Abstract
Background and Aims
Adaptation to different pollinators has been hypothesized as one of the main factors promoting the formation of new species in the Cape region of South Africa. Other researchers favour alternative causes such as shifts in edaphic preferences. Using a phylogenetic framework and taking into consideration the biogeographical scenario explaining the distribution of the group as well as the distribution of pollinators, this study compares pollination strategies with substrate adaptations to develop hypotheses of the primary factors leading to speciation in Lapeirousia (Iridaceae), a genus of corm-bearing geophytes well represented in the Cape and presenting an important diversity of pollination syndromes and edaphic preferences.
Methods
Phylogenetic relationships are reconstructed within Lapeirousia using nuclear and plastid DNA sequence data. State-of-the-art methods in biogeography, divergence time estimation, character optimization and diversification rate assessments are used to examine the evolution of pollination syndromes and substrate shifts in the history of the group. Based on the phylogenetic results, ecological factors are compared for nine sister species pairs in Lapeirousia.
Key Results
Seventeen pollinator shifts and ten changes in substrate types were inferred during the evolution of the genus Lapeirousia. Of the nine species pairs examined, all show divergence in pollination syndromes, while only four pairs present different substrate types.
Conclusions
The available evidence points to a predominant influence of pollinator shifts over substrate types on the speciation process within Lapeirousia, contrary to previous studies that favoured a more important role for edaphic factors in these processes. This work also highlights the importance of biogeographical patterns in the study of pollination syndromes.
Keywords: Biogeography, diversification, edaphic factors, Iridaceae, Lapeirousia, phylogenetics, pollinator, shift, speciation
INTRODUCTION
The role of pollinators in the diversification of angiosperms has been highlighted and debated ever since Charles Darwin's seminal work on orchid pollination (Darwin, 1877; Stebbins, 1970; Crepet, 1984; Kay and Sargent, 2009; van der Niet and Johnson, 2012). Many studies have focused on species (or species complexes) with particular pollination syndromes or with trait adaptations to different pollinators (floral morphology, scent chemistry, phenology, etc.). Several examples of these microevolutionary studies can be found in the pages of this Special Issue (e.g. Boberg et al., 2014; de Jager and Ellis, 2014). On the other hand, macroevolutionary studies (i.e. evolution of pollination syndromes above the species level) using phylogenetics and comparative methods to, for example, estimate correlations among floral traits and pollinators remain less common, but are increasing (Smith, 2010). The influence of geographical variation in floral traits and in the availability of various pollinators has been extensively explored at the microevolutionary level. However, the integration of biogeographical processes in the study of pollination-driven speciation above the species level is a rare occurrence. In this study, we examine the contribution of pollination syndromes and substrate shifts on speciation events in a group of African geophytes, the genus Lapeirousia, by integrating a biogeographical dimension and taking into account the distribution of pollinators.
The adaptation to different pollinators has been hypothesized as one of the main factors promoting the formation of new species in the Greater Cape Floristic Region (GCFR) of South Africa (Johnson, 1996, 2010), a small area home to more than 9000 species of vascular plants of which about 70 % are endemic (Goldblatt and Manning, 2002), and identified as one of the planet's 34 biodiversity hotspots (Mittermeier et al., 2005). The presence of a large number of specialized pollination systems in the GCFR has been perceived as supporting the importance of pollinator-driven diversification in the evolutionary history and diversification of this region (Johnson and Steiner, 2000, 2003; Johnson, 2010). In sub-Saharan representatives of the family Iridaceae alone, no fewer than 17 different pollination syndromes have been reported, with pollination by long-proboscid anthophorine bees assumed to be the ancestral condition in most genera of the subfamily Crocoideae (if not for the whole subfamily) with the other pollination systems derived and having potentially evolved multiple times (Goldblatt and Manning, 2006). Johnson (2010) identified five diversification modes involving pollinators: (1) the divergent use of a pollinator by various species such as the different placement of pollen on a pollinator; (2) coevolution between pollinator and plant, such as the covariation in floral tubes and the proboscid of pollinating flies; (3) trait tracking in which a plant species (e.g. non-rewarding) is required to follow the changes caused by the coevolutionary process in place between its pollinator and another plant species; (4) mimicry of different model flowers within a single species due to the presence of different pollinators in parts of its range; and (5) pollination syndrome shifts. The last mode is the main focus of the present study.
Until recently, Lapeirousia has been treated as comprising two subgenera (subgenera Lapeirousia and Paniculatae) each split in two sections (Goldblatt and Manning, 1990). A recent molecular phylogenetic study has resulted in its dismemberment: subgenus Paniculatae is now treated as three separate genera, the largely tropical Psilosiphon, and the Cape Codonorhiza and Schizorhiza (Goldblatt and Manning, 2014). Lapeirousia, now narrowly circumscribed, includes 27 species: 24 of them occur along the west coast and near-interior of southern Africa while two of the three remaining species are widespread in southern tropical Africa; the last is a narrow endemic of the Upper Karoo in central southern Africa. Despite comprising a relatively small number of species, Lapeirousia in its narrow sense has a diverse range of specialized pollination strategies (Goldblatt et al., 1995). These include large-bodied bees (mainly Anthophorinae), both sphinx and settling moths, bee flies (Bombyliidae), two separate guilds of long-proboscid flies (families Tabanidae and Nemestrinidae) and even generalist systems. This diversity of pollination syndromes is coupled to an equally diverse floral morphology (e.g. symmetry, tepal orientation and pigmentation, perianth tube length) and preference for a range of substrate types (e.g. sand, clay, granite, shale). This hypervariability in pollination and edaphic preferences makes this group of attractive corm-bearing geophytes an ideal case study for exploring various hypotheses about the causes of speciation in the mega-diverse winter-rainfall region of southern Africa.
A phylogenetic analysis based on morphological characters has shown that speciation in Lapeirousia was primarily allopatric and that edaphic diversity played an important role in the diversification of the genus (Goldblatt and Manning, 1996; Goldblatt et al., 1995; Procheş et al., 2006). These analyses also indicated that large-bodied bee pollination was derived in the genus, with long-proboscid fly pollination evolving repeatedly, resulting from ‘repeated entry into pre-existing pollination guilds’ (Goldblatt and Manning, 1996). Goldblatt and Manning (1996) concluded that there was no evidence of pollinator-driven speciation in Lapeirousia regardless of the great variability in floral morphology in the genus.
Here, we present the first comprehensive phylogenetic analysis of Lapeirousia based on molecular DNA sequence data from both the plastid and the nuclear genomes and with a near-complete species sampling for the clade now recognized as the genus Lapeirousia. Using this phylogenetic framework, combined with a biogeographical scenario, as well as character optimization and diversification analyses, we compare pollination strategies with substrate adaptations to develop hypotheses regarding the influence of pollination syndromes and edaphic factors in speciation within the genus. Specifically, we address the following topics: (1) the identity of the ancestral condition of the pollination systems and substrate types in Lapeirousia and its links with the biogeographical processes in the group, as well as the distribution of pollinators, particularly the dominant long-proboscid fly system; (2) the potential association of pollination strategies and edaphic preferences with increases or decreases in diversification rates; and (3) the predominance of shifts in either pollination or substrate types associated with species divergence. We are thus seeking evidence for the prevalence of either pollinator or substrate type shifts as triggers of speciation in Lapeirousia.
MATERIAL AND METHODS
Taxon sampling
Of the 27 species now assigned to Lapeirousia (Goldblatt and Manning, 2014), 25 were sampled as part of this study with only L. purpurea and L. kalahariensis missing. Three species have subspecies: L. pyramidalis (both sampled, subsp. pyramidalis and subsp. regalis), L. plicata (one sampled, subsp. plicata; subsp. effurcata and subsp. foliosa not included) and L. fabricii (one sampled, subsp. fabricii; subsp. purpurascens and subsp. compressa not included). Eight of the 15 species of Psilosiphon and six of the seven species of Codonorhiza, as well as the monotypic genera Schizorhiza, Cyanixia and Savannosiphon were also included in our analysis. The genus Zygotritonia, thought to be closely related to Cyanixia and Savannosiphon (Goldblatt and Manning, 2008), was not included in the present study; amplification of the material at hand remained unsuccessful. Outgroup taxa from other members of tribe Watsonieae were selected based on the results of a previous phylogenetic study of Iridaceae (Goldblatt et al., 2008); these include Thereianthus racemosus, Watsonia tabularis, Pillansia templemanii and Micranthus junceus. Included species and associated voucher information are provided in Supplementary Data Table S1. Hereafter, we refer to the clade corresponding to the ingroup comprising the genera Lapeirousia, Cyanixia, Savannosiphon, Codonorhiza, Schizorhiza and Psilosiphon (and most likely Zygotritonia) as the ‘Lapeirousia clade’, while Lapeirousia is used to identify what was previously Lapeirousia subgenus Lapeirousia (Goldblatt and Manning, 2014).
DNA sequencing
Total genomic DNA was extracted from 0·03–0·3 g of silica gel-dried plant material collected in the wild using a modified version of the 2× CTAB method (Doyle and Doyle, 1987) and followed by a caesium chloride/ethidium bromide gradient (1·55 g mL−1) and a dialysis procedure, to yield material suitable for long-term storage in the DNA & Tissue Collections at Royal Botanic Gardens, Kew (http://apps.kew.org/dnabank/homepage.html).
Phylogenetic relationships within the Lapeirousia clade were reconstructed using 11 DNA markers, including ten plastid, of which three are coding (matK, ndhJ, ycf5), two are introns (rpl16, trnL) and five are intergenic spacers (trnQ-5'-rps16, trnL-trnF, ndhF-rpl32, rpl32-trnL, accD-psa1). The nuclear marker is the low-copy gene RPB2, coding for the RNA polymerase II subunit. Only a portion of the matK coding region (about 850 bp in length) was amplified using primers XF and 5R (see www.kew.org/barcoding). Primers c and f were generally used to amplify the trnL intron and trnL-trnF spacer in one reaction, but these regions were sometimes amplified separately using primers c and f in combination with primers e and d (all primers from Taberlet et al., 1991). Primers for trnQ-5'-rps16, ndhF-rpl32, rpl32-trnL and accD-psa1 were obtained from Shaw et al. (2007). Amplification of the rpl16 intron was performed using primers designed by Shaw et al. (2005). The coding regions ndhJ and ycf5 were amplified using the primer pairs ndhJ-1F/ndhJ-4R and ycf5-1F/ycf5-4R, respectively (see www.kew.org/barcoding). The nuclear RBP2 region was amplified using a set of primers specifically designed for Iridaceae (P. Rymer, Royal Botanic Garden, Sydney, Australia, pers. comm.): RPB2-Irid-F (5′-GCACATATGGGGAAAGAAGG) and RPB2-Irid-R (5′-TTATCCACCTGAGATGATTGC).
The PCR amplifications for all plastid markers were conducted in 25-μL reactions, using 22·5 µL of Reddy PCR Master Mix (2·5 mm MgCl2; Thermo Fisher Scientific, Waltham, MA, USA), 0·5 µL of 0·4 % bovine serum albumin (BSA), 0·5 µL of each primer (100 ng μL−1) and 3 µL of template DNA. PCR conditions were as follows: initial denaturation at 80 °C for 5 min, followed by 35 cycles of 1 min at 95 °C, 1 min at 50 °C and 5 min at 65 °C, ending with a single final elongation of 4 min at 65 °C. The RBP2 marker was amplified using the following protocol: 1·5 µL of 50 mm MgCl2, 2 µL of 0·4 % BSA, 0·5 µl dNTP (10 µm), 0·75 µL of each primer (100 ng μL−1), 0·8 µL dimethyl sulfoxide (DMSO), 1·5 µL of template DNA, 2·5 units of Taq polymerase (Promega, Southampton, UK), and completed to a 20-μL volume reaction with water. PCR conditions were as follows: initial denaturation at 94 °C for 4 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 54 °C, 2 min at 72 °C, ending with a single final elongation of 7 min at 72 °C.
PCR amplifications were performed on a 9700 GeneAmp thermocycler (ABI, Warrington, UK) and resulting PCR products were purified with the Nucleospin Extract II kit (Machery-Nagel, Düren, Germany), following the manufacturer's protocol. Cycle sequencing reactions were performed in 10-μL reactions using 1 µL of BigDye® Terminator cycle sequencing chemistry (v3.1) and the same primers as for PCR. Complementary strands were sequenced on an ABI 3730 automated sequencer and then assembled; software base-calling was verified using Sequencher 4·5 (Gene Codes Corp., Ann Arbor, MI, USA). All DNA regions were aligned by eye in PAUP* (version 4·0b10; Swofford, 2002). Sequences are available from GenBank (Table S1).
Phylogenetic reconstructions
A combined Bayesian analysis was performed using a Markov-Chain Monte-Carlo (MCMC) approach, as implemented in MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003) and following recommendations by Nylander et al. (2004). Two partitions were defined corresponding to the plastid and nuclear genomes. Separate analyses of these partitions resulted in topologically similar trees with no well-supported incongruence; thus, only results from combined analyses are presented here and used in subsequent analyses. Each partition was assigned specific model parameters and the parameters were fully unlinked (except the topology). The best-fit model of DNA substitution for each partition was determined using MrModeltest 2·2 (Nylander, 2004) and the Akaike Information Criterion (Akaike, 1974). The General Time Reversible (GTR) model with a proportion of invariable sites and a gamma shape to account for rate heterogeneity among sites (GTR + I + G) was chosen for the plastid data, while an HKY85 model with a proportion of invariable sites was selected for the nuclear RPB2 partition. Two Metropolis-coupled MCMCs with incremental heating temperature of 0·2 were run for 10 million generations, with the parameters and the resulting phylogenetic trees being sampled every 1000 generations. The analysis was repeated three times, starting with random trees and performed on the Bioportal cluster at the University of Oslo (www.bioportal.uio.no). The MCMC sampling was considered sufficient when the effective sampling size (ESS) was higher than 200, as verified with Tracer v1.5 (Rambaut and Drummond, 2007). A burn-in period of one million generations per run was applied and the remaining trees were used to reconstruct an ‘allcompat’ consensus tree with posterior probabilities (PP) for each node.
Divergence time estimates were obtained using the Bayesian inference approach implemented in the package BEAST v.1.5.4 (Drummond and Rambaut, 2007), applying the same partition delimitation and evolutionary models as those used for the MrBayes analysis. We used an uncorrelated relaxed molecular clock with a lognormal distribution of rates and a Yule speciation model. The analysis was run on the Bioportal cluster (University of Oslo; www.bioportal.uio.no) for 20 million generations, sampling one tree every 1000 generations. Parameter convergence was confirmed following the same approach as in the MrBayes analysis (see above). Following a burn-in period of two million generations, a maximum clade credibility tree with median branch lengths and 95 % highest posterior density (HPD) interval on nodes was reconstructed using TreeAnnotator 1.5.4 (Drummond and Rambaut, 2007).
Calibration of the tree to obtain absolute age estimates was not possible for this group. No reliable fossils have been described for Iridaceae before the Miocene and only a few pollen records have been reported, all of uncertain assignation or assigned to other clades of Iridaceae, too distant from Watsonieae to be useful in the present study (Goldblatt et al., 2008). The only option remaining is to use secondary calibration points, but these have additional problems making them unattractive for our purpose (Forest, 2009; Graur and Martin, 2004). Nevertheless, an ultrametric tree was required for subsequent analyses and thus we assigned a value of 1·0 to the root of the Lapeirousia clade using a uniform prior.
Biogeographical analyses
Geographical areas were defined based on the current taxa distributions and areas as characterized by Linder et al. (2012) and Born et al. (2007) for the GCFR. We recognize six areas: (A) GCFR, (B) Southern African Region, (C) Zambezian Region, (D) Ethiopian/Somalian Region, (E) Sudanian/Sub-Saharan Region and (F) Congolian Region (see Fig. 1).
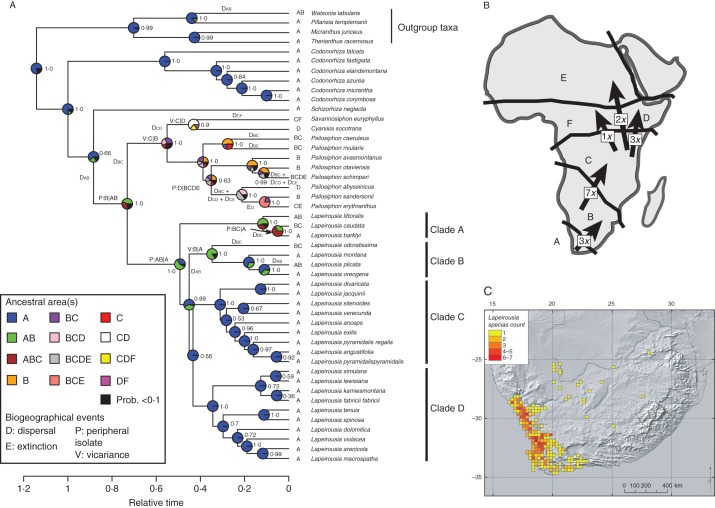
Fig. 1.
(A) Biogeographical scenario of the Lapeirousia clade inferred using the DEC model and displayed on the BEAST maximum credibility clade tree. (B) Area circumscriptions; the number of dispersal events inferred by the DEC analysis between areas is indicated with arrows: (A) GCFR, (B) Southern African Region, (C) Zambezian Region, (D) Ethiopian/Somalian Region, (E) Sudanian/Sub-Saharan Region, and (F) Congolian Region. (C) Distribution and species richness of genus Lapeirousia in South Africa.
The dispersal–extinction–cladogenesis (DEC) likelihood model implemented in Lagrange v.2.0.1 (Ree et al., 2005; Ree and Smith, 2008) was used to investigate the biogeographical history of this clade (further details on this method are presented in Buerki et al., 2011). The Lagrange analysis was performed on the BEAST maximum clade credibility tree (excluding the outgroup taxa) with the maximum number of areas at nodes constrained to two. Ancestral area reconstructions for each node were plotted on the BEAST tree using pie charts and the biogeographical scenario was produced using a collection of R scripts following Buerki et al. (2012). This latter procedure (i.e. the type and frequency of transition events between ancestral and descendant nodes along the dated phylogenetic tree) was inferred according to the Q matrix implemented in the DEC model (Ree et al., 2005; Ree and Smith, 2008).
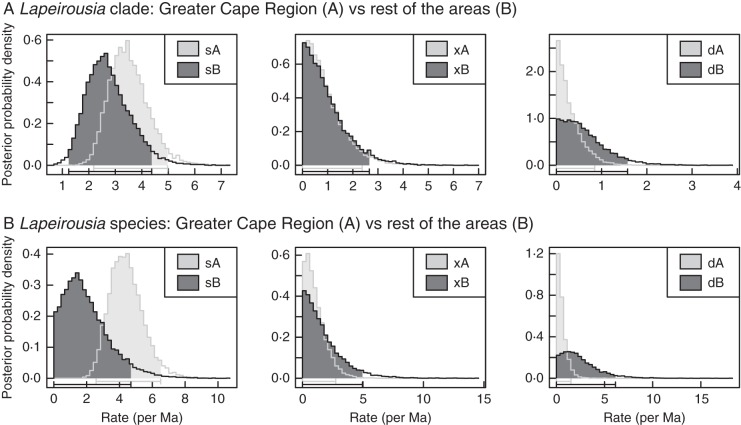
We assessed the effects of biogeography on the diversification of this clade using the geographical state speciation and extinction model (GeoSSE) implemented in the R package diversitree (FitzJohn, 2012). This method is an extension of the marginal ancestral state reconstruction for discrete characters (BiSSE) developed for biogeographical purposes (Goldberg et al., 2011). The GeoSSE method was applied separately on the whole Lapeirousia clade and on Lapeirousia itself. The BEAST maximum clade credibility tree was used because these methods require an ultrametric fully bifurcated tree. Only the patterns of diversification between the GCFR and the other areas were investigated. This method simultaneously features the characteristics of the constant-rates birth–death model with a three-state Markov model and allows the estimation of region-dependant rates of speciation, extinction and range evolution (Goldberg et al., 2011). Seven parameters can be estimated by the model: speciation within regions A (sA) and B (sB), between-region speciation (sAB), extinction from regions A (xA) and B (xB), dispersal from A to B (dA) and dispersal from B to A (dB) (see fig. 1 in Goldberg et al., 2011). Maximum-likelihood (ML) parameter estimation and model comparison were conducted followed by Bayesian parameter estimation through MCMC (as done in Buerki et al., 2012). To reduce the complexity of the analysis, two GeoSSE models – the full model and the model without between-region speciation (sAB) – were estimated under an ML framework and compared using a likelihood ratio test as implemented in diversitree (FitzJohn, 2012). For all analyses, the model without sAB constantly fitted the data better, suggesting that there are regional differences in diversification. Subsequently, an MCMC approach was used to perform a Bayesian analysis based on the six-parameter GeoSSE model. ML rate estimates were used as priors to seed the MCMC analysis. The MCMC was run for 10 000 generations and posterior probability distributions for the GeoSSE parameters were summarized using the function profiles.plot implemented in diversitree.
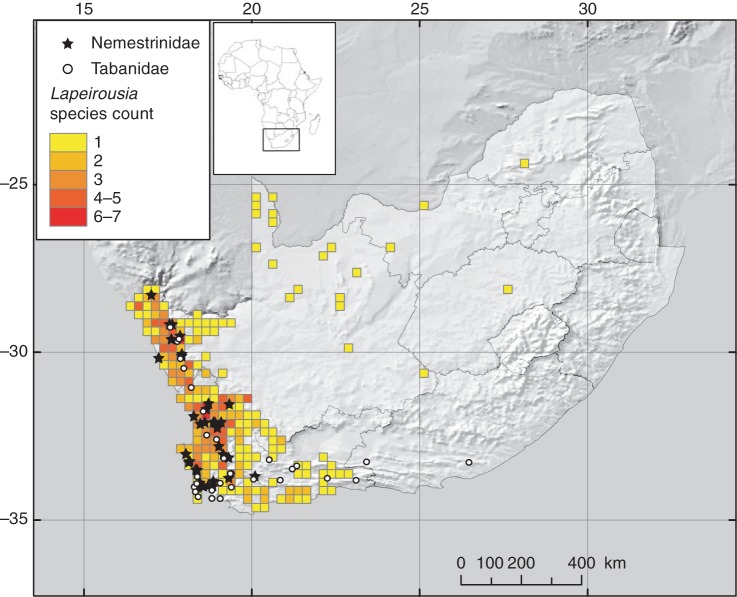
The specimen records of long-proboscid flies (families Nemestrinidae and Tabanidae), the ancestral state for the genus (see below), were mapped over the species richness of Lapeirousia. Biogeographical data for South African species of Lapeirousia was extracted from the PRECIS plant database (Computerised Information System of the National Herbarium in Pretoria, PRE; Germishuizen and Meyer, 2003). The database comprises over 1·7 million geo-referenced specimen records for over 22 000 plant taxa. Species distribution data for the two long-proboscid fly families (Nemestrinidae and Tabanidae) that are associated with pollination of Lapeirousia were extracted from taxonomic revisions (Usher, 1972; Barraclough, 2006) and specimen records housed in the Iziko Museum (Cape Town, South Africa), Natal Museum (Pietermaritzburg, South Africa) and the Durban Natural Science Museum (Durban, South Africa). These museums are the three main repositories of long-proboscid fly material. For Nemestrinidae, locality data for the genera Prosoeca Schiner (P. peringueyi Lichtwardt; P. sp. nov.; Manning and Goldblatt, 1996) and Moegistorhynchus Macquart (M. braunsi Bequaert; M. brevirostris Wiedemann; M. longirostris Wiedemann) were extracted and mapped. For Tabanidae, locality data for the genus Philoliche Hardwicke (P. rostrata Linnaeus; P. gulosa Wiedemann) were extracted and mapped.
Optimization of pollinator and substrate types
For each species of Lapeirousia, we scored the associated pollinator and substrate type(s) and reconstructed the ancestral character using the ML method implemented in the R package ape and the function ace, by setting the type argument to discrete (Paradis et al., 2004). Information on pollinator and substrate types (observed and inferred) for pollinator types was compiled from the literature and complemented by field observation and expertise of the group (see Table 1 for details). This analysis was performed only on Lapeirousia as the information for the other genera of the Lapeirousia clade is incomplete. The ancestral reconstructions were performed on the BEAST maximum clade credibility tree and results were displayed on the tree using the ‘thermo’ argument from the nodelabels function in ape (Paradis et al., 2004). Four pollinator types were defined and scored: (1) large-bodied bee, (2) sphingid moth (Sphingidae), (3) long-proboscid fly (including Tabanidae and Nemestrinidae) and (4) generalist. The long-proboscid fly families Tabanidae and Nemestrinidae are considered as different pollinator guilds (Goldblatt and Manning, 2006) and we adopt this view here, although we scored them as a single syndrome in the present study, as the ML optimization method does not allow the use of polymorphic characters. The distinction between the Tabanidae and Nemestrinidae is made clear in Fig. 3. Two other pollinator types are recorded in Lapeirousia, bee flies (Bombyliidae) and settling moths; these are each found in only one species and in combination with one of the four pollinator types scored (see above). As mentioned above, the ML method employed to optimize pollinator and substrate types does not allow the use of polymorphism, and thus we removed bee fly and settling moth syndromes from the reconstruction. The five substrate types used were defined as follows: (1) sand, (2) clay, (3) quartzite, (4) granite and (5) shale. We also accounted for polymorphism in substrate preference by including two states consisting of two of the above types: (6) sand/granite and (7) sand/clay. The shifts in pollinator and substrate types were mapped onto a lineage-through-time (LTT) plot obtained using the maximum credibility clade tree from BEAST and the R package ape (Paradis et al., 2004). A shift in either substrate or pollination syndrome was recognized when a change occurred between a state with a probability >0·5 and a different state with a probability >0·5; these shifts can take place either between two nodes or between a node and a tip.
Table 1.
Pollinator types, substrate types, phenology and distribution for each species included in the phylogenetic analysis of the Lapeirousia clade and used in the optimization analyses
| Species | Pollinator type | Substrate type | Distribution |
|---|---|---|---|
| Codonorhiza azurea | Apidae (1) | Granite (3,4) | GCFR |
| Codonorhiza corymbosa | Generalist (1) | Granite (3,4) | GCFR |
| Codonorhiza elandsmontana | Tabanidae (3) | Sand (3) | GCFR |
| Codonorhiza falcata | Apidae/Tabanidae/Nemestrinidae (3,6) | Sandstone (4) | GCFR |
| Codonorhiza fastigiata | – | Clay/Sandstone (3,4) | GCFR |
| Codonorhiza micrantha | Settling moth (1,3,6) | Sandstone (4) | GCFR |
| Cyanixia socotrana | – | – | Ethiopian–Somalian Region |
| Lapeirousia anceps | Tabanidae/Nemestrinidae (1,8) | Sand (2,3,4) | GCFR |
| Lapeirousia angustifolia | Generalist (3) | Clay (3) | GCFR |
| Lapeirousia arenicola | Tabanidae (1,3,6) | Sand (2,4) | GCFR |
| Lapeirousia barklyi | Apidae (1) | Sand (2,4) | GCFR |
| Lapeirousia caudata | Sphingidae (3,5) | Sand (5) | Southern African & Zambezian Regions |
| Lapeirousia divaricata | Generalist (1) | Sand (4) | GCFR |
| Lapeirousia dolomitica | Nemestrinidae (2) | Quartzite (3) | GCFR |
| Lapeirousia exilis | Apidae/Bombyliidae (1) | Sand (2,4) | GCFR |
| Lapeirousia fabricii subsp fabricii | Tabanidae/Nemestrinidae (1) | Sand/Granite (3) | GCFR |
| Lapeirousia jacquinii | Nemestrinidae (1) | Sand (2,4) | GCFR |
| Lapeirousia kamiesmontana | Nemestrinidae (3) | Granite (3) | GCFR |
| Lapeirousia lewisiana | Nemestrinidae (1) | Granite (3) | GCFR |
| Lapeirousia littoralis | Sphingidae (3,5) | Sand (5) | GCFR & Southern African Region |
| Lapeirousia macrospatha | Nemestrinidae (1) | Sand (3) | GCFR |
| Lapeirousia montana | Generalist (1) | Clay (4) | GCFR |
| Lapeirousia odoratissima | Sphingidae (5) | Sand (5) | Southern African & Zambezian Regions |
| Lapeirousia oreogena | Nemestrinidae (1) | Clay (2,4) | GCFR |
| Lapeirousia plicata susp. plicata | Generalist (1) | Clay (3,4) | GCFR & Southern African Region |
| Lapeirousia pyramidalis subsp. pyramidalis | Sphingidae (1,7) | Clay (7) | GCFR |
| Lapeirousia pyramidalis subsp. regalis | Nemestrinidae (1,7) | Sand (7) | GCFR |
| Lapeirousia silenoides | Nemestrinidae (1) | Granite (2,4) | GCFR |
| Lapeirousia simulans | Tabanidae (6) | Sand (7) | GCFR |
| Lapeirousia spinosa | Apidae/Settling moth (7) | Sand/Clay (7) | GCFR |
| Lapeirousia tenuis | Apidae (6) | Clay (7) | GCFR |
| Lapeirousia verecunda | Tabanidae/Nemestrinidae (1) | Shale (2,4) | GCFR |
| Lapeirousia violacea | Nemestrinidae (1) | Sand (2,4) | GCFR |
| Psilosiphon abyssinicus | Generalist (5) | – | Ethiopian–Somalian Region |
| Psilosiphon avasmontanus | Apidae (5) | – | Southern African Region |
| Psilosiphon coeruleus | Apidae (3) | – | Southern African & Zambezian Regions |
| Psilosiphon erythranthus | Generalist (5) | – | Zambezian & Saharan-Sudanian Regions |
| Psilosiphon otaviensis | Tabanidae/Nemestrinidae? (5) | – | Southern African Region |
| Psilosiphon rivularis | Apidae (5) | Granite (5) | Southern African & Zambezian Regions |
| Psilosiphon sandersonii | Generalist (5) | – | Southern African Region |
| Psilosiphon schimperi | Sphingidae (5) | – | Southern African, Zambezian, Ethiopian–Somalian & Saharan–Sudanian Regions |
| Savannosiphon euryphyllus | Sphingidae (3; inferred) | – | Zambezian & Congolian Regions |
| Schizorhiza neglecta | Generalist (3) | Sandstone (3) | GCFR |
| Outgroups | |||
| Micranthus junceus | – | – | GCFR |
| Pillansia templemanii | – | – | GCFR |
| Therianthus racemosus | – | – | GCFR |
| Watsonia tabularis | – | – | GCFR and Southern African Region |
References: (1) Goldblatt et al. (1995); (2) Goldblatt and Manning (2014); (3) P. Goldblatt and J. C. Manning, pers. obs.; (4) Goldblatt (1972); (5) Goldblatt (1990); (6) Goldblatt and Manning (2006); (7) Goldblatt and Manning (1994); (8) Pauw et al. (2008).
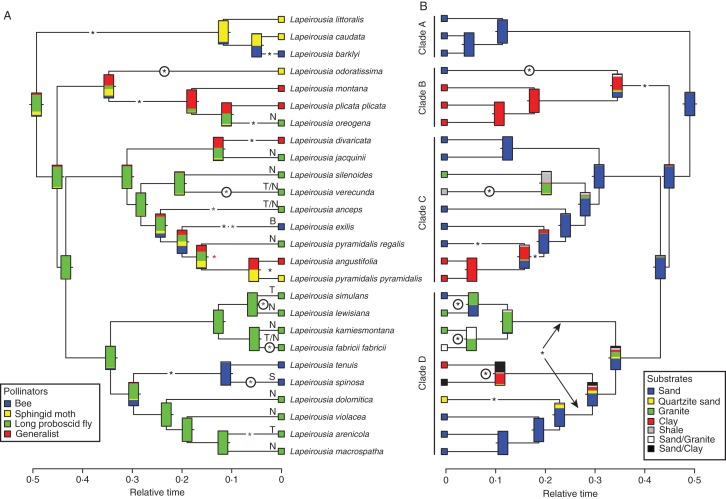
Fig. 3.
Maximum-likelihood optimizations of pollination syndromes (A) and substrate types (B) for Lapeirousia. The shifts in pollination syndromes and substrate types are displayed on the branches (or between branches if the exact position is uncertain between two sister lineages) by asterisks (*): black asterisks denote shifts between two nodes assigned a particular majority state (>0·5); green asterisks indicate assumed shifts on terminal branches due to the presence of polymorphic states; the red asterisk indicates the shift in pollination syndrome discussed in the text. The two alternative positions of a particular substrate type shift are indicated by an arrow. Species with two pollination systems are identified in A by an ‘S’ (settling months) and a ‘B’ (Bombyliidae). The two long-proboscid pollination systems are also specified: Tabanidae (T) and Nemestrinidae (N). The scale bars under the trees represent relative time (see text for details).
For biogeography, we examined the effects of pollinator types on the diversification of Lapeirousia using the multistate speciation and extinction model (MUSSE), implemented in the R package diversitree (FitzJohn, 2012). As for GeoSSE (see above), this method is an extension of the marginal ancestral state reconstruction for discrete characters (BiSSE) developed to accommodate multiple state characters in diversification analyses (FitzJohn, 2010). Again, the BEAST maximum clade credibility tree was used because this method requires an ultrametric, fully bifurcated tree. The same approach as presented for the GeoSSE method was applied here, but the MCMC was run for 1000 generations. It was not possible to apply the same approach to examine the effect of substrate types on diversification rates, as the number of states for substrate type is too high, which resulted in the analysis failing to converge properly.
RESULTS
Phylogenetic relationships
The combined matrix comprises 8204 characters, of which 547 are potentially parsimony-informative (7·9 %; Table 2). The most variable region in terms of potentially parsimony-informative characters is the nuclear RBP2 (13·3 %), while the most variable plastid region is the intergenic spacer ndhF-rpl32 (11·6 %; Table 2). The phylogenetic analyses performed here produced a generally well-supported topology. The genera Lapeirousia, Psilosiphon and Codonorhiza are all well supported (PP = 1·0; Fig. 1). Codonorhiza is retrieved as sister to the remainder of the Lapeirousia clade, in which the monotypic genus Schizorhiza is sister to a pair of clades, the first one formed by Lapeirousia and the second formed by genera Savannosiphon and Cyanixia, sister to Psilosiphon. Within Lapeirousia, section Lapeirousia is paraphyletic, with three of its species (L. littoralis, L. caudata and L. barklyi) retrieved as sister to the rest of the genus, in which section Sophronia is sister to section Chasmatocallis and the rest of section Lapeirousia. The BEAST analysis produced an identical tree with very similar posterior probabilities for each node, except for the relationship between L. silenoides and L. verecunda. In the BEAST analysis, these two species are found as sister (Fig. 1), while in the MrBayes analysis L. silenoides is sister to a clade comprising L. verecunda and five other taxa (Supplementary Data Fig. S1). In both cases, these relationships are not well supported, although the sister relationship of these two species is slightly better supported in the BEAST analysis. We used the dated tree obtained from BEAST in subsequent analyses, and thus consider L. silenoides and L. verecunda as sister species in the following discussion.
Table 2.
Characteristics of the ten DNA regions used in the phylogenetic analysis of the Lapeirousia clade
| matK | ndhJ | ycf5 | rpl16 | trnL-F | trnQ-rps16 | ndhF-rpl32 | rpl32-trnL | ACCD-psa1 | RPB2 | Combined | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aligned characters | 884 | 443 | 284 | 1072 | 863 | 1177 | 802 | 1072 | 944 | 663 | 8204 |
| Included characters | 859 | 374 | 212 | 1005 | 684 | 1069 | 612 | 783 | 804 | 512 | 6914 |
| Constant characters | 743 | 339 | 182 | 876 | 610 | 888 | 479 | 620 | 731 | 360 | 5828 |
| Variable characters | 116 | 35 | 30 | 129 | 74 | 181 | 133 | 163 | 73 | 152 | 1086 |
| PPI characters | 52 (6·1 %) | 8 (9·4 %) | 12 (5·7 %) | 67 (6·7 %) | 38 (5·6 %) | 106 (9·9 %) | 71 (11·6 %) | 90 (11·5 %) | 35 (4·4 %) | 68 (13·3 %) | 547 (7·9 %) |
PPI, parsimony-informative characters; percentage of PPI characters in the total number of aligned characters is indicated in parentheses.
Biogeographical analyses
The biogeographical scenario is represented on the BEAST maximum clade credibility tree using pie charts and by specifying on which branches/nodes each biogeographical event occurs (Fig. 1A), with the inferred dispersal events displayed on a map depicting the area circumscription used in the analysis (Fig. 1B). The biogeographical patterns observed in the Lapeirousia clade are explained using 16 dispersal events, one extinction, four peripheral isolations and three vicariance events (Fig. 1A, B). This analysis indicates that the Lapeirousia clade originated and was always present in the GCFR (area A) and subsequently dispersed elsewhere in Africa (areas B–F; Fig. 1). Most dispersal events take place in the sister clade of Lapeirousia (ten events), which has no species in the GCFR. Furthermore, the GCFR was not recolonized by lineages that dispersed out of the GCFR (clades A and B), a result supported by the null probability of dispersal towards the GCFR, as inferred by the GeoSSE analysis (Fig. 2).
Fig. 2.
Posterior probability distributions for the speciation (sA, sB), extinction (xA, xB) and dispersal (dA, dB) rates inferred by the GeoSSE model between the GCFR and the other areas. The analysis was performed on the Lapeirousia clade (A) and genus Lapeirousia (B). Abbreviations: A = GCFR and B = outside the GCFR, s = speciation, x = extinction, d = dispersal.
The biogeographical patterns within the clade sister to Lapeirousia (comprising genera Savannosiphon, Cyanixia and Psilosiphon) are shaped by peripheral isolation and vicariance events in their early evolutionary history, followed by several subsequent dispersals (Fig. 1). The first dispersal outside of the GCFR occurred on the stem lineage of Lapeirousia and its sister clade, followed by a peripheral isolation event where the sister clade inherits only area B, whereas Lapeirousia remains widespread in areas A and B. The most recent common ancestor of extant Lapeirousia species had a widespread distribution in areas A and B, where a peripheral isolation event resulted in clade A being distributed in areas A and B and where the remainder of the genus (clades B, C and D) is restricted to the GCFR (area A). Clade B then dispersed to area B and subsequently speciated by vicariance followed by two additional dispersals (Fig. 1). Clades C and D remained restricted to the GCFR and, within Lapeirousia, have more species restricted to the GCFR than there are species found in this area in clades A and B, i.e. clades that retain the widespread ancestral state have fewer species than those found only in the GCFR.
The GeoSSE analyses indicate an increase of diversification rates in the GCFR region compared with the remaining areas (i.e. a higher rate of speciation was recorded in the GCFR, but similar rates of extinction were found in this region and the rest of Africa; Fig. 2A). This result is even more significant when the analysis is conducted only on Lapeirousia (Fig. 2B). The GeoSSE analyses confirmed the biogeographical scenario obtained by the DEC model in inferring a very limited rate of dispersal to the GCFR (dA; Fig. 2).
Shifts in pollinator and substrate types
The ML optimization of pollinator types inferred the long-proboscid fly type as the ancestral state for Lapeirousia (Fig. 3A). Seventeen pollinator shifts are inferred during the evolution of Lapeirousia. In the case of substrate types, the analysis recovered a smaller number of shifts (n = 10; Fig. 3B). The shifts in pollinator and substrate types occurred mainly during the second half of the evolutionary history of the group, found closer to the terminals in the phylogenetic tree (Fig. 4). This could be due to the larger number of branches towards the present, but what is probably more relevant to the present argument is that the number of shifts in pollination systems is higher than the number of shifts in substrate types closer to the present (Fig. 4). There are six simultaneous shifts of pollinators and substrate, all assigned to the most recent common ancestors of extant species (Figs 3 and 4). In five of these six cases, the exact position of one or both shifts is ambiguous, potentially taking place on one of two sister lineages (e.g. shifts in the sister species pair L. simulans and L. lewisiana). In only one instance, on the crown node of L. pyramidalis subsp. pyramidalis, L. pyramidalis subsp. regalis and L. angustifolia, a shift was inferred despite absence of a change occurring between a state with a probability >0·5 and a different state with a probability >0·5 (see red asterisk in Fig. 3). None of the states inferred on this node has a probability >0·5, but given the states observed in the three species, a shift necessarily occurred on one of the two sister lineages arising from this node (Fig. 3). Finally, the pollinator MUSSE reconstruction inferred similar speciation rates for all pollinator types, with the 95 % confidence intervals of speciation rate densities for each state overlapping (Fig. 5).
Fig. 4.
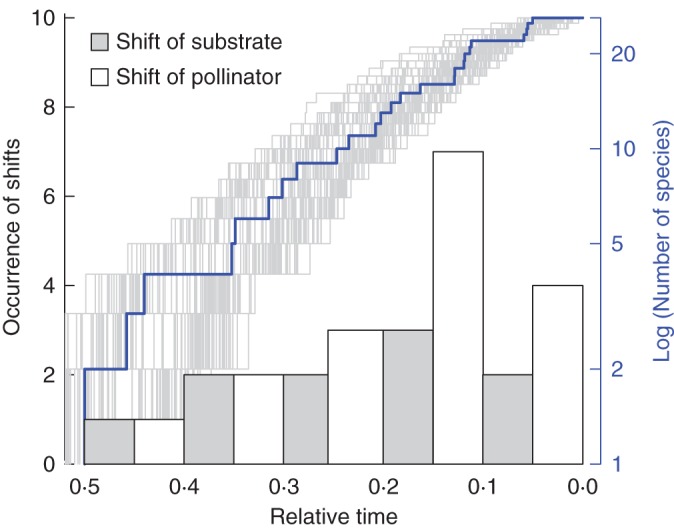
Lineage through time plots of genus Lapeirousia based on the BEAST maximum credibility clade tree (in blue) and 100 randomly selected trees (in grey) from the BEAST analysis. The distribution of shifts in pollinator (in white) and substrate types (in grey) are displayed as histograms. The position of these shifts is indicated in Fig 3.
Fig. 5.
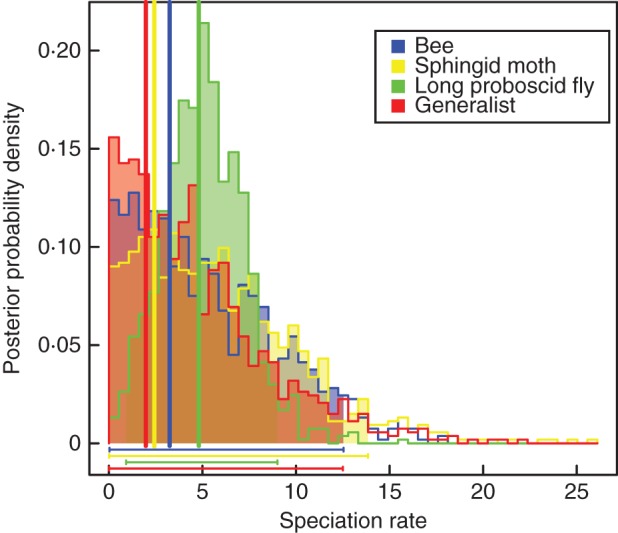
MUSSE reconstructions of the pollination syndromes inferred for genus Lapeirousia. The vertical lines represent ML estimations for each type and the horizontal lines represent the 95 % confidence intervals of speciation rate densities for each state.
DISCUSSION
Seventeen pollinator shifts and ten changes in substrate types were inferred during the evolution of Lapeirousia. All nine sister species pairs examined within the genus show divergence in pollination syndromes, while only four pairs are found on different substrate types. This evidence alone points to a predominant influence of pollinator shifts over substrate types on the speciation process within Lapeirousia, contrary to previous studies that favoured a more important role for edaphic factors. The long-proboscid fly pollination syndrome is the ancestral state in Lapeirousia, an unusual result in itself. Furthermore, the inferred distribution range on the crown node of Lapeirousia (i.e. GCFR) coincides with the adoption of the long-proboscid fly pollination system in this group and mirrors the foraging range of these pollinators. These results demonstrate the importance of integrating a biogeographical approach in studies of pollination syndromes, while taking into account the distribution of pollinators.
A link between biogeography and ecological shifts
The biogeographical reconstruction performed on the Lapeirousia clade revealed that the ancestral condition at the root node of the group is the GCFR (area A), with subsequent dispersals northward (with no return southward) to the Southern African region (area B), then the Zambezian region (area C) and the other regions to the north, as defined for the purpose of this study (Fig. 1). This northward migration scenario with an origin in the GCFR has been postulated in several other plant groups (e.g. Galley et al., 2006). Ten of the 16 dispersal events inferred from the Lagrange analysis involve the Southern Africa region (area B), of which seven are dispersals out of this region to the Zambezian region (area C). This pattern indicates that the GCFR, home to most of the diversity of Lapeirousia, and Codonorhiza and Schizorhiza, is the source of dispersal events rather than being a recipient. The GeoSSE analyses performed on the Lapeirousia clade as a whole, and on Lapeirousia alone, both show that speciation rates in the GCFR are higher than in the other regions combined (i.e. the rest of sub-Saharan Africa; Fig. 2), although more convincingly in the case of the former. Extinctions and dispersal rates on the other hand were similar between the GCFR and the rest of Africa.
Within Lapeirousia, for all species not restricted to the GCFR (which are found in clades A and B), shifts in pollination syndromes and substrate types are linked to vicariance and peripheral isolation events (Figs 1 and 3). The shift from long-proboscid fly to sphingid moth on the stem of clade A is associated with a peripheral isolation event and remained widespread between areas A and B (Fig. 1). A second peripheral isolation event is associated with the shift from sphingid moth to bee pollination on the terminal branch leading to L. barklyi, a species restricted to the GCFR, whereas the sister species, L. caudata, remains widespread in areas B and C. A vicariance event at the crown node of clade B is linked to four ecological shifts, two linked to pollinators and two to substrate types (Figs 1 and 3). A shift from generalist to the more specialized long-proboscid fly pollination syndrome on the terminal branch subtending L. oreogena is not associated with a biogeographical event as this species is restricted to the GCFR, where long-proboscid flies mostly occur (Fig. 6). Contrary to clades C and D, which are restricted to the GCFR and in which most species are pollinated by range-restricted long-proboscid flies, the dispersal success of species from clades A and B is associated with pollinator guilds of wider ranges (e.g. generalist, sphingid moths). The most recent common ancestor of Lapeirousia is estimated as having been pollinated by long-proboscid flies and, interestingly, constrained to areas A and B where these pollinators are also restricted.
Fig. 6.
Distribution of long-proboscid fly families Nemestrinidae (stars) and Tabanidae (circles) involved in the pollination of South African species of Lapeirousia, mapped over the distribution and species richness of Lapeirousia in South Africa.
The MUSSE analysis performed on the pollination systems did not identify one (or more) of the strategies as associated with high speciation rates (Fig. 5). Valente et al. (2012) found increased diversification rates associated with derived pollination systems in Gladiolus (Iridaceae), i.e. not the predominant (and ancestral) bee pollination syndrome. We found no such increased diversification rates when we compared the ancestral long-proboscid fly pollination system in Lapeirousia with the other systems (not shown), but the ML value for this pollinator type is the highest of those examined (vertical lines in Fig. 5). This indicates that the high diversification rates in Lapeirousia are linked to its presence in the GCFR, but does not identify unequivocally a potential causal factor to explain these increased diversification rates. The presence of a diversity of specialized pollination syndromes (Johnson and Steiner, 2000, 2003) rather than the shift to a particular system could, however, have influenced the diversification rates. The macroevolutionary history of species of Lapeirousia restricted to the GCFR (i.e. clades C and D) could only be inferred here using ecological shifts, without a biogeographical approach; this would be feasible, however, using a biogeographical analysis of the GCFR at a finer scale.
Pollination versus substrate shifts
The variety of pollinator guilds in the GCFR has unquestionably contributed to the high plant diversity of this region (Johnson, 1996, 2010), but evidence presented in many studies indicates that substrate diversity may have had a greater impact on plant diversity in general in the GCFR (e.g. Linder, 1985; Linder and Vlok, 1991) while the pollinators would have had a secondary role (e.g. Goldblatt and Manning, 1996, 2006). Further supporting this claim is the fact that many of the most species-rich groups in the GCFR (e.g. the Cape clades of Linder, 2003) show limited or no variation in pollination systems and have conserved floral morphology [e.g. Aspalathus (Leguminosae), Agathosma (Rutaceae), Muraltia (Polygalaceae), Phylica (Rhamnaceae), Oxalis (Oxalidaceae); Goldblatt and Manning, 2006]. These groups, therefore, would have probably been influenced more by the mosaic of soils and local climatic conditions than the diversity of pollinator guilds. As shown by Schnitzler et al. (2011), using a comparative analysis of four groups with and without varied pollination syndromes, the diversity of edaphic factors has played an important role in speciation in the GCFR in all these groups. On the other hand, a sister-species pair comparison in several Cape-centred groups revealed that more species divergence was potentially associated with pollinator differences than edaphic factors. These analyses were, however, based on a relatively small sample and focused on orchid groups (van der Niet and Johnson, 2009). Our study of Lapeirousia seems to contradict many previous studies as the available evidence points towards a greater involvement of pollinator shifts in speciation in the genus than changes in edaphic preferences.
Our analysis shows that the ancestral pollination system in genus Lapeirousia is long-proboscid flies and that other pollination strategies evolved subsequently, including that by large-bodied bees (Fig. 3). This is an unusual pattern as previous studies of GCFR groups with varied pollination systems show that long-proboscid fly pollination is a derived syndrome evolving from large-bodied bee or generalist systems (Goldblatt and Manning, 2006). One argument put forward to explain this situation is that once a species has evolved a flower with a long tube adapted to long-proboscid flies or moth pollination, bees no longer forage for nectar on these flowers and no longer visit them (see van der Niet and Johnson, 2012; Whittall and Hodges, 2007). In Gladiolus and Babiana (also Iridaceae), one pollination shift has been inferred for every five to six species (Goldblatt and Manning, 2006). Valente et al. (2012) confirm this estimate for Gladiolus, in which pollinator shifts have taken place every 5·4 species (based on their ML optimization of pollination systems onto a molecular phylogeny comprising 148 of the 265 species of the genus). In Lapeirousia, the presence of 17 shifts in pollination systems results in one shift every 1·5 species, almost four times as much as that reported for Babiana and Gladiolus. This is compiled by taking into account as shifts cases in which species have two pollinators (L. spinosa by bees and settling moths; L. exilis by large-bodied bees and Bombyliidae; three species pollinated by both Tabanidae and Nemestrinidae). One shift every 1·5 species is a remarkably high frequency by all accounts and surpasses all estimates presented to date.
Such a high rate of shift in pollination strategy could reasonably lead to the conclusion that speciation in this group of corm-bearing monocots is tightly linked to pollination. The same approach (i.e. character optimization on the phylogenetic tree) applied to substrate types reveals that edaphic factors also show a relatively high number of shifts (n = 10), although less than pollination syndromes (Fig. 3). The most likely interpretation regarding substrate types is that Lapeirousia originated on sandy substrates and then diversified multiple times to other substrate types. With ten shifts in substrate types within the genus, or one shift every 2·6 species, this suggests that pollination syndromes and environmental factors both played an important role in the diversification of the genus. Based on these observations alone, the available evidence points to a predominant influence of pollinator shifts on the speciation process within the genus, but does not rule out the hypothesis previously put forward that pollinator shifts would act more secondarily on the speciation process, which would potentially be led by edaphic preferences.
The LTT plot on Lapeirousia indicates that diversification rates were relatively constant throughout its history (Fig. 4), consistent with several other GCFR-centred groups (e.g. Linder, 2005; Schnitzler et al., 2011). The relative climatic stability of the region in the past two million years has been identified as one of the main factors responsible for these stable rates of diversification, a combination of more or less constant speciation rates coupled with low extinction rates (Cowling and Lombard, 2002). Others, however, have shown that both speciation and extinction rates are high in the GCFR (Buerki et al., 2012), assigning these high rates of extinction to the presence of large numbers of narrow endemics in this region. When pollination and substrate type shifts are mapped on the LTT, shifts in pollination strategies appear slightly more concentrated towards the tips of the tree, later in the evolutionary history of Lapeirousia, while shifts in substrate types are more evenly distributed (histogram in Fig. 4). Although this concentration of shifts towards the tips may be caused by an increase in the available number of branches where these shifts can occur, the number of shifts in pollination syndromes remains higher than the number of shifts in substrate types towards the tip of the tree. This concentration of pollination shifts towards the terminals, and the more even distribution of substrate type shifts, suggests that speciation may be triggered primarily by edaphic factors throughout the history of the genus and that reproductive isolation was achieved through shifts in pollination strategies later in the evolution of the group, as postulated by Goldblatt and Manning (1996). These authors concluded that speciation in Lapeirousia was either allopatric or resulting from microgeographical differentiation combined with ecological diversification triggered by the diverse substrate mosaic of the GCFR. They also found no evidence of pollinator-driven speciation. However, our sister species pairs comparisons provide evidence contradicting this conclusion.
A role for reinforcement in pollination shifts?
Two main competing hypotheses have been put forward to explain the manner in which changes in pollination syndromes take place and their influence in the speciation process (e.g. Van der Niet et al., 2006): (1) reproductive isolation is an indirect consequence of adaptation to different environmental settings (e.g. Johnson, 1996); and (2) shifts in pollination systems take place through reinforcement after secondary contact involving newly formed species that diverged primarily on different substrate types (e.g. Goldblatt and Manning, 2006). Using a comparative analysis of 41 sister species pairs from plant groups in the GCFR, including Iridaceae, Van der Niet et al. (2006) showed that pollination shifts are significantly linked to changes in substrate types in sympatric sister species pairs, while the contrary is evident for allopatric species pairs, i.e. no significant connection between edaphic and pollination shifts. The results of Van der Niet and colleagues provide support for a role of reinforcement in pollination syndrome shifts, which is also in accord with the conclusions of Goldblatt and Manning (1996) for Lapeirousia.
We established that pollination shifts occurred several times (n = 17) in the evolutionary history of Lapeirousia, while shifts in substrate types were less common (n = 10), at odds with the conclusions of Goldblatt and Manning (1996) regarding the importance of substrate in the speciation of this group. To evaluate this further, we used a sister species pair comparison for Lapeirousia and identified nine pairs in the phylogenetic tree (Fig. 3; Table 3). The small sampling size is too limited to justify the use of particular statistical tests (as in Van der Niet et al., 2006) but, nevertheless, provides additional information allowing us to elucidate this problem. All these nine pairs of sister species present a shift in pollination systems, either a comprehensive change (e.g. L. arenicola pollinated by tabanid flies and L. macrospatha by nemestrinid flies) or involving the adoption of a second pollination syndrome by one of the two species (e.g. L. tenuis is pollinated by bees, while L. spinosa is pollinated by bees and settling months), but not all pairs present shifts in substrate types (only four of the nine). For a given area in the GCFR, the co-occurrence of different pollinators might be more likely than the co-occurrence of different substrate types (or vice versa), which would introduce a spatial bias in pollinator versus substrate types. To our knowledge, this has not been explored in any great detail and should be considered when making inferences about shifts in pollination syndromes or substrate types between sister species pairs. Of the nine pairs examined, six occur either in sympatry or in parapatry and only one of these presents a clear divergence in substrate type (L. silenoides occurs on granite, while L. verecunda is restricted to shale); the other two cases involve the presence of one of the species on two substrate types, one of which is the same as its sister. Thus, all species pairs in Lapeirousia that are sympatric (or parapatric) have different pollinators while only half of them occur on different substrate types, suggesting that shifts in pollination syndrome are not often associated with shifts in substrate types in sympatric (or parapatric) species. This is contrary to the conclusions of Van der Niet et al. (2006) and presents the possibility that pollination shifts may have been more important than substrate shifts in speciation in Lapeirousia and that factors such as competition for pollinators and phenological shifts may play a greater role (Ollerton et al., 2003; Warren et al., 2011; Johnson et al., 2012). However, of these nine sister species pairs, only one presents a clear difference in flowering time (L. barklyi flowers in September and October, while L. caudata produces flowers from December to April; these species have allopatric distributions). Moreover, all the sympatric or parapatric sister species pairs have, to various extents, overlapping phenology. This situation indicates that phenological shifts are uncommon in Lapeirousia. Finally, the absence in our sampling of two species and three subspecies (see above) could possibly affect some of the sister species pair comparisons, but given the dominance of pollination syndrome shifts over substrate shifts in the nine observed pairs, it is reasonable to assume that these potential changes would not drastically alter the overall conclusions.
Table 3.
Species pairs examined; these species pairs were determined using the phylogenetic tree produced by the Bayesian inference (see Fig. 1)
| Sister species pair | Support | Pollinator type | Substrate type | Phenology | Distribution |
|---|---|---|---|---|---|
| Lapeirousia barklyi | 1·0 | Apidae | Sand | Sep–Oct | Allopatric |
| Lapeirousia caudata | Sphingidae | Sand | Dec–Apr | ||
| Lapeirousia oreogena | 1·0 | Nemestrinidae | Clay | Aug–Sep | Parapatric |
| Lapeirousia plicata subsp plicata | Generalist | Clay | Jul–Sep | ||
| Lapeirousia divaricata | 1·0 | Generalist | Sand | Sep–Oct | Parapatric |
| Lapeirousia jacquinii | Nemestrinidae | Sand | Aug–Sep | ||
| Lapeirousia silenoides | 0·67 | Nemestrinidae | Granite | Jul–Aug | Sympatric |
| Lapeirousia verecunda | Tabanidae/Nemestrinidae | Shale | Aug–Sep | ||
| Lapeirousia angustifolia | 0·92 | Generalist | Clay | Jul–Sep | Parapatric |
| Lapeirousia pyramidalis subsp. pyramidalis | Sphingidae | Clay | Jul–Sep | ||
| Lapeirousia lewisiana | 0·59 | Nemestrinidae | Granite | Sep–Oct | Allopatric |
| Lapeirousia simulans | Tabanidae | Sand | Aug–Sep | ||
| Lapeirousia fabricii subsp. fabricii | 0·36 | Tabanidae/Nemestrinidae | Sand / Granite | Sep–Oct | Sympatric |
| Lapeirousia kamiesmontana | Nemestrinidae | Granite | Oct | ||
| Lapeirousia spinosa | 1·0 | Apidae/Settling moth | Sand / Clay | Aug–Sep | Sympatric |
| Lapeirousia tenuis | Apidae | Clay | Jul–Aug | ||
| Lapeirousia arenicola | 0·99 | Tabanidae | Sand | Aug–Sep | Allopatric |
| Lapeirousia macrospatha | Nemestrinidae | Sand | Sep–Oct |
The type of distributions for each pair is based on field observations. Shifts in either pollinator or substrate types are marked in bold; non-overlapping phenologies are also marked in bold. Support values for each sister species pair are provided (BEAST Bayesian posterior probabilities; see Fig. 2).
CONCLUSIONS
Comparative phylogenetic approaches, such as those used here, provide an insight into the role that pollinators and other ecological factors have on the speciation process. These methods may also reveal general macroevolutionary patterns such as the direction of the evolution of a particular feature and its possible correlation with other ecological factors, but they have some limitations when it comes to understanding the processes responsible for these patterns. Investigations at the microevolutionary scale (at the population level for example) are needed to provide the necessary information regarding these evolutionary processes (Johnson, 2010; Smith, 2010; Van der Niet and Johnson, 2012). Many examples of this type of study are presented in this Special Issue (e.g. Peter and Johnson, 2014; Van der Niet et al., 2014). The current study of genus Lapeirousia reveals the general patterns in this group in terms of pollinator shifts and edaphic preferences and provides some evidence for the evolutionary processes involved in species divergence by showing that speciation events appear more likely to be primarily linked to shifts in pollination systems than substrate types for this genus. The phylogenetic relationships within Lapeirousia uncovered here will be crucial for the selection of species pairs or species complexes that could be the target of more detailed microevolutionary studies involving ecological and population genetics tools.
The importance of combining studies of pollination syndromes with a biogeographical approach, while taking into account the distribution of pollinators, proved to be a useful tool to investigate macroevolutionary processes. This was particularly well demonstrated in the present study, where the unusual optimization of long-proboscid fly pollination on the crown node of Lapeirousia coincides with the biogeographical scenario inferring a distribution range corresponding to the one occupied by the long-proboscid fly pollinators. Macroevolutionary studies making use of a suite of phylogenetic approaches hold promising avenues for our understanding of the processes that shape speciation.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Ingrid Nänni, Timo van der Niet, Len Porter, Eugene Marinus, Nick Helme, Nicola Bergh, Edith Kapinos and Laszlo Csiba for providing important material, field assistance or laboratory support. We thank Paul Rymer for providing primer sequences for the RPB2 region. We thank the Handling Editor, Jeffrey Karron, and two anonymous reviewers for constructive comments on an earlier version of this work. This work was supported by a grant from the Committee for Research and Exploration of the National Geographic Society (grant number 824307) and the Royal Botanic Gardens, Kew.
LITERATURE CITED
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Barraclough DA. An overview of the South African tangle-veined flies (Diptera: Nemestrinidae), with an annotated key to the genera and a checklist of species. Zootaxa. 2006;1277:39–63. [Google Scholar]
- Boberg E, Alexandersson R, Jonsson M, Maad J, Ågren J, Nilsson LA. Pollinator shifts and the evolution of spur length in the moth-pollinated orchid Platanthera bifolia. Annals of Botany. 2014;113 doi: 10.1093/aob/mct217. 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born J, Linder HP, Desmet P. The greater Cape Floristic Region. Journal of Biogeography. 2007;34:147–162. [Google Scholar]
- Buerki S, Forest F, Alvarez N, Nylander JAA, Arrigo N, Sanmartin I. An evaluation of new parsimony-based versus parametric inference methods in biogeography: a case study using the globally distributed plant family Sapindaceae. Journal of Biogeography. 2011;38:531–550. [Google Scholar]
- Buerki S, Jose S, Yadav SR, Goldblatt P, Manning JC, Forest F. Contrasting biogeographic and diversification patterns in two Mediterranean-type ecosystems. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0039377. e39377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RM, Lombard AT. Heterogeneity, speciation/extinction history and climate: explaining regional plant diversity patterns in the Cape Floristic Region. Diversity and Distributions. 2002;8:163–179. [Google Scholar]
- Crepet WL. Advanced (constant) insect pollination mechanisms – Patterns of evolution and implications vis-a-vis angiosperm diveristy. Annals of the Missouri Botanical Garden. 1984;71:607–630. [Google Scholar]
- Darwin CR. The various contrivances by which orchids are fertilised by insects. London: John Murray; 1877. [Google Scholar]
- de Jager ML, Ellis AG. Floral polymorphism and the fitness implications of attracting pollinating and florivorous insects. Annals of Botany. 2014;113 doi: 10.1093/aob/mct189. 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. EAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7 doi: 10.1186/1471-2148-7-214. 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzJohn RG. Quantitative traits and diversification. Systematic Biology. 2010;59:619–633. doi: 10.1093/sysbio/syq053. [DOI] [PubMed] [Google Scholar]
- FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution. 2012;3:1084–1092. [Google Scholar]
- Forest F. Calibrating the Tree of Life: fossils, molecules and evolutionary timescales. Annals of Botany. 2009;104:789–794. doi: 10.1093/aob/mcp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galley C, Bytebier B, Bellstedt DU, Linder HP. The Cape element in the Afrotemperate flora: from Cape to Cairo? Proceedings of the Royal Society of London B Biological Sciences. 2006;274:535–543. doi: 10.1098/rspb.2006.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germishuizen G, Meyer NL. Plants of southern Africa: an annotated checklist. Strelitzia. 2003;14:1–1231. [Google Scholar]
- Goldberg EE, Lancaster LT, Ree RH. Phylogenetic inference of reciprocal effects between geographic range evolution and diversification. Systematic Biology. 2011;60:451–465. doi: 10.1093/sysbio/syr046. [DOI] [PubMed] [Google Scholar]
- Goldblatt P. A revision of the genera Lapeirousia Pourret and Anomatheca Ker in the winter rainfall region of South Africa. Cape Town: The Bolus Herbarium, University of Cape Town; 1972. [Google Scholar]
- Goldblatt P. Systematics of Lapeirousia (Iridaceae-Ixioideae) in tropical Africa. Annals of the Missouri Botanical Garden. 1990;77:430–484. [Google Scholar]
- Goldblatt P, Manning JC. Leaf and corm tunic structure in Lapeirousia (Iridaceae—Ixioideae) in relation to phylogeny and infrageneric classification. Annals of the Missouri Botanical Garden. 1990;77:365–374. [Google Scholar]
- Goldblatt P, Manning JC. New taxa and revisions to the taxonomy of southern African Lapeirousia subgenus Lapeirousia (Iridaceae: subfamily Ixioideae) Novon. 1994;4:339–346. [Google Scholar]
- Goldblatt P, Manning JC. Phylogeny and speciation in Lapeirousia subgenus Lapeirousia (Iridaceae: Ixioideae) Annals of the Missouri Botanical Garden. 1996;83:346–361. [Google Scholar]
- Goldblatt P, Manning JC. Plant diversity of the Cape Region of southern Africa. Annals of the Missouri Botanical Garden. 2002;89:281–302. [Google Scholar]
- Goldblatt P, Manning JC. Radiation of pollination systems in the iridaceae of sub-Saharan Africa. Annals of Botany. 2006;97:317–344. doi: 10.1093/aob/mcj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt P, Manning JC. The Iris family: natural history and classification. Portland, OR: Timber Press; 2008. [Google Scholar]
- Goldblatt P, Manning JC. Systematics and biology of the sub-Saharan African Lapeirousia Pourr., and its segregates, the new genera Codonorhiza, Psilosiphon and Schizorhiza (Iridaceae: Crocoideae) Strelitzia. 2014 in press. [Google Scholar]
- Goldblatt P, Manning JC, Bernhardt P. Pollination biology of Lapeirousia subgenus Lapeirousia (Iridaceae) in Southern Africa – Floral divergence and adaptation for long-tongued fly pollination. Annals of the Missouri Botanical Garden. 1995;82:517–534. [Google Scholar]
- Goldblatt P, Rodriguez A, Powell MP, et al. Iridaceae ‘out of Australasia’? Phylogeny, biogeography, and divergence time based on plastid DNA sequences. Systematic Botany. 2008;33:495–508. [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends in Genetics. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Johnson SD. Pollination, adaptation and speciation models in the Cape flora of South Africa. Taxon. 1996;45:59–66. [Google Scholar]
- Johnson SD. The pollination niche and its role in the diversification and maintenance of the southern African flora. Philosophical Transactions of the Royal Society B-Biological Sciences. 2010;365:499–516. doi: 10.1098/rstb.2009.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SD, Hollens H, Kuhlmann M. Competition versus facilitation: conspecific effects on pollinator visitation and seed set in the iris Lapeirousia oreogena. Oikos. 2012;121:545–550. [Google Scholar]
- Johnson SD, Steiner KE. Generalization versus specialization in plant pollination systems. Trends in Ecology & Evolution. 2000;15:140–143. doi: 10.1016/s0169-5347(99)01811-x. [DOI] [PubMed] [Google Scholar]
- Johnson SD, Steiner KE. Specialized pollination systems in southern Africa. South African Journal of Science. 2003;99:345–348. [Google Scholar]
- Kay KM, Sargent RD. The role of animal pollination in plant speciation: integrating ecology, geography, and genetics. Annual Review of Ecology, Evolution and Systematics. 2009;40:637–656. [Google Scholar]
- Linder HP. Gene flow, speciation, and species diversity patterns in a species-rich area: the Cape flora. In: Vrba ES, editor. Species and speciation. Pretoria: Transvaal Museum; 1985. pp. 53–57. [Google Scholar]
- Linder HP. The radiation of the Cape flora, southern Africa. Biological Review. 2003;78:597–638. doi: 10.1017/s1464793103006171. [DOI] [PubMed] [Google Scholar]
- Linder HP. Evolution of diversity: the Cape flora. Trends in Plant Science. 2005;10:536–541. doi: 10.1016/j.tplants.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Linder HP, de Klerk HM, Born J, Burgess ND, Fjeldsa J, Rahbek C. The partitioning of Africa: statistically defined biogeographical regions in sub-Saharan Africa. Journal of Biogeography. 2012;39:1189–1205. [Google Scholar]
- Linder HP, Vlok JH. The morphology, taxonomy and evolution of Rhodocoma (Restionaceae) Plant Systematics and Evolution. 1991;175:139–160. [Google Scholar]
- Manning JC, Goldblatt P. The Prosoeca peringueyi (Diptera: Nemestrinidae) pollination guild in southern Africa: long-tongued flies and their tubular flowers. Annals of the Missouri Botanical Garden. 1996;83:67–86. [Google Scholar]
- Mittermeier RA, Gil PR, Hovman M, et al. Hotspots revisited: Earth's biologically richest and most threatened terrestrial ecoregions. Mexico: Agrupacion Sierra Madre; 2005. [Google Scholar]
- Nylander JAA. v2. 2004. MrModeltest. Evolutionary Biology Centre, Uppsala University, Program distributed by the author. [Google Scholar]
- Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Systematic Biology. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Ollerton J, Johnson SD, Cranmer L, Kellie S. The pollination ecology of an assemblage of grassland asclepiads in South Africa. Annals of Botany. 2003;92:807–834. doi: 10.1093/aob/mcg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pauw A, Stofberg J, Waterman RJ. Flies and flowers in Darwin's race. Evolution. 2008;63:268–279. doi: 10.1111/j.1558-5646.2008.00547.x. [DOI] [PubMed] [Google Scholar]
- Peter CI, Johnson SD. A pollinator shift explains floral divergence in an orchid species complex in South Africa. Annals of Botany. 2014;113 doi: 10.1093/aob/mct216. 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procheş Ş, Cowling RM, Goldblatt P, Manning JC, Snijman DA. An overview of the Cape geophytes. Biological Journal of the Linnean Society. 2006;87:27–43. [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1·5. 2007 Available from http://beast.bio.ed.ac.uk/Tracer . [Google Scholar]
- Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schnitzler J, Barraclough TG, Boatwright JS, et al. Causes of plant diversification in the Cape biodiversity hotspot of South Africa. Systematic Biology. 2011;60:343–357. doi: 10.1093/sysbio/syr006. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Smith SD. Using phylogenetics to detect pollinator-mediated floral evolution. New Phytologist. 2010;188:354–363. doi: 10.1111/j.1469-8137.2010.03292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Adaptive radiation of reproductive characteristics in angiosperms, I: Pollination mechanisms. Annual Review of Ecology and Systematics. 1970;1:307–326. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinaur Associates; 2002. 4·0b10. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Usher PJ. A review of the South African horsefly fauna (Diptera: Tabanidae) Annals of the Natal Museum. 1972;21:459–507. [Google Scholar]
- Valente LM, Manning JC, Goldblatt P, Vargas P. Did pollination shifts drive diversification in Southern African Gladiolus? Evaluating the model of pollinator-driven speciation. American Naturalist. 2012;180:83–98. doi: 10.1086/666003. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Johnson SD. Patterns of plant speciation in the Cape floristic region. Molecular Phylogenetics and Evolution. 2009;51:85–93. doi: 10.1016/j.ympev.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Johnson SD. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends in Ecology & Evolution. 2012;27:353–361. doi: 10.1016/j.tree.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Johnson SD, Linder HP. Macroevolutionary data suggest a role for reinforcement in pollination system shifts. Evolution. 2006;60:1596–1601. doi: 10.1554/05-705.1. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Pirie MD, Shuttleworth A, Johnson SD, Midgley JJ. Do pollinator distributions underlie the evolution of pollination ecotypes in the Cape shrub Erica plukenetii? Annals of Botany. 2014;113 doi: 10.1093/aob/mct193. 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren BH, Bakker FT, Bellstedt DU, et al. Consistent phenological shifts in the making of a biodiversity hotspot: the Cape flora. BMC Evolutionary Biology. 2011;11:39. doi: 10.1186/1471-2148-11-39. doi:10.1186/1471-2148-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall JB, Hodges SA. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447:706–709. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.