type cells (Figure 7D), whereas similarly increased activities were observed in vacuole fractions from wild-type and atg1-mutant cells. In marked contrast, lipase activity remained at a very low level in vacuoles from atg15-mutant cells, independent of growth conditions (Figure 7E). Of note, we never observed internalization of GFP-tagged variants of the major cytosolic TAG lipases Tgl3 and Tgl4 into the vacuole, indicating that these lipases are stripped off during LD autophagy. Thus we conclude that vacuolar lipase activity is, for the most part, executed by Atg15. Furthermore, analysis of LD turnover in atg15 cells using Faa4-GFP or Erg6-GFP as markers also showed only a very minor vacuolar GFP band (Figure 7F), indicating that the overall turnover rate of LDs is drastically reduced in atg15-mutant cells. Of interest, deletion of Atg15 led to lumenal vacuolar staining by the FM4-64 dye, indicating that it may interact with nondegradable (membrane) lipids inside the vacuole. To corroborate the physiological relevance for degradation of LDs by the vacuole, we grew atg1, atg15, and wild-type cells in the presence of the de novo fatty acid synthesis inhibitor soraphen A. Whereas wild-type and atg1 mutants showed the same level of resistance, growth of atg15 mutants was significantly reduced (Figure 7G). Thus internalization of LDs into the vacuole, in the absence of the Atg15 lipase, limits the availability of fatty acids to sustain growth; atg1 mutants, on the other hand, retain LDs in the cytosol, where they remain accessible to lipolytic degradation by Tgl3 and Tgl4 lipases.
DISCUSSION
Triacylglycerol accumulation and its turnover by lipases are of great biomedical interest in view of the pandemic dimensions of lipid (storage)-associated disorders. The discovery in recent years of major metabolic triacylglycerol lipases and steryl ester hydrolases in mammals (Zechner et al., 2009, 2012; Ghosh, 2012) and yeast (Athenstaedt and Daum, 2005; Köffel et al., 2005; Kurat et al., 2006; Kohlwein et al., 2013) has led to a fairly defined picture of the key players in neutral lipid turnover in metabolically active cells. Major questions remain, however, regarding the regulation of these processes and the specific role and metabolic channeling of lipid degradation products. Lipid droplets play a key role in neutral lipid homeostasis, and their formation and mechanisms of lipid deposition and turnover are subjects of intensive study (Walther and Farese, 2012). Recent evidence from mouse model systems suggested that LDs may be degraded by autophagy, indicating that, in addition to the existing and highly efficient set of LD-resident cytosolic lipases, complete degradation of the organelle in lysosomes/vacuoles may contribute to lipid homeostasis as well (Singh et al., 2009a). Some controversy, however, exists about the role of a key autophagy protein, LC-3, and its conjugation system (orthologue of yeast Atg8), which was also suggested to contribute to LD formation (Shibata et al., 2009, 2010). Furthermore, several other atg-knockout mouse mutants show lean phenotypes, which contradicts an essential function of autophagy in organismal neutral lipid homeostasis (Zhang et al., 2009; Singh et al., 2009b). Nevertheless, the recent implication of lipophagy in Huntington's disease and in reverse cholesterol transport from foam cells during development of atherosclerosis (Martinez-Vicente et al., 2010; Ouimet et al., 2011) has greatly stimulated biomedical interest in LD autophagy (Singh and Cuervo, 2011; Dugail, 2014).
This is the first report to show that in the yeast S. cerevisiae, LDs are engulfed and degraded by vacuoles via an autophagic process morphologically resembling microautophagy. We demonstrate that LD autophagy in yeast relies on the core autophagy machinery, with some exceptions, making LD-phagy distinct from ER-phagy or other organelle-specific degradation processes. In mammalian cells, LD autophagy is augmented in response to external stimuli that promote LD accumulation, such as addition of oleate (Singh et al., 2009a). Similarly, incubation of yeast cells in the presence of oleate also stimulated vacuolar LD uptake. We assume that the presence of oleate triggers a starvation response, which promotes LD autophagy, or leads to a sequestration of neutral lipids away from cytosolic lipases. Of note, under starvation conditions, cytosolic lipase activity governed by Tgl3 and Tgl4 lipases dropped significantly, with a concomitant increase in vacuolar lipase activity. This stimulation of lipolytic activity in the vacuole was not dependent on Atg1 but was dependent on the vacuolar lipase Atg15. We observed rather broad substrate specificity for this enzyme, which harbors a putative catalytic triad consisting of His-435, Asp-387 (or Asp-421), and Ser-332 (Epple et al., 2001; Teter et al., 2001). The yeast enzyme worked equally well on steryl esters and triacylglycerols, which is consistent with observations for other members of the acid lipase family, such as lysosomal lipase, endothelial lipase, and carboxyl ester hydrolases, some of which additionally hydrolyze phospholipids (Hui and Howles, 2002; McCoy et al., 2002).
What is the physiological relevance of LD autophagy in yeast? Given that the known yeast triacylglycerol lipases Tgl3, Tgl4, and Tgl5 and steryl ester hydrolases Tgl1, Yeh1, and Yeh2 are dispensable for growth and long-term survival (Athenstaedt and Daum, 2005; Köffel et al., 2005; Kohlwein, 2010b), we propose that autophagic degradation of LDs may be a potential mechanism to support viability in the absence of carbon sources. Mutants lacking cytosolic lipases remain viable for >12 d under starvation conditions in buffered media. It is likely that these mutants benefit from accumulated TAG stores, which may be accessible to autophagic degradation in the absence of other carbon sources. Even in proliferating cells, vacuolar degradation of LDs clearly provides an advantage under conditions of attenuated de novo fatty acid synthesis: inhibition of de novo fatty acid synthesis renders cells that are unable to express vacuolar lipase more sensitive than wild-type cells or atg1 cells that are unable to undergo autophagy. This observation clearly demonstrates that LD autophagy and vacuolar breakdown of the neutral lipid stores contribute significantly to fatty acid and lipid homeostasis in growing cells. In the absence of the key autophagy protein Atg1, LDs remain in the cytosol and, therefore, accessible to cytosolic lipolysis. In the absence of Atg15, vacuolar LD uptake leads to a shortage of TAG degradation products presumably required for membrane lipid synthesis and cell proliferation (Kurat et al., 2006 2009).
A major question remains to be solved, namely the export from the vacuole of massively accumulating free fatty acids and sterols resulting from phospholipid, triacylglycerol, and steryl ester breakdown. So far, no fatty acid or sterol export proteins have been identified. Some evidence derived from electron microscopic investigation of mutant strains accumulating lipids in the vacuole suggests that Atg22 might be a candidate in that process, which, however, requires further biochemical confirmation. Of note, absence of Atg17, which plays a role in LD internalization into the vacuole, renders cells sensitive to the presence of oleic acid (Lockshon et al., 2007), further supporting the physiological significance of LD autophagy in yeast to maintain fatty acid and neutral lipid homeostasis.
MATERIALS AND METHODS
Yeast strains and media
All strains used in this study were derived from S. cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). The kanamycin selection marker in strains expressing Faa4-GFP, Erg6-GFP, and Sec63-GFP from the O'Shea collection (Huh et al., 2003) was swapped for the clonNAT marker, selected for nourseothricin resistance, and subsequently used for synthetic genetic array technology (Tong and Boone, 2006). In-frame insertion was checked by colony PCR and fluorescence microscopy. Cells were grown at 30˚C on standard YPD medium containing 1% yeast extract, 2% glucose, and 2% peptone or on minimal medium (YNB) containing 0.17% yeast nitrogen base without ammonium sulfate (Difco, Franklin Lakes, NJ) at pH 6.0. When required, media were supplemented with 30 mg/l leucine, 20 mg/l histidine, and 30 mg/l uracil.
For growth on glucose, YNB medium was supplemented with 0.5% ammonium sulfate and 0.5% glucose. Oleate medium consisted of YNB supplemented with 0.5% ammonium sulfate, 0.05% yeast extract, 0.1% oleic acid, and 0.05% Tween 80. SD N− medium contained 0.17% YNB without amino acids and ammonium sulfate, 2% glucose. SD C− contained 0.17% YNB and 0.5% ammonium sulfate. For GFP-ATG8 expression, pUG36-Ura/ATG8 was transformed into cells; positive transformants were selected on plates containing uracil-free minimal medium with 0.67% YNB, 0.5% ammonium sulfate, and 2% glucose supplemented with the required amino acids (Eisenberg et al., 2009).
Biochemical methods
SDS–PAGE and Western blotting were performed according to established procedures. Blots were decorated using monoclonal GFP antibody (Roche Diagnostics, Mannheim, Germany) and polyclonal rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody. Protein concentration was determined using the Pierce BCA Protein assay kit (Pierce Biotechnology, Rockford, IL), according to the manufacturer's instructions.
Vacuoles were isolated essentially according to Zinser and Daum (1995), followed by trypsin treatment and an additional centrifugation step. Spheroplasts were washed with 1.2 M sorbitol, 20 mM K-PO4 buffer, pH 7.4, resuspended in breakage buffer containing 12% Ficoll, 0.2 mM EDTA, and 10 mM Mes/Tris, pH 6.9, supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), and homogenized using a Dounce homogenizer with a loose pestle (Wheaton, Millville, NJ). The suspension was overlaid with one volume of breakage buffer with 1 mM PMSF and centrifuged for 1 h at 100,000 × g (SW28 rotor; Beckman, Fullerton, CA). The floating top layer was gently resuspended in breakage buffer with 1 mM PMSF using a homogenizer with a loose pestle, overlaid with one-half volume of 8% Ficoll, 0.2 mM EDTA, and 10 mM Mes/Tris, pH 6.9, with 1 mM PMSF, overlaid with one-half volume of 4% Ficoll, 0.2 mM EDTA, and 10 mM Mes/Tris, pH 6.9, with 1 mM PMSF, and centrifuged for 1 h at 100,000 × g. The top layer was resuspended in 4% Ficoll, 0.6 M sorbitol, 0.2 mM EDTA, and 10 mM Mes/Tris, pH 6.9, and overlaid with one volume of 0.25 M sorbitol, 0.2 M EDTA, and 10 mM Mes/Tris, pH 6.9, and centrifuged for 30 min at 100,000 × g. The floating lipid droplet fraction was collected and the pellet resuspended in 500 μl of 4% Ficoll, 0.6 M sorbitol, 0.2 mM EDTA, and 10 mM Mes/Tris, pH 6.9, and 200 μg/ml trypsin was added, with incubation on ice for 15 min. The same buffer, 14 ml, was added, overlaid with one volume of 0.25 M sorbitol, 0.2 M EDTA, and 10 mM Mes/Tris, pH 6.9, with centrifugation for 30 min at 100,000 × g. The pellet containing purified vacuoles was resuspended in 0.25 M sucrose, 1 mM EDTA, and 1 mM dithiothreitol (DTT).
Lipid analysis
For lipid analysis of vacuole fractions, lipids were extracted with chloroform/methanol 2:1 (vol/vol) and analyzed by TLC on silica gel plates (Merck, Darmstadt, Germany), as described (Schneiter and Daum, 2006), using chloroform/methanol/water 32.5:12.5:2 (vol/vol/vol) as solvent for phospholipids and petrolether/diethylether/acetic acid 32:8:2 (vol/vol/vol) for neutral lipids. Lipids were visualized by dipping the plates in 0.5% MnCl2 and 3.2% H2SO4 and charring at 120ºC and were quantified by densitometric scanning at 450 nm (TLC Scanner 3; CAMAG, Wilmington, NC).
Lipase activity assay
Lysates were incubated in a final volume of 100 μl buffer A (0.25 M sucrose, 1 mM EDTA, 1 mM DTT, 20 μg/ml leupeptin, 2 μg/ml antipain, 1 μg/ml pepstatin, pH 7.0) with TAG substrate in a water bath for 60 min at 37°C. Substrate was prepared by emulsifying 0.33 μM triolein (Sigma-Aldrich, St. Louis, MO), 0.45 μM phosphatidylcholine/phosphatidylinositol (3:1, Sigma-Aldrich), 0.5% defatted bovine serum albumin (Carl Roth, Karlsruhe, Germany) and [9,10-3H]triolein (10,000 cpm/μl; Perkin Elmer Life Sciences, Waltham, MA) as a radioactive tracer, as described (Holm et al., 2001). Reactions were terminated by addition of 3.25 ml of methanol/chloroform/heptane (10:9:7) and 1 ml of 0.1 M potassium carbonate and 0.1 M boric acid, pH 10.5, and free fatty acids were extracted by vortexing. After centrifugation (800 × g, 15 min), radioactivity in 1 ml of the upper phase was determined by liquid scintillation counting.
Microscopy
Wide-field fluorescence microscopy (Figures 1 and 2) was performed using a Zeiss Axioskop microscope (Carl Zeiss, Sliedrecht, Netherlands) with a Princeton Instruments 1300Y digital camera. The GFP signal was detected using a 470/40-nm bandpass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm bandpass emission filter. Vacuoles were stained by adding FM4-64 (final concentration 10 μM) to the cultures. FM4-64 was visualized with a 546/12-nm bandpass excitation filter, a 560-nm dichromatic mirror, and a 575/640-nm bandpass emission filter. Confocal fluorescence microscopy was performed on a Leica SP5 confocal microscope (Leica Microsystems, Mannheim, Germany) with spectral detection and a Carl Zeiss LSM510 (Carl Zeiss, Jena, Germany) with photomultiplier tubes (Hamamatsu Photonics, Hamamatsu City, Japan). GFP was excited at 488 nm with an argon laser, and emission was detected using a 500- to 550-nm bandpass emission filter. FM 4-64 (Invitrogen, Carlsbad, CA) was excited at 543 nm using a helium neon laser (Lasos, Jena, Germany), and emission was detected using a 565- to 615-nm bandpass emission filter. BODIPY 493/503 (Invitrogen) was excited at 488 nm and emission detected between 500 and 530 nm (spectral detector). CARS images were acquired on a Leica SP5 confocal microscope, using a High Q picoEmerald laser (High Q, Rankweil, Austria) with optical parametric oscillator (APE, Berlin, Germany) and nondescanned detector in forward-CARS mode tuned to 2845 cm−1.
Deconvolution of fluorescence images was performed using Huygens Pro 4.0 (Scientific Volume Imaging). Images were adjusted for brightness and contrast and assembled using Photoshop CS5 (Adobe).
For electron microscopy, cells were fixed in 1.5% KMnO4 and further processed as detailed (Waterham et al., 1993).
Supplementary Material
Acknowledgments
We thank the members of the van der Klei and Kohlwein laboratories for helpful discussions. Soraphen A was a kind gift of Klaus Gerth, Helmholtz-Zentrum für Infektionsforschung, Braunschweig, Germany. This work was supported by grants from the Netherlands Organisation for Scientific Research/Earth and Life Sciences to T.v.Z. M.K. and H.F.H. were supported by the PhD program “Molecular Enzymology” funded by the Austrian Science Fund, which also funded project F3005 SFB Lipotox to S.D.K.
Abbreviations used:
- CARS
coherent anti-Stokes Raman scattering
- GFP
green fluorescent protein
- LD
lipid droplet
- TAG
triacylglycerol
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0448) on November 20, 2013.
*Present address: Department of Pediatrics, Center for Liver, Digestive, and Metabolic Diseases, University Medical Center Groningen, University of Groningen, 9700 RB Groningen, Netherlands.
The authors declare no conflict of interest.
REFERENCES
- Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–37309. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- Backues SK, Klionsky DJ. Atg11: a Rab-dependent, coiled-coil membrane protein that acts as a tether for autophagy. Autophagy. 2012;8:1275–1278. doi: 10.4161/auto.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLOS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–426. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- Cusi K. Role of obesity and lipotoxicity in the development of nonalcoholic steatohepatitis: pathophysiology and clinical implications. Gastroenterology. 2012;142:711–725. doi: 10.1053/j.gastro.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Dugail I. Lysosome/lipid droplet interplay in metabolic diseases. Biochimie. 2014;96:102–105. doi: 10.1016/j.biochi.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. Early steps in reverse cholesterol transport: cholesteryl ester hydrolase and other hydrolases. Curr Opin Endocrinol Diabetes Obes. 2012;19:136–141. doi: 10.1097/MED.0b013e3283507836. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillitsch K, Connerth M, Köfeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta. 2011;1811:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Guthrie BA, Wickner W. Yeast vacuoles fragment when microtubules are disrupted. J Cell Biol. 1988;107:115–120. doi: 10.1083/jcb.107.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Olivecrona G, Ottosson M. Assays of lipolytic enzymes. Methods Mol Biol. 2001;155:97–119. doi: 10.1385/1-59259-231-7:097. [DOI] [PubMed] [Google Scholar]
- Horst M, Knecht EC, Schu PV. Import into and degradation of cytosolic proteins by isolated yeast vacuoles. Mol Biol Cell. 1999;10:2879–2889. doi: 10.1091/mbc.10.9.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hui DY, Howles PN. Carboxyl ester lipase. structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res. 2002;43:2017–2030. doi: 10.1194/jlr.r200013-jlr200. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Jerome WG. Lysosomes, cholesterol and atherosclerosis. Clin Lipidol. 2010;5:853–865. doi: 10.2217/clp.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger PC, Oberer M, Lass A, Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009;(Suppl):S63–S68. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köffel R, Tiwari R, Falquet L, Schneiter R. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol Cell Biol. 2005;25:1655–1668. doi: 10.1128/MCB.25.5.1655-1668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD. Obese and anorexic yeasts: experimental models to understand the metabolic syndrome and lipotoxicity. Biochim Biophys Acta. 2010a;1801:222–229. doi: 10.1016/j.bbalip.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Kohlwein SD. Triacylglycerol homeostasis: insights from yeast. J Biol Chem. 2010b;285:15663–15667. doi: 10.1074/jbc.R110.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD, Veenhuis M, van der Klei IJ. Lipid droplets and peroxisomes—key players in cellular lipid homeostasis. Or: A matter of fat—store ‘em up or burn‘ em down. Genetics. 2013;193:1–50. doi: 10.1534/genetics.112.143362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci USA. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D, Surface LE, Kerr EO, Kaeberlein M, Kennedy BK. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics. 2007;175:77–91. doi: 10.1534/genetics.106.064428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Jain S, Farré JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol. 2010;189:303–310. doi: 10.1083/jcb.200909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MG, Sun G-S, Marchadier D, Maugeais C, Glick JM, Rader DJ. Characterization of the lipolytic activity of endothelial lipase. J Lip Res. 2002;43:921–929. [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in nonselective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–106. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. Endoplasmic reticulum and Golgi complex: contributions to, and turnover by, autophagy. Traffic. 2006;7:1590–1595. doi: 10.1111/j.1600-0854.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- Nguyen LN, Bormann J, Le GT, Stärkel C, Olsson S, Nosanchuk JD, Giese H, Schäfer W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet Biol. 2011;48:217–224. doi: 10.1016/j.fgb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Daum G. Extraction of yeast lipids. Methods Mol Biol. 2006;313:41–45. doi: 10.1385/1-59259-958-3:041. [DOI] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- Shibata M, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Commun. 2010;393:274–279. doi: 10.1016/j.bbrc.2010.01.121. [DOI] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Titorenko VI, Swaving GJ, Harder W, Veenhuis M. Peroxisomes in the methylotrophic yeast do not necessarily derive from pre-existing organelles. EMBO J. 1993;12:4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124:3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jina S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Nat Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.