Abstract
Smoothened (Smo) is a member of the Frizzled (FzD) class of G-protein-coupled-receptors (GPCRs), and functions as the key transducer in the Hedgehog (Hh) signalling pathway. Smo has an extracellular cysteine-rich domain (CRD), indispensable for its function and downstream Hh signalling. Despite its essential role, the functional contribution of the CRD to Smo signalling has not been clearly elucidated. However, given that the FzD CRD binds to the endogenous Wnt ligand, it has been proposed that the Smo CRD may bind its own endogenous ligand. Here we present the NMR solution structure of the Drosophila Smo CRD, and describe interactions between the glucocorticoid budesonide (Bud) and the Smo CRDs from both Drosophila and human. Our results highlight a function of the Smo CRD, demonstrating its role in binding to small molecule modulators.
Smoothened (Smo), a member of the Frizzled (FzD) class of G-protein coupled receptors (GPCRs) is an integral member of the Hedgehog (Hh) signal transduction cascade, first identified through genetic analysis in fruit fly Drosophila melanogaster1–4. The Hh pathway plays an important role during embryogenesis controlling cell maturation, differentiation and proliferation5,6. Disruption of Hh signalling during development leads to a plethora of developmental anomalies7,8. Conversely, ectopic Hh signalling during adult life contributes to tumour development and progression, particularly in cancers including medulloblastoma, basal cell carcinoma and rhabdomyosarcoma9. Consequently, the pathway is an important therapeutic target in oncology and regenerative medicine.
The Hh signal is transduced in a sequential manner that is initiated by the endogenous ligand–Indian, Sonic or Desert Hh binding to the twelve trans-membrane protein Patched (Ptc) to attenuate its housekeeping inhibition of Smo10–13. Smo then translocates to the cell membrane in Drosophila, or the primary cilium in vertebrates, and induces downstream signalling to the Gli/Cubitus interruptus (Ci) transcription factors5,13. The crystal structure of the seven trans-membrane (7TM) domain of human Smo was recently elucidated14. This structure revealed the canonical GPCR 7TM helical fold along with the conformation of the extra-cellular loop (ECL) and an extracellular linker domain (ECLD), both of which are stabilized by disulphide bonds between the conserved cysteine residues14,15. However, the major portion of the Smo extracellular domain (ECD) is the cysteine-rich domain (CRD) which was not included in the crystal structure of the Smo receptor14.
The CRDs of FzD family GPCRs possess modest homology. Nevertheless, the cysteines in this domain are highly conserved16,17. While the CRD of FzD plays an essential role in Wnt ligand binding and receptor dimerization16,18 the role of the Smo CRD is not clear although it has been shown that the Smo CRD is indispensable for Hh signalling in both Drosophila and vertebrates19,20. Drosophila and vertebrate Smo lacking the CRD fail to cycle to the plasma membrane or primary cilium to induce maximum signalling19,20. Furthermore, in Drosophila CRD deletion mutants fail to dimerize, suggesting that the CRD may govern Smo dimerization21. Based upon its functional importance, and the observation that the related FzD CRD binds to Wnt, it has been speculated that the Smo CRD may facilitate binding of an as yet unidentified, endogenous small molecule that modulates Smo signalling activity in the presence of Hh19. Recent studies have identified two major groups of small molecules capable of modulating Smo signalling activity: hydroxyl-sterols and glucocorticoids, both of which modulate Hh signalling through governing sub-cellular localization and activity of Smo22,23. Twenty hydroxycholesterol (20-OHC) and budesonide (Bud) have been reported to bind to a Smo domain distinct from the orthosteric site present in the trans-membrane core, previously demonstrated to bind the inhibitory small molecule cyclopamine22–24. Recent reports also indicate that 20-OHC binds the vertebrate Smo CRD25,26. Therefore, we hypothesized that the CRD of Smo may act as an allosteric binding site for Bud and additional small molecule Smo modulators.
To test this hypothesis, and to gain more insight into the functional significance of the Smo CRD, we performed structure-function studies. Here, we present the first solution structure of the Drosophila Smo CRD. We demonstrate that the CRD possesses a conserved pocket that contributes to the binding and allosteric regulation by the small molecule Bud.
Results
The Smo CRD is essential for Hh signalling
The Smo CRD is essential for regulating its subcellular localization and signalling19–21. The cysteines in this domain are conserved from Drosophila through vertebrates (Fig.1). Despite this essential role, the functional contribution of the CRD to Smo regulation has remained unclear. To gain functional insight into the role of the CRD, we generated a mutant lacking this domain (ΔCRD; V85-K202), and tested its ability to rescue Hh-dependent reporter gene induction following knockdown of endogenous smo in cultured Clone 8 (Cl8) cells27. Consistent with previous in vivo studies, we found that deletion of this domain ablates Smo signalling capacity in vitro (Fig 2). In order to better dissect this essential functional domain, we studied the structure of the Smo CRD using NMR (Nuclear Magnetic Resonance) methods.
Figure 1. Sequence alignment of the Smo CRD.
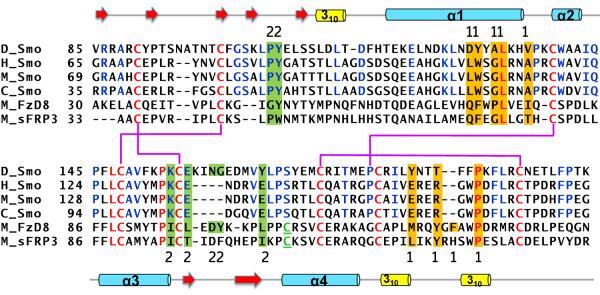
Primary sequence alignment of the Drosophila (D), human (H), mouse (M) and chicken (C) Smo CRD with that of mouse FzD8 CRD and mouse secreted Frizzled Related Protein 3 (sFRP3). The residues conserved in Smo and FzD CRD are shown in red, whereas the residues conserved only in the Smo CRD are shown in blue. The cysteine in FzD not conserved in Smo is underlined and in green. The disulphide bond pattern for the Smo CRD is shown in thick purple lines. The secondary structure elements are shown above and below the primary sequence. The residues highlighted in orange indicate the “site 1” residues of mouse FzD8 that interact with the palmitate modification on the ligand Wnt. The residues highlighted in green indicate the “site 2” on mouse FzD8 that interact with the amino acid sidechains on the opposite side of Wnt. The sequence alignment was generated using ClustalW2.
Figure 2. Identification of cysteines essential for Smo signalling.
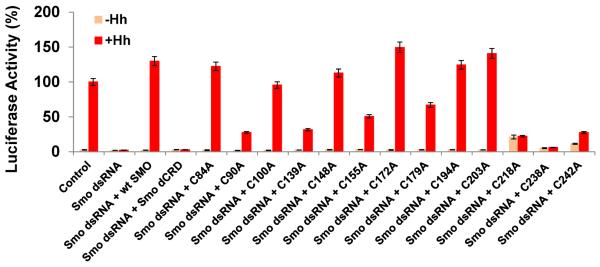
CRD residues C90, C139, C155, C179 and the Smo ECLD residues C218, C238 and C242 are required for proper Hh reporter gene induction. Mutation of C84, 100,148,172,194,203 to alanine did not compromise the ability of Smo protein to induce the Hh reporter gene activity, and behaved similarly to the wild type protein. Percent activity for each of the experimental assays is shown relative to control. The control level of Hh-induced, ptcΔ136-luciferase activity for control dsRNA was set to 100%. For all conditions, luciferase activity is normalized to pAc-renilla control. Experiments were performed a minimum of two times in duplicate or triplicate and all data pooled. Error bars indicate standard error of the mean (s.e.m.).
Solution structure of the Smo CRD
A recombinant Drosophila Smo CRD (V85-K202) was expressed and purified using the E. coli expression system. For the purpose of NMR studies, 15N and 13C labelled protein was prepared in 10 mM deuterated acetic acid buffer and 10% D2O (volume/volume) at pH 5. Resonance assignments were performed using the standard triple resonance strategy for 13C, 15N labelled proteins28. In the 2D 1H-15N HSQC spectra, all the amide resonances of the 118 residues of the CRD could be observed except D116. D116 is in the loop region preceding the first helix and hence could be undergoing motions in the intermediate time scale resulting in peak broadening for the backbone amide; nevertheless, all the side chain resonances of this residue were observed. Using the structural information obtained from the NMR studies, one hundred structures were calculated and the 20 structures with the lowest energy are represented as an ensemble (Fig. 3a). The statistical details for the NMR structure calculation are given in Table 1. The structure has no distance violation > 0.03 Å and no angle violations > 1°. The structure with the lowest energy is used to describe the secondary structure elements (Fig. 3b). The tertiary fold of the Smo CRD is very similar to that of the FzD CRD16,18 (Fig.3c, cyan compared to red).
Figure 3. Solution structure of the Smo CRD.
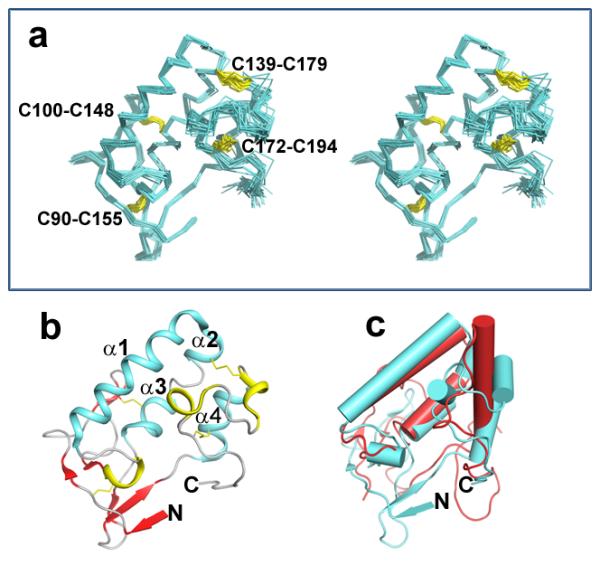
(a) Stereo view of the backbone atoms (N, Cα, C') of the 20 superimposed structures of Smo CRD with the lowest energy. The disulphides are indicated in yellow and are labelled for clarity. (b) Ribbon diagram of the Smo CRD showing the secondary structure elements. The structure with the lowest energy is used to describe the secondary structure elements. The colour scheme is as follows: Cyan: Alpha helices; Red: β strands; Yellow: 310 helices Grey: random coil. (c) Superimposition of the Smo CRD with FzD CRD. The Smo CRD is represented in cyan and the FzD8 CRD is represented in red. The helices in both proteins are shown as cylinders and the beta-strands as arrowheads. All figures were generated using PyMol.
Table 1.
NMR and refinement statistics used for calculating the structure of Smo CRD
| Protein | |
|---|---|
| NMR distance & dihedral constraints | |
| Distance constraints | |
| Total NOE | 1421 |
| Intra-residue | 430 |
| Inter-residue | |
| Sequential (|i−j| = 1) | 422 |
| Medium-range (|i−j < 4) | 248 |
| Long-range (|i−j > 5) | 321 |
| Hydrogen bonds | 22 |
| Total dihedral angle restraints | 139 |
| phi | 71 |
| psi | 68 |
| Structure Statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.0176±0.0019 |
| Dihedral angle constraints (°) | 0.7607±0.0849 |
| Max. dihedral angle violation (°) | 0.9547±0.0849 |
| Max. distance constraint violation (Å) | 0.0238±0.0152 |
| Average pairwise r.m.s.d.** (Å) | |
| V85-K202 | |
| Heavy | 1.28±0.19 |
| Backbone | 0.7±0.15 |
| Average pairwise r.m.s.d.** (Å) | |
| V85-E177 | |
| Heavy | 1.08±0.19 |
| Backbone | 0.4±0.15 |
Average r.m.s deviation from the lowest energy structure was calculated among 20 structures with the lowest energy.
The cysteines in the Smo CRD
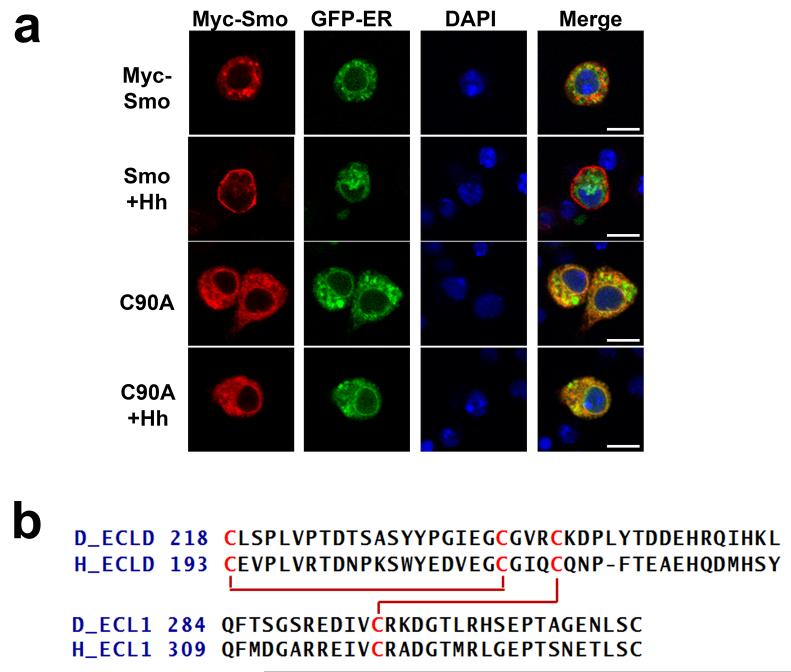
Structure analysis shows that the eight conserved cysteines in Smo CRD form four disulphide bonds. The chemical shifts observed for the cysteine C-alpha and C-beta carbons are suggestive that all the cysteines are oxidized29. The disulphide bond pattern was determined based on the intermolecular NOEs observed between the β-protons of the cysteine residues and is as follows: C90-C155, C100-C148, C139-C179 and C172-C194 (Fig. 3a). In order to study the structural and biological relevance of these disulphide bonds, we mutated the cysteines individually to alanine, and checked the ability of the cysteine to alanine Myc-Smo mutants to rescue Hh induced reporter gene in a smo knockdown background. C100A, C148A, C172A and C194A were able to rescue Hh-induced reporter gene induction to wild-type levels (Fig. 2) indicating that the C100-C148 and C172-C194 disulphide bonds are not essential for a functional CRD. Conversely, C90A, C139A, C155A and C179A showed attenuated rescue of the Hh-induced reporter gene induction (Fig. 2) suggesting that the C90-C155 and C139-C179 disulphide play critical roles in maintaining the integrity of the CRD structure. Indeed, additional indirect immunofluorescence revealed that Smo cysteine to alanine mutants that were compromised in their ability to signal were retained in the endoplasmic reticulum (ER), overlapping with the ER marker protein Calreticulin-GFP-KDEL (Fig. 4a and Supplementary Fig. S1a). ER retention is likely due to altered protein folding resulting from loss of essential disulphide bridges. However, such misfolding did not trigger obvious degradation of Smo protein, as the cysteine to alanine mutants were present at protein levels similar to and greater than that of wild type Smo (Supplementary Fig. S1b lane 2 compared to 3–6). Though it is difficult to understand the cause of higher protein levels for cysteine to alanine mutants, we speculate that it may be due to the misfolded Smo mutants failing to exit the ER and cycle to the plasma membrane to be phosphorylated and desensitized like the wild type protein30,31.
Figure 4. Sub cellular localization of the cysteines essential for Smo signalling.
(a) CRD mutants with compromised signalling activity have altered sub cellular localization. Cl8 cells expressing wild type or the indicated Myc-Smo mutant protein, in the presence of Hh (+) or empty vector control, were examined by indirect immunofluorescence. Wild type Smo translocates to the plasma membrane in response to Hh, whereas Smo CRD cysteine to alanine mutant C90A that was required for maximal Hh reporter gene induction was largely retained in the ER. Smo was detected using anti-Myc (red), Calreticulin-GFP-KDEL marks the ER (green) and DAPI (blue) marks the nucleus. Scale bar: 10uM (b) Sequence alignment of the Drosophila (D) Smo ECLD and ECL1 with the human (H) Smo ECLD and ECL1. The cysteines engaged in disulphide bond formation in human Smo are conserved in Drosophila. The red lines indicate the disulphide bond pattern.
The cysteines in the ECLD of Smo
In addition to the cysteines of the CRD (V85-K202), the Drosophila Smo ECD has five other conserved cysteines, at positions 84, 203, 218, 238 and 242. In the 3D structure of the Drosophila Smo CRD the N- and the C-termini are in close proximity (Fig. 3b); implying that C84 and C203 may form a disulphide bond. Nevertheless, mutation of C84 and C203 to alanine does not alter the ability of Smo to rescue reporter gene induction in the smo knockdown background (Fig.2), suggesting that they are either not engaged in a disulphide bond or the disulphide bond is not crucial in defining the 3D fold of the CRD. However, when C218, C238 and C242 of the ECLD were mutated individually to alanine, they expressed at near-normal levels (Supplementary Fig. S1c), but failed to rescue Hh-induced reporter gene induction following endogenous smo knockdown (Fig.2). C218, C238 and C242 of Drosophila Smo are homologous to C193, C213 and C217 of human Smo (Fig. 4b). The crystal structure of the human Smo shows that the ECLD is stabilized towards the ECL by disulphide bonds between C193 and C213 of the ECLD and C217 of the ECLD and C295 ECL1 loop14 (Fig. 4b). Hence, C218 and C238 in the ECLD of Drosophila Smo may form a disulphide bond and C242 in the ECLD is likely to form a disulphide bond with C320 in ECL1 of Drosophila Smo. Thus, cysteines both in the Smo CRD and ECD linker play an important role in maintaining the conformation of Smo essential for downstream signalling.
Drosophila Smo CRD binds to the glucocorticoid Bud
Smo activity can be modulated by small molecule inhibitors such as cyclopamine, which binds in the orthosteric binding site located within a cavity in the 7TM bundle14,24. The FzD CRD binds to its endogenous ligand Wnt underscoring the ligand binding capability of the CRDs16,18. Therefore we decided to test whether the Drosophila CRD might bind to 20-OHC or Bud. Due to a solubility issue with 20-OHC, we carried out all experiments with Bud. We used NMR chemical-shift perturbations (CSPs) to identify the residues that are involved in binding32 since this method allows us to determine precisely the amino acid residues involved in the interaction33. Despite recent reports stating the inability of 20-OHC to bind to the Drosophila Smo protein we tested if Bud bound to Drosophila Smo CRD25. In the 2D 1H-15N HSQC spectra of Smo CRD in the absence and presence of an increasing concentration of the ligand Bud, CSPs induced by the binding of Bud are clearly observed indicating Bud bound to the CRD (Supplementary Figs. S2a and S3). The normalized CSP for each residue of Smo in the presence of Bud is shown in Fig. 5a. The residues that have significant CSP are further shown in the back-bone “ribbon” representation of the Smo CRD, wherein the thickness of the ribbon is proportional to CSP values observed on binding of Bud to Smo CRD (Fig. 5b). The results show that the residues of Smo CRD that have the largest CSPs induced by the bound Bud are located between the first helix and the C-terminus 310 helical domain as it folds back between the first and second helix. As a control we tested the ability of cyclopamine to bind and induce CSPs within the Smo CRD and Smo CRD did not bind to cyclopamine at a ratio of 1:2 of protein to ligand24 (Supplementary Fig. S2b).
Figure 5. Drosophila Smo CRD binds to the glucocorticoid Bud.
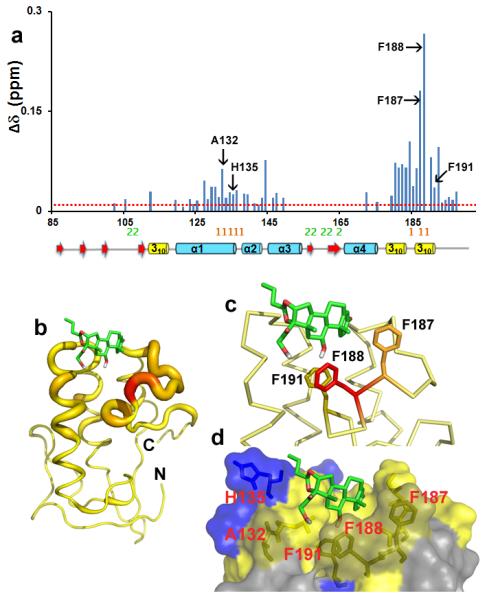
(a) CSPs of Smo CRD upon addition of Bud are plotted versus residue numbers. The red line indicates CSP greater than 0.01 ppm. The residues labelled in black form the Bud-binding pocket on Smo CRD as analysed from the HADDOCK docking experiments. The mouse FzD8-Wnt interacting “site 1” and “site 2” residues are shown in orange and green respectively. The corresponding secondary structure elements of the Drosophila Smo CRD are represented below the plot. (b) “Ribbon” representation of the Smo CRD. The backbone thickness of the ribbon is directly proportional to the weighted sum (in ppm) of the 1H and 15N chemical shifts upon binding to the ligand Bud. (c and d) Results of the HADDOCK docking of Bud on Smo CRD. (c) The aromatic side chains of the Bud contacting Smo CRD residues are shown. (d) Surface representation of the residues that interact with Bud are shown in yellow and the positively charged H135 is shown in blue.
Based on the NMR studies, we generated the docked structure of Bud bound to Smo CRD using the program HADDOCK34. The CSP data on the Smo CRD and the bound Bud were used as ambiguous restraints (Supplementary Table S1) for docking and generating one thousand binding poses. The structure with the lowest binding energy was selected and this agrees with the CSP data on the Smo CRD (Fig. 5b). The modelled structure showed that Bud is buried into a hydrophobic surface formed by the residues A132, H135, F187, F188 and F191 on the Smo CRD (Fig. 5c and 5d). F191 is conserved in the vertebrate Smo family whereas F188 is a tryptophan in vertebrate Smo (Fig.1). The conservation of these residues in the vertebrate family, and the recent demonstration that the domain engages 20-OHC25,26, suggests that this pocket may have an important role in binding an as yet unidentified allosteric regulator of Drosophila Smo. To determine whether Bud would attenuate signalling by the Drosophila Smo protein, we treated Cl8 cells with increasing concentrations of Bud, and observed a ~20–40% reduction in Hh-induced reporter gene activity (Supplementary Fig. S4). Thereby we speculate that Bud acts as a weak, synthetic mimic of an endogenous ligand for the Drosophila Smo CRD, displacing it from this binding pocket.
The human Smo CRD binds to the glucocorticoid Bud
Recent reports indicate that 20-OHC binds the vertebrate Smo CRD25,26. We therefore tested the binding of Bud to human Smo CRD. The recombinant human Smo CRD (G65-G177) was expressed and purified using the E. coli expression system. Similar to the studies of Drosophila Smo CRD, 15N and 13C labelled protein was prepared in 10 mM deuterated acetic acid buffer and 10% D2O (volume/volume) at pH 5. In the 2D 1H-15N HSQC spectra of human Smo CRD, all the amide resonances of the 113 residues were assigned except two residues, L73 and R74. NMR titration experiments were performed to determine the specific residues involved in ligand binding. The 2D 1H-15N HSQC spectra of human Smo CRD in the absence and presence of an increasing concentration of Bud showed that Bud bound to human Smo CRD as well (Supplementary Figs. S5 and S6). The normalized CSP for each residue of human Smo CRD in the presence of Bud is shown in Fig. 6a. Comparing with Drosophila Smo CRD (Fig. 5a), Bud binds to human CRD in a similar fashion. However, there are some clear differences. For example, a smaller number of residues in human Smo CRD had CSPs induced by the binding of Bud, and the absolute values of CSPs observed in human Smo CRD are smaller. Moreover, the binding affinities measured from different residues in human Smo CRD are clustered around 45 μM whereas those obtained from residues in Drosophila Smo CRD range from 200–1000 μM indicating that Bud binding induced backbone conformational stabilization in Drosophila CRD35. Nevertheless, judged by the NMR titration data, it seemed that Bud binds to human Smo CRD with higher affinity (Supplementary Figs. S3 and S6). Although NMR is a useful method for determining specific protein ligand interactions in solution, it is not the ideal method for calculating dissociation constants36. The associated systemic error in calculating Kd values from NMR is likely caused by the averaging effect during NMR experiments37. Kd values calculated by this method can only be treated as the upper limit for interaction. Therefore we validated the binding affinity measurements using additional experiments. Indeed, using Bio-Layer Interferometry (BLI) we found that Bud bound to Drosophila Smo CRD with a Kd value of 120 ± 98 μM and bound to human Smo CRD with a Kd value of 52 ± 16 μM (Fig. 6b and c). Both NMR and BLI data suggest that Bud binds to human Smo CRD better than Drosophila Smo CRD.
Figure 6. Analysis of the binding of Bud to human Smo CRD.
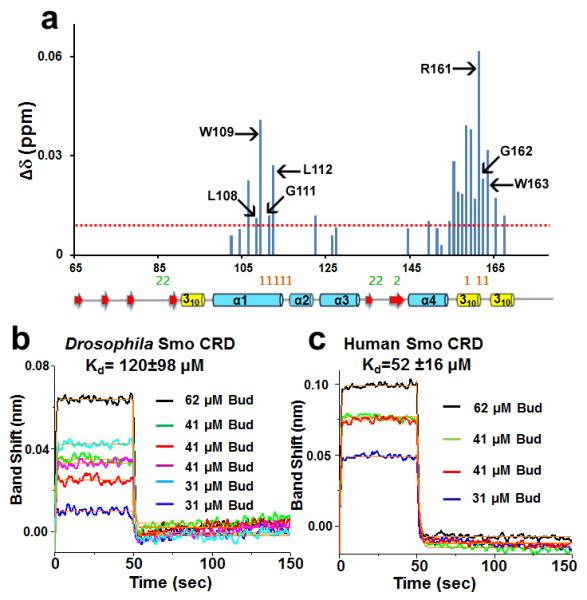
(a) CSPs of human Smo CRD upon addition of Bud are plotted versus residue numbers. Dotted red line indicates CSP greater than 0.01 ppm. L108, W109, G111, L112 and R161 are homologous to the mouse Smo residues that interact with 20-OHC. G162 is homologous to Drosophila F187 and W163 is homologous to Drosophila Smo F188 that interacts with Bud. All these residues map to the “site 1” of mouse FzD8-Wnt interaction. “Site 1” and “site 2” residues of mouse FzD8-Wnt interaction are shown in orange and green respectively. The secondary structure elements as in Drosophila Smo CRD structure are shown below the plot. (b) BLI binding assays show that Bud binds to Drosophila Smo CRD. The super streptavidin sensors with biotinylated Drosophila Smo CRD were exposed to three different concentrations of Bud (62, 41, and 31 μM). The processed data were fitted locally with the integrated fitting function by a 1:1 binding model (orange line). The respective Kd values obtained by curve fitting were 89 μM (62 μM Bud, black line), 74 μM (41 μM Bud, green line), 59 μM (41 μM Bud, red line), 318 μM (41 μM Bud, magenta line), 93 μM (31 μM Bud, cyan line), and 85 μM (31 μM Bud, blue line). The average Kd value of Drosophila Smo CRD for Bud was 120 ± 98 μM. (c) BLI binding assays show that Bud binds to human Smo CRD. The experimental and data analysis procedure were same as described above. The respective Kd values obtained by curve fitting were 74 μM (62 μM Bud, black line), 37 μM (41 μM Bud, green line), 44 μM (41 μM Bud, red line) and 54 μM (31 μM Bud, blue line). The average Kd value of human Smo CRD for Bud was 52 ± 16 μM.
Discussion
Classical GPCR signalling involves binding of ligands to the extracellular regions of the receptor which induces a conformational change in the core of the 7TM domains to modulate signalling events in the cytoplasm38,39. Several studies have reported that GPCRs are regulated by natural ligands, as well as exogenous small molecules38–41. Furthermore, GPCRs have been reported to possess allosteric binding sites distinct from their orthosteric sites, that when bound by small molecule modulators, induce distinct signal outputs42,43. In the FzD GPCR the CRD functions to bind to the endogenous ligand Wnt, which interacts with the CRD at two sites16,18. The palmitate modification on Wnt engages a groove on the FzD8 CRD termed site 1, whereas the second Wnt binding site on FzD8 CRD (site 2) is located on the opposite end and involves protein-protein interactions18. In the current studies, we showed that Bud interacts with Smo CRD residues analogous to site 1. Within the site 1 of human Smo R161 demonstrated the highest observed CSP. L108, W109, G111, L112, G162 and W163 also demonstrated CSPs. Consistent with these findings, a recent report shows that mutating the homologous murine Smo CRD residues “WGL” (W113, G115, and L116) and R165 to alanine blocked the specific binding of 20-OHC to the Smo CRD and attenuated its signalling ability26. Furthermore, mutating murine L112 (human L108) and W113 (human W109) to equivalent Drosophila Smo CRD residues blocked 20-OHC binding25, suggesting that the site 1 is involved in the binding of hydroxyl-sterols and glucocorticoids. Drosophila Smo protein does not bind to 20-OHC in vitro25, however that does not preclude the fact that there could be an as yet unidentified endogenous ligand for Drosophila Smo CRD binding to this pocket. Consistent with this hypothesis, genetic experiments suggest that phospho-lipids could serve as the physiological molecules regulating Drosophila Smo activity44.
The cache of identified Smo modulators dock on distinct sites on Smo to regulate signalling14,24,45,46. The extracellular region of Smo may be flexible (Fig.7a). Small molecules may bind in the cavity of the 7TM domains (Fig. 7b right) and modulate signalling irrespective of CRD binding14,24. In the present study, we provide structural evidence identifying the specific residues in the Smo CRD that interact with the small molecule modulator Bud. We speculate that these same residues bind the endogenous allosteric activator, and that activator binding to the CRD may cause a conformational shift that bridges the extracellular domains and the 7TM domains (Fig. 7b middle), similar to what is observed for class B GPCRs40,41,47. These shifts may enhance communication between the CRD and the 7TM domains to trigger structural alterations in the core and the cytoplasmic domains to regulate downstream signalling (Fig. 7).
Figure 7. A novel model for Smo allosteric regulation.
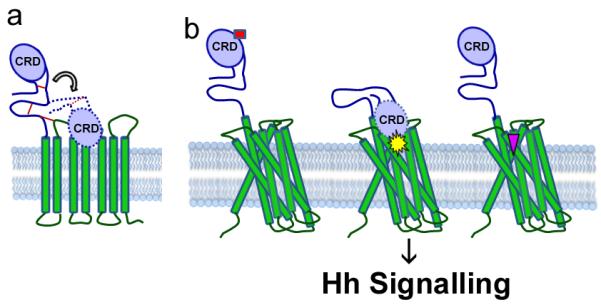
(a) Smo CRD may be flexible. The disulphide bonds stabilizing Smo extracellular linker and extracellular loop are shown in red lines. (b) Smo has more than one binding site. We propose that molecules like Bud bind to the Smo CRD (left, red rectangle) to alter its conformation and attenuate its signalling activity. Cyclopamine and vismodegib are known to bind near the orthosteric binding site located in the cavity of the Smo 7TM domains (right, pink inverted triangle). We speculate that there is the possibility of a class of molecules (middle, yellow star) which would bind to the CRD and cause a conformational change of the Smo extracellular structures to bring it closer to the 7TM domains. This in turn might change the conformation of the trans-membrane domains to regulate signalling.
Currently, the only FDA approved therapeutic aimed at targeting Smo is the antagonist Vismodegib, which is likely to bind in the cavity of the 7TM domains and also make contacts with the ECLD and ECL14,24,48,49. Despite initial clinical success, resistance to this compound has been observed48. Thus, the need exists for developing second-generation drugs that can overcome the acquired drug resistance. Based upon our data, we speculate that the Bud binding cleft in the Smo CRD could be evaluated for the development of novel antagonists of Smo. Bud is a clinically approved anti-inflammatory glucocorticoid steroid and has the advantage of acting on wild type as well as Vismodegib resistant Smo23,50. Thus future investigation and identification of Bud-like molecules which bind to the Smo CRD with high affinity may represent a novel class of Smo regulators.
Methods
Vector construction
The Drosophila Smo CRD (V85-K202) was amplified from pET-SmoN3. The forward and the reverse primers used were 5'- ATA TAT CAT ATG GTC CGA CGT GCC CGT TGC- 3' and 5'- ATA TAC TCG AGT TAT TTC GTC GGA AAG AGT G-3'. The PCR amplified product was cloned in pET-28a (+) and validated by nucleotide sequencing. To generate cysteine to alanine mutants, Smo mutant expression vectors pAc-myc-smo51 was mutated using QuickChange II Site Directed Mutagenesis Kit (Stratagene) using primers generated per manufacturer's instructions. All mutations were verified by sequencing. Smo 5'UTR dsRNA was generated using the Megascript T7 kit (Life Technologies)15. The human Smo CRD (G65–G177) was amplified from the smo cDNA purchased from Origene. The forward and the reverse primers used were 5'-TAT ATC ATA TGG GCC GGG CTG CCC CCT GC -3' and 5'- ATA TAT CTC GAG TTA GCC TTC AGG GAA GCG -3' respectively. The PCR amplified product was cloned in pET-28a (+) and validated by nucleotide sequencing.
Protein expression and purification
For the expression of both the Drosophila and human recombinant protein pET28a-smoCRD was transformed in to BL21 (DE3) cells. The cells were grown in MOPS (3-(N-morpholino) propane sulfonic acid) minimal media supplemented with 15N ammonium chloride and 13C glucose as the sole source of nitrogen and carbon respectively. Cells were grown to an optical density (OD600) of 0.6 and induced with 1mM IPTG (isopropyl β-D-1-thiogalactopyrasonide). The protein of interest accumulated in the inclusion bodies and was refolded from the same by the method of rapid dilution. The protein was further purified by reverse phase HPLC using a C18 column (Hitachi) and gel filtration (AKTA) on a Hiload 16/60 Superdex 200 preparative grade column (GE Healthcare).
NMR Spectroscopy
A 200 μM pure monomeric sample of the recombinant Drosophila (V85–K202) or human (G65–G177) Smo CRD was prepared in 10 mM deuterated acetic acid buffer and 10% D20 (v/v) for NMR experiments. All spectra were recorded using either 1H, 15N labelled or 1H, 15N, and 13C labelled protein on Bruker Avance 600 MHz or 800 MHz NMR spectrometers equipped with 1H/15N /13C detect, TCI triple resonance cryogenic probes at 25°C. All spectra were processed using Topspin (Bruker Biospin) 3.0 NMR software and analysed using the program, CARA (computer aided resonance assignment)52. The backbone chemical shift assignment were obtained using the standard triple- resonance assignment strategy using the two dimensional 1H-15N HSQC and three dimensional HNCA, HNCACB, CBCA(CO)NH, HN(CA)CO and HNCO NMR experiments. The aliphatic side chain resonance assignments were obtained using HBHA(CBCACO)NH and (H)CCH-TOCSY. Distance constraints for the structure calculation were derived from 15N; 13C resolved 1H-1H NOESY with a mixing time of 100 ms. Chemical shift perturbation experiments were carried out using 15N labelled Drosophila or human Smo CRD. 1H-15N HSQC spectra were recorded in the absence and presence of an increasing concentration of Bud. Kd values were calculated by a nonlinear least-squares analysis in the program Origin using the equation [1]
| [1] |
where [L] is the concentration of the ligand, [P] is the concentration of the protein, Δδ is the observed chemical shift change and Δδmax is the normalized chemical shift change at saturation, calculated using equation [2]
| [2] |
where Δδ is the chemical shift in p.p.m.
A stock solution of 50 mM Bud (Sigma) was made in deuterated DMSO for use in NMR titration experiments. The concentration of DMSO in the NMR titration experiment was maintained at or below 1%. A control experiment was done by titrating 1% DMSO and no CSPs were observed.
Structure calculation and refinement
An automated program UNIO53 was used for the NOE assignment and all these assignments were manually checked. Integrated NOE peaks were calibrated and converted to distance constraints with the program CALIBA54. A total of 1421 meaningful distance constraints were derived from the NMR data and used as input for structure calculation using the program CYANA 2.154. The structure was refined using 139 dihedral angle constraints and 22 hydrogen bond constraints. The dihedral angles were generated by the program TALOS+55 which uses six kinds (HN, HA, CA, CB, CO, N) of chemical shift values for a given residue as input to give the phi, psi angles. The Ramachandran statistics are as follows: most favourable 72.5 %, additionally allowed 25.7 %, generously allowed 1.6%, and disallowed 0.2 %. Also, four disulphide bonds were added as additional constraints between residues, C90–C155, C100–C148, C139–C179 and C172–C194. A total of 100 conformers were initially generated and 20 conformers with the lowest target function were used to represent the 3-D NMR structure.
Structural modelling of Smo CRD in complex with Bud
To calculate the structure of the CRD-Bud complex, we used the simulated annealing protocol in the program CNS34. All calculations were done using the program HADDOCK 2.1(High Ambiguity Driven protein-protein Docking)34. Topology and parameter files for Bud were generated by using xplo2d, and the charge parameters of Bud were calculated by using the antechamber module in the AMBER10 software package56. We used ambiguous distance restraints resulting from the CSPs of 1H-15N Smo CRD (Supplementary Table S1). To define the active and passive residues we also calculated a relative accessible surface area by NACCESS57. One thousand initial structures of the Smo CRD-Bud complex were generated and the final ten structures of the complex with the lowest energy were selected manually that were consistent with the CSPs on Smo CRD following Bud binding.
Lysate Preparation and Functional Assays
Expression vectors and dsRNA were transfected using Lipofectamine 2000 according to the manufacturer's instructions. For rescue reporter experiments ~1.5 × 106 Cl8 cells were co-transfected with 500 ng Smo 5 'UTR dsRNA, 100 ng ptcΔ136-luciferase27 10 ng pAc-Renilla51, and unless otherwise indicated, 100 ng of pAc-Hh (+Hh) and 50 ng of the indicated pAc-myc-smo expression vector. DNA and RNA content were normalized with control dsRNA and/or pAc5.1A. Forty-eight hours post-transfection, cells were lysed and luciferase activity measured using the Dual Luciferase Assay System (Promega)15. For all reporter assays, experiments were performed a minimum of two times in duplicate or triplicate and all data pooled. Error bars indicate standard error of the mean (s.e.m.).
For the Bud treatment, Cl8 cells were treated with the indicated concentration of Bud (Sigma) or DMSO vehicle. Bud was added in serum-free M3 media (Sigma) 24 hours post transfection. Cells were treated with Bud for 24 hours prior to measuring luciferase activity. Experiments were performed at least three times in duplicate and all data pooled.
For biochemical analysis, ~6 × 106 Cl8 cells were transfected with 5 μg of the indicated pAc-myc-smo expression vector and 3 μg of pAc-Hh, as indicated. DNA content was normalized with empty pAc5.1A. Forty-eight hours post transfection, cells were lysed in NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, 50 mM NaF, 0.5 mM DTT and 1X PIC (Roche), pH 8.0), and centrifuged for 10 minutes at 2000xg. The resulting supernatants were analysed by SDS-PAGE and western blot using anti-Myc (Roche, Cat # 11667149001 diluted 1:5000) and anti-Kinesin (Cytoskeleton, AKIN01-A, diluted 1:10,000). Full scans of the western blots are represented in Supplementary Figure S7. Antibodies were diluted in Tris-buffered saline with 0.1% Tween and 5% powdered milk.
Immunofluorescence and microscopy
For immunofluorescence experiments, ~6 × 106 Cl8 cells were transfected with 2 μg of Calreticulin-GFP-KDEL58, 2 μg pAc Hh, and 2 μg of the indicated pAc-myc-smo expression vector. DNA content was normalized with pAc5.1A. Forty-eight hours post-transfection, cells were re-plated on chamber slides, fixed using 4% formaldehyde and immunostained using anti-Myc (Roche, Cat # 11667149001)15, Calreticulin-GFP-KDEL was used to mark the endoplasmic reticulum, and DAPI was used to mark the nucleus. Confocal images were collected using a Zeiss LSM 510 NLO Meta Microscope.
Bio-Layer Interferometry Assay
The Octet RED instrument (FortéBio) was used to measure the interaction of Drosophila Smo CRD (or human Smo CRD) and Bud. Super streptavidin (SSA) sensors were used to catch the biotinylated target protein. Biotinylation of the target protein was achieved by incubating 200 μl of 1.0 mg/ml EZ-Link NHS-LC-Biotin (Thermo Sci.) with 100 μl of 0.2 mM target protein in 0.1 M potassium phosphate buffer, pH 6.5 for 15 hours on a rocking platform at room temperature. Excess unreacted biotin was removed by three successive dialysis under the same buffer conditions. Prior to the start of the assay the SSA sensors were pre-wet in 0.1M potassium phosphate buffer, pH 6.5 with 0.01 % Trion-X100 for 10 minutes. For the binding affinity assay 3 ~ 5 μM biotinylated protein was immobilized on the SSA sensor by incubating for 30 minutes. Subsequently the free streptavidin sites were quenched by incubating with 0.5 mM biocytin (Thermo Sci.) for 10 minutes. Excess protein and biocytin were removed by washing with 0.1M potassium phosphate, pH 6.5 for 10 minutes. To determine the binding affinity of Drosophila Smo CRD (or human Smo CRD) to Bud, three different concentrations of Bud (62, 41, and 31 μM) were used. The association step for 50s was followed by the dissociation step for 100s. The assay buffer used was 0.1 M potassium phosphate, pH 6.5 with 0.5% DMSO. Biocytin-loaded SSA sensor without loading biotinylated protein was used as a control to correct the systematic optical artefacts and baseline drifts. All steps were performed at 30 °C with 1000 rpm rotary shaking. The processed data were fitted locally with the integrated fitting function using the 1:1 binding model in FortéBio analysis software (v6.4). The kinetic constants kon (on-rate constant), koff (off-rate constant) and Kd were calculated from curve fitting.
Supplementary Material
Acknowledgements
This work was supported by NIH research grants R01 GM100909 (J.J.Z.), R01 GM081492 (J.J.Z.), R01GM101087 (S.K.O.), March of Dimes MOD5-FY10-6 (S.K.O.), NIH Cancer Center grant P30 CA021765, and ALSAC. C.C. was supported by a UNCF/Merck Postdoctoral Science Research Fellowship.
Footnotes
Author contributions J.J.Z and S.K.O conceived the study, designed the experiments and serve as corresponding authors. R.R performed protein purification, NMR data acquisition and analysis, structure determination and refinement. R.R cloned, purified and assigned the human Smo CRD and performed both Drosophila and human NMR titration experiments. H.L determined the Kd from the NMR experiments and performed the BLI assays. H.L and J.B performed HADDOCK docking experiments. C.E.C. and S.M. performed all cell based assays. C.R.R.G helped with NMR data acquisition and structure calculation. C.D.G made the clone of the Drosophila protein for NMR studies. R.R., S.K.O., J.J.Z coordinated the experimental work and wrote the manuscript with contribution from all authors.
Accession code: Chemical shifts of Drosophila Smo CRD have been deposited at the BioMagResBank under accession code 19354. The coordinates for the structure are deposited in the Protein Data Bank under the accession codes 2MAH.
Supplementary Information accompanies this paper at http://www.nature.com/naturecommunications.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 3.Alcedo J, Ayzenzon M, VonOhlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 4.Ogden SK, et al. G protein G alphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456:967–976. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Hui C. Hedgehog signaling in development and cancer. Dev. Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen MM., Jr. Hedgehog signaling update. Am. J. Med. Genet. A. 2010;152A:1875–1914. doi: 10.1002/ajmg.a.32909. [DOI] [PubMed] [Google Scholar]
- 8.Bale AE. Hedgehog signaling and human disease. Annu. Rev. Genomics Hum.Genet. 2002;3:47–65. doi: 10.1146/annurev.genom.3.022502.103031. [DOI] [PubMed] [Google Scholar]
- 9.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol. Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 11.Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- 12.Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 13.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat. Cell Biol. 2007;9:1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll CE, Marada S, Stewart DP, Ouyang JX, Ogden SK. The extracellular loops of Smoothened play a regulatory role in control of Hedgehog pathway activation. Development. 2012;139:612–621. doi: 10.1242/dev.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dann CE, et al. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 17.Pei J, Grishin NV. Cysteine-rich domains related to Frizzled receptors and Hedgehog-interacting proteins. Protein Sci. 2012;21:1172–1184. doi: 10.1002/pro.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano Y, et al. Functional domains and sub-cellular distribution of the Hedgehog transducing protein Smoothened in Drosophila. Mech. Dev. 2004;121:507–518. doi: 10.1016/j.mod.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Aanstad P, et al. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh Signaling. Curr. Biol. 2009;19:1034–1039. doi: 10.1016/j.cub.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 22.Nachtergaele S, et al. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 2012;8:211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Glucocorticoid compounds modify Smoothened localization and Hedgehog pathway activity. Chem. Biol. 2012;19:972–982. doi: 10.1016/j.chembiol.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol. 2013;9:557–564. doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers BR, et al. Hedgehog pathway modulation by multiple lipid binding sites on the Smoothened effector of signal response. Dev. Cell. 2013;26:346–357. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CH, von Kessler DP, Park WJ, Beachy PA. Nuclear trafficking of cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 28.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc. Chem. Res. 1993;26:131–138. [Google Scholar]
- 29.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 30.Molnar C, et al. Role of the Drosophila non-visual beta-arrestin kurtz in Hedgehog signalling. PloS Genet. 2011;7 doi: 10.1371/journal.pgen.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia JH, Tong C, Wang B, Luo LP, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 32.Wuthrich K. Protein recognition by NMR. Nat. Struct. Biol. 2000;7:188–189. doi: 10.1038/73278. [DOI] [PubMed] [Google Scholar]
- 33.Zheng J, et al. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J. Mol. Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez C, Boelens R, Bonvin A. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 35.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Mag. Res. Sp. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 36.London TBC, Lee HJ, Shao YM, Zheng J. Interaction between the internal motif KTXXXI of Idax and mDvl PDZ domain. Biochem. Biophys. Res. Commun. 2004;322:326–332. doi: 10.1016/j.bbrc.2004.07.113. [DOI] [PubMed] [Google Scholar]
- 37.Shan JF, Zheng J. Optimizing Dvl PDZ domain inhibitor by exploring chemical space. J Comput Aided Mol Des. 2009;23:37–47. doi: 10.1007/s10822-008-9236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 39.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 40.Grace CRR, et al. NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2004;101:12836–12841. doi: 10.1073/pnas.0404702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koth CM, et al. Molecular basis for negative regulation of the glucagon receptor. Proc. Natl. Acad. Sci. USA. 2012;109:14393–14398. doi: 10.1073/pnas.1206734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 43.Hudson BD, Ulven T, Milligan G. The therapeutic potential of allosteric ligands for free fatty acid sensitive GPCRs. Curr. Top. Med. Chem. 2013;13:14–25. doi: 10.2174/1568026611313010004. [DOI] [PubMed] [Google Scholar]
- 44.Yavari A, et al. Role of lipid metabolism in Smoothened derepression in hedgehog signaling. Dev. Cell. 2010;19:54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu VM, Chen SC, Arkin MR, Reiter JF. Small molecule inhibitors of Smoothened ciliary localization and ciliogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:13644–13649. doi: 10.1073/pnas.1207170109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong MQ, Li ZJ, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between the amino terminus of a peptide agonist and the top of the sixth transmembrane segment of the secretin receptor. J. Biol. Chem. 2004;279:2894–2903. doi: 10.1074/jbc.M310407200. [DOI] [PubMed] [Google Scholar]
- 48.Von Hoff DD, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 49.Robarge KD, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg. Med. Chem. Lett. 2009;19:5576–5581. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 50.Balanag VM, Yunus F, Yang PC, Jorup C. Efficacy and safety of budesonide/formoterol compared with salbutamol in the treatment of acute asthma. Pulm. Pharmacol. Ther. 2006;19:139–147. doi: 10.1016/j.pupt.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Ogden SK, et al. Smoothened regulates activator and repressor functions of Hedgehog signaling via two distinct mechanisms. J. Biol. Chem. 2006;281:7237–7243. doi: 10.1074/jbc.M510169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller RLJ. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag; Goldau.Switzerland: 2004. [Google Scholar]
- 53.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 54.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 55.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 56.Case DA, et al. AMBER 10. University of California; San Francisco: 2008. [Google Scholar]
- 57.Hubbard SJ, Thornton JM. NACCESS. Department of Biochemistry and Molecular Biology; University College London: 1993. [Google Scholar]
- 58.Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB. Drosophila signal peptide peptidase is an essential protease for larval development. Genetics. 2005;170:139–148. doi: 10.1534/genetics.104.039933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.