Abstract
The emerging field of “metabolomics,” in which a large number of small molecule metabolites from body fluids or tissues are detected quantitatively in a single step, promises immense potential for early diagnosis, therapy monitoring and for understanding the pathogenesis of many diseases. Metabolomics methods are mostly focused on the information rich analytical techniques of nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Analysis of the data from these high-resolution methods using advanced chemometric approaches provides a powerful platform for translational and clinical research, and diagnostic applications. In this review, the current trends and recent advances in NMR- and MS-based metabolomics are described with a focus on the development of advanced NMR and MS methods, improved multivariate statistical data analysis and recent applications in the area of cancer, diabetes, inborn errors of metabolism, and cardiovascular diseases.
Keywords: Metabolomics, metabonomics, NMR spectroscopy, mass spectrometry, multivariate statistical analysis, early detection, cancer, diabetes, inborn errors of metabolism, coronary heart disease
While vast progress in the fields of genomics and proteomics has occurred, additional evidence of biological end points of human diseases is highly desired for disease diagnosis, prognosis and therapeutic development. Thus detection of metabolites that are involved in human diseases or that can be used to help develop new drugs, using cells, tissue, organs or biological fluids has risen in prominence over the past several years [1–6]. The promising field of metabolomics, and the closely related areas of metabonomics and metabolite profiling, involve the quantitative detection of multiple small molecule metabolites in biological systems. An improved understanding of biological systems at the molecular level, i.e., systems biology, is anticipated to result from the metabolomics approach, especially when combined with genomics and proteomics information. Perhaps even more importantly, truly “personalized medicine” is anticipated to become a reality through the advancement of metabolomics and other ‘omic’ sciences [7]. A major advantage in the application of metabolomics stems from an improved ability to detect up to many hundreds of metabolites in parallel, which provides an efficient method for monitoring altered biochemistry. It is thought that the human body contains approximately 3000 to 5000 detectable metabolites, a sizable fraction of which have already been identified [8]. In addition, metabolite concentration alterations are often amplified when compared to those of gene expression or protein levels, making the detection of metabolite profiles a relatively sensitive measure of biological status. Changes in biological status is then based on the detection of perturbations in the concentrations and fluxes of specific endogenous metabolites involved in a number of key disease-related or other specific cellular pathways. Thus, metabolomics can reveal crucial information that is closely related to the current disease or theraputic status. More generally, the metabolic profile of biological specimens is affected by a numerous factors such as diet, age, ethnicity, drugs, lifestyle or gut microfloral populations, and these factors need to be either controlled or deconvoluted in order to obtain information specific to disease [2]. A number of articles providing information on the background of NMR- and MS-based metabolomics and related areas, its various applications and technologies, as well as the advantages and limitations of the metabolomics approach have appeared [6, 9–19].
Among the analytical techniques that can be employed for metabolomics applications, nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) are the most common. A number of methodologies within these two technological areas are currently being developed specifically to deal with the types of complex samples analyzed in metabolomics studies. NMR spectroscopy is known as one of the premier methods for the analyses of multi-component mixtures as it requires little or no sample preparation; is rapid, non-destructive, and non-invasive; and provides highly reproducible results (coefficient of variation ~1–2%). Peaks in the NMR spectra can be reliably assigned to specific metabolic species, based on their chemical shifts and multiplet patterns, and thus NMR provides a wealth of information on the identity and quantity of a large number of metabolites in parallel from a single experiment. With advanced high-throughput NMR methodology, up to 200 samples can be measured within a day with the assistance of flow-injection probes and automated liquid handlers. The detection limit can also be decreased to 10’s of ng by the use of high field magnets, cryogenically cooled probes, microcoil probes equipped to handle very small samples, and methodologies that couple NMR to liquid chromatography and solid phase extraction [20–22]. On the other hand, the intrinsic high sensitivity (typically pg level) of MS detection makes it an important method for measuring metabolites in complex biofluids. A variety of MS methods in combination with separation techniques such as gas-chromatography (GC) and liquid chromatography (LC) and their variants have been used in numerous metabolomics investigations [19, 23–27]. Recently, a variety of promising atmospheric sample introduction MS methods have been developed that require essentially no sample separation or preparation [28–30].
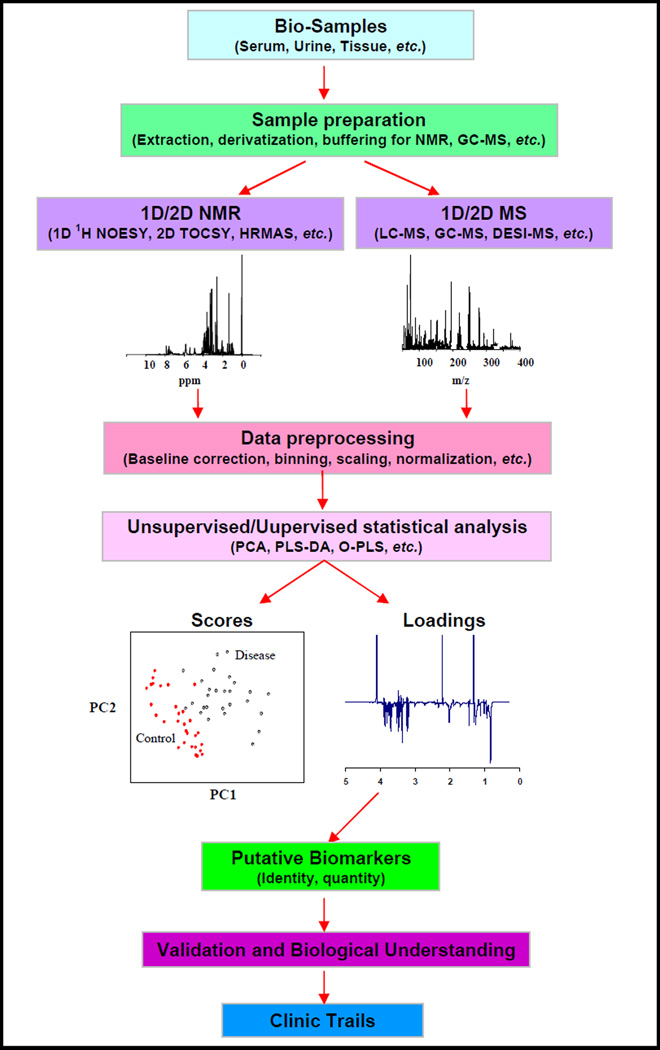
Data from NMR and MS experiments are generally complex since they contain qualitative/quantitative information on upwards of several hundreds of metabolites. Multivariate statistical analyses are thus used for data reduction and in particular for differentiating biofluids samples into “disease” and “control” populations based on the differences in signals of multiple metabolites (Figure 1). A variety of statistical methodologies exist and many are now easily accessible via commercial software or “freeware,” and these methods provide extremely helpful tools for filtering the large amounts of data and for accessing the often-subtle biochemical perturbations latent in the spectra [31–35]. In addition, these approaches are used to extract single biomarkers or sets of biomarkers with the best properties for the assessment of disease status. Validation of such putative biomarkers is of great importance, as is the biological understanding of the disease that can provide additional validation in the application of metabolomics.
Figure 1.
Schematic representation illustrating the NMR/MS based metabolomics workflow.
Biological specimens
Urine and blood serum or plasma are the most commonly used biofluids for metabolomics-based studies for the simple reasons that they both contain hundreds to thousands of detectable metabolites and can be obtained non- or minimally invasively. A number of other fluids such as cerebrospinal fluid, bile, seminal fluid, amniotic fluid, synovial fluid, gut aspirate and saliva have also been studied [36–38]. More recently, metabolic profiling of intact tissue and its lipid and aqueous metabolites extracts is gaining more importance for biomarker detection [39].
Compared with other biofluids, the analysis of urine provides certain obvious advantages. The relatively low concentrations of proteins and high concentrations of low molecular weight compounds minimize sample preparation and result in high quality measurements due to the narrow line widths of the spectral peaks in the NMR spectra. These characteristics enhance the process of biomarker identification by NMR for both diagnostic and monitoring applications. However, the high salt content of urine is more challenging for MS measurements which typically require some sample pretreatment.
Blood maintains a normal homeostasis in the human body by constant regulatory mechanisms and hence metabolic profiling of serum/plasma provides a global view of the instantaneous metabolic status. Moreover, blood perfuses essentially all living cells in the human body and thus is anticipated to carry vital information on virtually every cell. Unlike urine, the NMR spectrum of serum/plasma includes both narrow signals from small molecule metabolites and broad signals from proteins and lipids. A variety of spectral editing methods are used to selectively detect small or large molecules signals. MS analysis of serum is normally carried out using extracts, and in the case of GC/MS, derivatization procedures.
Metabolic profiling using intact tissue has gained momentum as an approach for understanding the molecular basis of diseases [39]. This interest stems from the fact that biomarkers due to pathophysiological stress are anticipated to be more highly concentrated in the pathological source for diseases such as cancer. The latest technological advancements in NMR have reduced the required sample quantity to as little as a few mgs so that even the biopsy tissue is sufficient to obtain good quality NMR spectra with resolution that is comparable to solution-state spectra. The rich metabolic profile of tissue is thought to be particularly useful for guiding the detection of biomarkers in more relatively easily accessible biofluids.
NMR Spectroscopy
A number of one and two-dimensional NMR methods are currently used for metabolomics applications. A recent article provides NMR experimental protocols for the common biological samples used in metabolomics along with protocols for sample preparation [40].
Currently, the simple one pulse sequence and one-dimensional nuclear Overhauser enhancement spectroscopy (NOESY) sequence with water suppression are the most commonly used NMR methods for metabolomics applications. Water suppression in the one pulse experiment depends more critically on good shimming. On the other hand, 1D NOESY is more robust and provides a flatter baseline under similar conditions. A number of pulse sequences are available all of which are designed to effectively suppress the high intensity water signal leaving the metabolites signals intact [41, 42]. A commonly used method for suppressing the broad signals from large molecules (such as in tissue or serum samples) is the Carr–Purcell–Mieboom–Gill (CPMG) sequence. This sequence is generally robust, and is widely used in a number of studies to date. In contrast, the so-called “diffusion edited” NMR experiment may be used for observing signals from large molecules such as lipids [43]. The 1D selective TOCSY experiment has been successfully applied to metabolomics studies to detect metabolites quantitatively even if they are found at concentrations 10–100 times below those of the major components [44]. This approach has been shown to be highly useful for detecting targeted metabolites in biological samples [45].
2D NMR methods are highly useful for reducing the spectral complexity and obtaining connectivity between the nuclei to make assignments and identify metabolites. However, these methods have not been widely used in metabolomics to date because of their increased acquisition time, data size, and complexity in data analysis. Nevertheless, a small but growing number of papers report using 2D approaches in metabolomics studies [46–49]. The most commonly used include 2D–J spectroscopy, correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), heteronuclear single quantum coherence (HSQC) spectroscopy and heteronuclear multiple bond correlation (HMBC) experiments. 2D J-resolved (JRES) spectroscopy is attractive for metabolomics studies [50] because it can lead to a substantial simplification of the spectra. One drawback of this method is that the integral of signals is strongly influenced by T2 relaxation during generally the long t1 evolution period and hence only a relative quantification of concentration of metabolites is possible.
The use of 13C NMR in metabolomics, while attractive from the standpoint of improved resolution, has been limited due to the low natural abundance (~1.1%) and low gyromagnetic ratio of 13C nuclei, and therefore 13C NMR requires unacceptably long data acquisition times. To improve this situation, a 13C labeling approach that can be carried out directly in aqueous solution at ambient temperature has been introduced [51]. The analysis of complex mixtures such as urine, serum or other bio-fluids is improved because the approach can be combined with fast 2D (1H-13C) heteronuclear experiments to yield spectra with good signal-to-noise ratios. The method has been applied to identify patients with inborn errors of metabolism.
For the non-invasive metabolic profiling of tissue specimens, a technique called high resolution magic angle spinning (HRMAS) NMR spectroscopy is utilized. In this approach, a rotor containing the tissue sample is spun typically at 3 to 6 kHz at an angle of 54.7° relative to the applied magnetic field, resulting in high-resolution, liquid-like NMR spectra [52]. Most of the common NMR pulse techniques can be used for tissue NMR applications. 1H HRMAS NMR spectra of tissue samples can generally be obtained using small quantity of intact tissue (~ 5 to 20 mg).
Mass spectrometry
MS methods coupled with prior separation modalities such as gas chromatography (GC), liquid chromatography (LC), and capillary electrophoresis (CE) provide enormous amounts of chemical information for metabolomics studies [19, 23–26,53]. A range of MS instruments, including quadrupoles, triple quads, ion traps, and time-of-flight mass analyzers are commonly used. Tandem MS (MS/MS or even MSn) methods are often used to validate the identity of unknown molecules. Fourier-transform ion cyclotron resonance (FT-ICR) provides an (expensive) alternative approach with extremely high resolution and a mass accuracy better than 1 ppm [23]. The introduction of Orbitrap MS [53], allows an alternative high resolution mass analyzer that detects ions with very high mass accuracy.
LC-MS is currently the most important MS-based approach for metabolomics application [26] because of its sensitivity and rich information content. Biofluids such as urine can be directly injected into the LC system while samples such as serum require minimal sample preparation such as protein precipitation. LC-MS is considered to be a moderately high throughput method. The recently developed ultrahigh pressure liquid chromatography (UPLC) approach has significantly improved the chromatic resolution, and reduced the limit of detection by 3–5 fold [54]. A common challenge for LC- (and GC-) MS measurements is the inter-batch-variation as well as the fact that the separation process makes the analysis time-consuming. In addition, the selectivity of LC/GC-MS to specific classes of analytes provides both benefits and complications.
The high separation efficiency and reproducibility of GC-MS makes it also a very useful tool for metabolomics [55]. Depending on the sample preparation conditions, GC-MS can be applied to the analysis of a wide range of metabolite classes including ketones, aldehydes, alcohols, esters, sulfides, sugars, sugar-phosphates, sugar-alcohols, organic acids, amino acids, lipids, peptides, alkaloids, amines and amides. Unlike LC-MS, GC-MS often requires rather extensive sample preparation steps such as chemical derivatization before the analysis. A new technology that incorporates a second GC column (2D GC-MS) is very promising for metabolomics applications because of the additional resolution it provides [27, 56].
As an alternative to chromatographic separation, sample effusion methods [57] and several recently introduced atmospheric sample introduction methods appear promising for fast screening purposes. Recently developed techniques such as EESI (extractive electrospray ionization)-MS [28], DESI (desorption electrospray atmospheric ionization)-MS [29], DART (direct analysis in real time)-MS [30] promise new avenues for metabolic profiling of human body specimens particularly since these methods require little or no sample preparation or extraction. DESI is carried out by directing a charged and nebulized solvent toward an analyte of interest (which could be a biofluid sample directly deposited on to a piece of filter paper). DART-MS, which uses a stream of excited metastable He gas and hot N2 to volatilize and ionize analytes of interest, also provides real-time information on analytes of interest and can be used to identify metabolites. Another atmospheric method is EESI, which utilizes two colliding spray sources for ionization and introduction into the MS. EESI has been used recently to study human breath samples [58], and to analyze rat urine to identify dietary changes [59]. MALDI (matrix-assisted laser desorption/ionization)-MS has been investigated for the simultaneous detection of several metabolites using a synthetic cocktail of 30 metabolites separately and after spiking into a microbial extract [60]. A recent study that compared several ionization methods quantitatively using serum found more metabolite species by laser desorption on silica compared to other methods, indicating its potential for biomarker detection [61].
A summary of potentially useful NMR and MS methods for metabolomics applications is shown in Table 1.
Table 1.
NMR and MS methods used in metabolomics applications.
| Techniques | Information content | Advantages | disadvantages |
|---|---|---|---|
| NMR Methods | |||
| 1D sequences: presaturation and noesy | Detects signals from both small and relatively large molecular weight metabolites | Highly quantitative and good for high-throughput screening | Spectra are generally complex because of the presence of both high and low concentration molecular signals and extensive J couplings |
| CPMG | Provides signals from small molecules by suppressing large molecule NMR signals | The spectra are easy to interpret due to the simplification achieved, and are good for high-throughput screening applications using samples such as serum/plasma. CPMG can also be used for HRMAS-NMR studies. | Does not show appreciable simplification for biofluids such as urine in which majority of the metabolites are small molecules, and is less quantitative |
| Diffusion-edited sequence | Provides information on relatively large molecules such as lipids | Simple, good for high throughput screening of large molecules in serum/plasma | Not useful for samples such as urine where there are no appreciable difference in diffusion coefficients, and is less quantitative |
| 2D J-RES | The spectra are highly simplified due to the elimination of multiplicity due to J coupling | Overlapping signals can be better resolved which is very useful to interpret and to accurately follow the metabolites variation corresponding to the otherwise overlapping signals in normal 1D spectra | Since the spectrum is derived from a 2D experiment, it is more time consuming, and is less quantitative. |
| MS Methods | |||
| LC-MS (HPLC/UPLC-MS) | Metabolites are separated using liquid chromatography and detected using MS | Most commonly used method in metabolomics because of the simplicity in sample preparation, and is fairly high throughput | Ion suppression in the presence of multiple cations or anions can be problematic and significantly affect the results |
| GC-MS | Similar to LC-MS, but the separation is based on gas chromatography | The method can be more sensitive and more reproducible than LC- MS. It has high separation efficiency and is more quantitative than LC-MS. | Requires rather tedious sample preparation steps, and not all compound classes can be detected by this method. Sample derivatization can increase the complexity of the data and analysis. |
| 2D GC-MS | Similar to GC-MS, but the metabolites separation occurs in two dimensions | Similar to GC-MS but additional resolution achieved by the second dimension makes it more suitable for the analysis of complex biological samples | Requires rather tedious sample preparation, and not all compound classes can be detected by this method |
| EESI-MS | Direct solution analysis method that utilizes two spray sources: One nebulizes and the other provides charged solvent droplets. | No sample preparation or extraction required | Less quantitative, and can also suffer from ion suppression in the complex mixture of biological samples |
| DESI-MS | Ambient ionization method that analyses the sample directly. | Sensitive, and has excellent tolerance to high salt concentrations, no sample preparation or extraction required | Less quantitative, and may suffer from ion suppression in the complex mixture of biological samples |
| DART-MS | Uses stream of excited metastable helium gas and hot nitrogen, provides real time information, direct sample analysis | Snsitive, and useful for very high throughput screening, no sample preparation or extraction required | Less quantitative, and may suffer from ion suppression in the complex mixture of biological samples |
Abbreviations: CPMG: Carr–Purcell–Mieboom–Gill; J-RES: J-resolved; LC-MS: Liquid Chromatography-Mass Spectrometry; HPLC: High Pressure Liquid Chromatography; UPLC: Ultrahigh Pressure Liquid Chromatography; 2D GC-MS: Two Dimensional Gas Chromatography-Mass Spectrometry; EESI: Extractive Electrospray Ionization; DESI Desorption ElectroSpray atmospheric Ionization; DART: Direct Analysis in Real Time.
Data preprocessing
Both NMR and MS data contain up to thousands of signals arising from the many hundreds of detected metabolites. Analysis of such complex data is extremely challenging, and thus a variety of pattern recognition methods are used to simplify the data. A number of data pre-treatments are generally required before statistical analysis can be performed to obtain meaningful information on healthy and disease samples.
For NMR data, baseline correction is used to reduce the effect of any non-ideal offsets in individual data. This is particularly important for low-abundance but potentially important metabolites that have small peaks and are more prone to baseline artifacts than high-abundance metabolites. Statistical analysis of spectral data sets requires each spectral peak (or variable) be compared throughout all observations (samples). Misalignment will jeopardize the construction of an appropriate model, which readily produces incorrect metabolic patterns and erroneous identification of potential biomarkers. NMR spectra can be aligned using a reference compound (for example trimethylsilylpropionic acid-d4 sodium salt, TSP or sodium 4,4-dimethyl-4-silapentane-1-sulphonate-d6, DSS). Metabolomic data analysis packages such as KnowItAll (BioRad, Philadelphia, PA), AMIX (Bruker, Billerica, MA) and others allow rapid analysis of biosamples with improved NMR data alignment. The alignment of LC- and GC-MS data is often more challenging because of the tendency of peaks to shift or even reverse in the chromatographic separation as well as the size of the data sets involved. A number of attempts at solving these challenges have been made [62–66], which are typically based on either pattern recognition, time warping, or similarity calculations. This important and challenging area is still evolving.
Data binning is often used and follows spectral alignment. An important advantage of data binning is that it reduces the effect of peak misalignments. At the extremes, binning size does affect biomarker exploration, so judicious use of binning is advised. In any event, the full resolution NMR or MS spectra are recoverable if necessary for metabolite identification.
Data scaling, which allows the emphasis of smaller concentration metabolites is often used. A number of scaling methods are popular, including variance scaling (division by the standard deviation of the peak intensities across the set of spectra) and Parato scaling (division by the square root of the standard deviations). Log scaling has been used to reduce the size of very large and dominant peaks. The data are then typically mean centered by subtracting the average of all the spectra. For samples such as urine, data normalization is also generally performed to reduce any dependence on overall concentration differences.
Statistical Analysis
Modern multivariate statistical methods have become an essential part of the metabolomics field. While feature selection through the use of p-values is of high utility, the need to build predictive models based on multiple biomarkers necessitates the use of multivariate methods. These methods are also quite useful to reduce the dimensionality of the NMR/MS data, and to extract the maximum information from the data. A variety of such methods are capable of analyzing several thousand inputs or “variables” and their corresponding intensities. These statistical approaches are broadly classified into two categories: “unsupervised” and “supervised” methods.
Unsupervised methods classify the spectra without the knowledge of the class of biological specimens (such as disease or control) by using the NMR frequencies or MS m/z values and their intensities for each sample as the sole inputs. Principal component analysis (PCA) is the most commonly used unsupervised method in multivariate analysis. PCA generates orthogonal and ranked principal components (PCs) that explain the variance in the data. The PCs are essentially a new set of data descriptors (basis set or axes) obtained from the linear combination of the variables (metabolite signals) from the NMR or MS data. Hierarchical cluster analysis (HCA) aims to define natural clusters based on comparing distances between pairs of samples (or variables): small distances between samples imply that the samples share similar metabolite content representing similar physiological properties, dietary habits or disease grades, etc. HCA represents analytical results in the format of dendrogram, and facilitates the visualization of different categories with a given similarity level. In biomarker discovery, HCA is usually used as a supporting method to more powerful methods such as PCA in order to target key individual metabolites or spectral regions which most correlate to the class membership. Proton NMR spectroscopy of sera coupled with PCA and HCA has been successfully used in discriminating 120 serum samples into three baseline clusters and two treatment clusters to detect variations in the metabolism of lipids resulting from statin treatments [67]. K nearest neighbor (KNN) analysis is a method of classification based on the similarity within classes. Each spectrum can be treated as a point in a multi-dimensional space. In KNN, the Euclidean distance between every pair of spectra is first calculated. The class assignment of one sample is based on the majority vote of its nearest neighbors.
Supervised methods require a training data set in which the outcome (i.e., disease or healthy) is known and used to build a (hopefully) predictive model. After training, the model can be used on a test set to classify unknown samples and measure the predictive accuracy of the model. Supervised methods are very useful for detecting subtle differences between similar samples in order to identify potential biomarker candidates. Cross validation [31] is used to test the robustness of putative biomarker candidates during the training process. Supervised techniques can be appropriate to force classification (such as in determining which metabolites distinguish between groups) or to regress a pattern against a trend (such as correlating a temporal progression with metabolic changes). Methods for supervised pattern recognition include partial least squares discriminant analysis (PLS-DA) [68] and soft independent modeling of class analogies (SIMCA), which are extensive tools for the classification of spectroscopic results. Other methods, including orthogonal signal correction (OSC), genetic programming and neural networks are also used. OSC and PLS-DA have recently been combined to extend the power of supervised methods into metabolomics analyses [35], and this approach has quickly gained wide usage. In general, it is extremely important to validate the findings of PCA, PLS or other methods using extensive cross validation and, in particular, a second set of samples (preferably blinded and from a second location). Ultimately, biological validation, involving a disease hypothesis specifically related to the discovered biomarkers, will likely be required before acceptance by the medical and scientific communities can be anticipated.
Correlation methods, either within one spectroscopic method [69] or used to combine NMR and MS data have shown to be effective in identifying metabolites based on NMR chemical shifts and m/z values [70,71]. STOCY has been useful in providing multiple metabolite peaks from a single metabolite, thus simplifying identification [69]. Statistical heterospectroscopy (SHY) operates through the analysis of the intrinsic covariance between signal intensities in the same and related molecules measured by different techniques across cohorts of samples [70]. Further, it is also possible to combine the results of PCA from NMR and MS. This is so because the principle components of NMR data can be treated as independent to those of MS data [72]. Such combined analysis may be useful for larger data sets where 2D score plots are insufficient to differentiate the samples.
Metabolomics Applications
There has been an explosive growth in the application of NMR- and MS-based metabolomics driven by the potential for earlier disease detection, therapy monitoring, and ultimately for reaching the goal of personalized medicine. In particular, metabolomics studies have been focused on the identification of metabolites associated with a number of diseases including cancer, diabetes, inborn errors of metabolism and cardiovascular diseases. In general, these early studies are promising, however, validation studies are critically needed to confirm the identity and generality of the putative biomarkers. Validation studies normally involve several independent sample sets for the same disease with even representation to such factors as gender, age, ethnicity, co-morbidity from other diseases, and geographical origin.
Cancer
Cancer is typically detected radiographically, and often in late stage when therapy options are limited. Therefore, there is a high demand for alternative, earlier and chemically based detection modalities. NMR- and MS-based metabolomics tools have the potential for early diagnosis and even therapy management. Recent developments include the exploration of cancer biomarkers and disease pathways in both humans and animal cancer models (Table 2).
Table 2.
NMR- and MS-based metabolomics studies of biological samples from various cancers.
| Cancer type | Sample type |
Analytical Methods |
Important outcome | Reference |
|---|---|---|---|---|
| Breast cancer | Tissue, cells, breath | 1H, 31P, (1D and 2D) NMR, HRMAS, GC-MS | 82% sensitivity and 100 specificity was achieved based on choline metabolites; detection of decanes and heptanes in breath | 73–79 |
| Ovarian cancer | Serum, tissue | 1H NMR, GC-TOF-MS | 100% classification accuracy between cancer and benign, 51 metabolites differentiated tumor from borderline tissue | 80,81 |
| Liver cancer | Tissue | 1H NMR, HR-MAS | High lactate and amino acids, and low carbohydrates in cancer | 52 |
| Pancreatic cancer | Plasma, tissue | 1H NMR, HRMAS, MS | Decreased levels of phosphocholine, glycerophosphocholine and phosphatidylinositol in cancer | 82, 83 |
| Cervical Cancer | Tissue | 1H NMR, HRMAS | Low glucose, and high cholines and amino acids levels in cancer | 84, 85 |
| Lung cancer | Urine, Serum, breath | 1H NMR, DESI-MS, LC-MS, GC-MS | Altered metabolites associated with the purine pathway, detection of hexanal and heptanal in serum, and a large number of volatile organic compounds with altered concentrations in breath | 72, 86–91 |
| Prostate cancer | Plasma, tissue | 1H NMR, HRMAS | Altered amino acids levels; increased lactate, phospholipids, choline, low citrate and polyamines; spermine regulation | 92–97 |
| Renal cancer | tissue | 1H, 13C NMR, 2D JRES, 2D TOCSY, HMQC, HRMAS | 100 % classification accuracy, high triglycerides and cholesteryl esters | 100–103 |
| Brain cancer | Tissue, cells | 1H, 13C, 1D and 2D HRMAS NMR | Alterations in Kennedy and Sonic Hedgehog pathways; apoptosis, altered lipids, inositol, N-acetyl aspartate to choline ratio and creatine to choline ratio. | 104–112 |
A number of investigations have been carried out to establish breast cancer biomarkers [73–78], with a majority of these focused on biomarker detection directly in breast cancer tumors. The correlation of multiple metabolites such as lactate, lipids, phosphocholine, choline and glycine with the cancer was observed using a variety of 1D and 2D 1H or 31P high resolution MAS NMR. Based on multivariate statistical analysis of the NMR data, tumor and non-involved tissues could be classified with a high specificity (100%) and sensitivity (82%) [73]. Differences in mammary epithelial cell lines such as upregulation of fatty acid synthesis have been detected from the combined 2D NMR and GC-MS methods [78]. Attempts to detect breast cancer from the analysis of exhaled breath have also been made. Volatile organic compounds were targeted in breath from women with abnormal mammograms and biopsies. In this study, cancer patients and controls have been distinguished with a sensitivity and specificity of 94.1% 73.8%, respectively [79].
The 1H-NMR study of preoperative serum specimens has been made to detect epithelial ovarian cancer [80]. NMR data were analyzed using PCA and SIMCA to classify the patients into ovarian cancer and non-cancer (benign ovarian cysts and healthy controls) subgroups. Statistical analysis distinguished cancer patients from benign and control samples with 97–100% accuracy. However, the putative biomarkers consisted of two non-specific signals emanating from the lipid region, and 3-hydroxybutarate, which has been seen in a number of other metabolomics studies and may result from gut microfloral metabolism. In another study, multivariate analysis of GC-MS data of ovarian tumor and the borderline tissue has shown classification accuracy of 88%. In this study, 51 metabolites were found to be significantly different between the two types of tissue [81].
Recently, NMR-based metabolomics has been applied to explore liver cancer biomarkers. Differentiation of both low-grade and high-grade tumors from adjacent non-involved tissue was obtained using 1H HRMAS NMR in combination with multivariate statistical analysis [52]. Interestingly, apart from showing metabolic differences between cancer and non-cancer tissue, the analysis showed distinct metabolic differences between low-grade and high grade tumors. Significant alteration in the levels of metabolites such as lactate, phosphorylethanolamine, phosphocholine, amino acids triglycerides, glucose, and glycogen were detected.
Altered metabolic profiles in pancreatic cancer have been studied using plasma and tissue samples [82, 83]. 1H NMR analysis of plasma [82] was based on the hypothesis that the reported altered insulin and glucose levels in pancreatic cancer result in an altered lipid profile in the blood. NMR spectra of the extracted plasma lipids were subjected to statistical analysis using PLS-DA. The sensitivity, specificity and the overall accuracy of detecting pancreatic cancer was reported to be 96%, 88% and 92%, respectively, when 4 NMR spectral regions were used for the discrimination, and 98%, 94% and 96%, respectively, when 5 regions were used. Mass spectrometric analysis indicated a decrease of phospholipids in the cancer samples. These results are complementary to the metabolic profile recently derived from the 1H HRMAS studies of tissue from animal models of pancreatic cancer [83]. In that study, phosphocholine and glycerophosphocholine were found to decrease in pancreatic cancer.
Metabolites characterization of cervical tumors has been shown using 1H HRMAS NMR and multivariate statistical analysis. Malignant tissue of the cervix showed higher levels of cholines and amino acids compared to non-malignant tissue [84]. Very recently, HRMAS NMR studies were performed to explore apoptosis in cervical cancer. The spectra were analysed for lipid and non-lipid metabolites using one pulse and spin-echo experiments, respectively [85]. Significant correlations were found between the tumor cell fraction and glucose concentration, between tumor cell density and glycerophosphocholine concentration, and in the ratio of glycerophosphocholine to choline. The results suggest an application of the method to explore the role of apoptosis in the course of the disease.
Advancements in metabolite-based detection of lung cancer have been made over a number of years [86–91]. The majority of these investigations focus on the detection of volatile organic compounds in breath samples using various collection strategies and highly sensitive GC-MS detection. A large number of volatile organic compounds have been shown to distinguish between lung cancer and controls [86, 88–91], however achieving a diagnostic accuracy above 90% has been challenging. Lung cancer biomarkers investigations have also been carried out using body fluids such as serum and urine [72,87]. From the GC-MS analysis of serum, higher concentrations of two aldehydes, hexanal and heptanal, have been shown to distinguish lung cancer patients from controls [87]. Metabolite profiles in urine from xenographic lung cancer mice models have been explored using both NMR and the new DESI-MS technique [72]. Urine from both cancerous and control mice were subjected to NMR and MS combined with multivariate statistical analysis. PCA of both the NMR and MS identified a large number of differentiating metabolites, many of which where localized to the purine metabolism pathway.
A combined metabolomic and proteomic approach was employed to study a mouse model of prostate cancer [92]. Multivariate analysis including O-PLS was applied to interpret the differences in plasma metabolomic and proteomic profiles. Correlations between a serotransferrin precursor and both tyrosine and 3-D-hydroxybutyrate, and between a decreased concentration of tyrosine and an increased presence of gelsolin were observed. Several metabolomic studies on prostate cancer focused on the analysis of tumors and seminal fluid [93–99] A combined 1H HRMAS NMR and quantitative histopathology study on same tumor specimens showed a linear correlation between the concentration of spermine measured by NMR and the volume percentage of normal prostatic epithelial cells quantified by histopathology [93]. These findings highlight the role of NMR as an effective tool for investigating the inhibitory mechanism of spermine in humans. A combined in vivo and in vitro NMR, and histopathology study indicate the potential utility of translating ex vivo derived biomarkers for improved clinical interpretation of prostate cancer using in vivo NMR [94]. In this study, healthy glandular tissue was discriminated from prostate cancer based on high citrate and polyamines, and low choline, phosphocholine and glycerophosphocholine. In addition, concentrations of taurine, myo-inositol, and scyllo-inositol were all higher in cancer compared to healthy glandular and stromal tissues. A computer model of tissue pathology based on metabolic profiles derived from HRMAS NMR was proposed for prostate cancer [95]. The results of NMR and computer aided tissue analysis showed a linear correlation between them for both normal epithelium and prostate cancer. The diagnostic capability of NMR spectroscopy for predicting prostate cancer was tested using the multivariate analysis of HRMAS NMR metabolic and quantitative histopathology data of 199 tissue samples from 82 cancer patients [96]. Recently, quantities of several prostate metabolite concentrations were determined using 1H HRMAS NMR and compared between normal and cancer tissue [97]. Concentrations of phosphocholine /glycerophosphocholine, total choline, lactate, and alanine were higher in prostate cancer than in healthy glandular and healthy stromal tissues, while citrate and polyamine concentrations were significantly higher in healthy glandular tissues than in healthy stromal or prostate cancer tissues. A 1H NMR study utilizing seminal fluid indicated that citrate based prostate cancer detection outperforms prostate specific antigen testing [98] and a more recent 1H NMR study highlights the use of myo-inositol and spermine, in addition to citrate, for detecting the presence of prostate cancer [99].
Exploring metabolite biomarkers for renal cancer based on tissue samples was explored soon after HRMAS was developed [100]. Subsequently, renal tumors have been extensively investigated employing the latest technological advancements in HRMAS NMR and multivariate analysis [101,102]. Conventional 1D NMR methods such as one pulse and spin-echo as well as 2D experiments such as J-resolved, TOCSY and 1H-13C HMQC have been utilized for the resonance assignments. Unsupervised and supervised multivariate statistical analyses of the HRMAS NMR data focused on classifying normal and renal carcinoma tissue, and showed a clear distinction based on NMR signal intensities. A linear discriminant analysis was used to classify normal and tumor tissues with 100% accuracy [101]. Recently, metabolic profiles of normal cortex and medulla samples were compared with malignant tissue also using HRMAS NMR [102]. Compared to the normal cells, renal carcinoma cells had lower organic osmolytes and higher lipid concentrations. On the other hand, in the papillary renal cell carcinoma the taurine concentration was higher and the lipid signals were absent. Kind et al. have evaluated three MS-based analytical methods to identify potential biomarkers for renal cancer. The combined approach gives a good coverage of the urinary metabolites, several of which the authors believe may be useful for diagnosis [103].
Quite a few recent studies have focused on the identification of differential metabolites in brain cancers [104–112]. Detailed assignments of the biochemical compounds in brain tumors have been made using the combination of in vivo and ex vivo analyses and employing several 1D and 2D NMR experiments [104]. A large number of metabolites have been shown to differentiate brain tumor from normal tissue, and metabolic ratios are used to achieve the highest sensitivity [105]. Considering the fact that brain in vivo MR spectroscopy is easily amenable for clinical applications, attempts have been made to establish the link between MR spectroscopy and neuropathological analysis utilizing the metabolic profile obtained using ex vivo high resolution NMR spectroscopy [104,106]. A recent study combined HRMAS NMR and micro scale genomics. It was shown that tissue samples as small as 2 mg could be successfully used for HRMAS experiments and minute mRNA amounts yielded high-quality genomic data. This is one of new interesting applications in which metabolomics and genomics have been combined. In this case, alterations in the expression of Kennedy pathway genes and dysregulation in Sonic Hedgehog pathway in the pathogenesis of cancer were observed [107]. Classification of patients based on metastasis and survival prediction of brain cancer patients were made by performing multivariate analysis of the HRMAS NMR data [108]. While the PCA results clearly showed a trend in clustering due to the origin of the metastases, PLS analysis indicated distinct clustering of the spectra of the patients who died less than 5 months after surgery. Although further validation is needed, these results indicate the potential for clinical applications to manage brain cancer patients.
Diabetes
Metabolomics is ideal for studying metabolic diseases, and has already been applied to both type 1 and type 2 diabetes utilizing a range of biological specimens including urine, serum/plasma and tissue (Table 3). 1H NMR analysis of urine has identified a number of significantly changing metabolites, including acetate, lactate, citrate, glycine, alanine, hippurate, trimethylamine-N-oxide, and dimethylamine [113,114]. Multivariate statistical analyses of the 1H NMR data from human, rat and mouse urine demonstrate metabolic similarities among the three species including responses associated with type 2 diabetes involving glucose metabolism, the TCA cycle, and the nucleotide and methylamine metabolisms [115]. Another recent study has used quantitative NMR-based metabolomics to correlate differentiating metabolites in induced diabetes in rats. Significant disturbances in several metabolic pathways including glucose metabolism and the TCA cycle, the alanine pathway, the Cori cycle, the acetate switch, and choline metabolism, as well as a contribution from gut microbial metabolism were identified from the analysis [116]. A dramatic loss in the correlation among the detected metabolites was observed. Studies on animal diabetic models aimed at understanding insulin-resistance induced by a high fat diet have shown increased concentrations of lipids, lactate, pyruvate, glucose, fucose, phosphatidylcholine, trimethylamine N-oxide and methylamines in plasma [117]. The effect of gut microbiota to the fatty liver phenotype in insulin-resistant mice has been studied and a model linking the impaired glucose homeostasis and nonalcoholic fatty liver disease (NAFLD) in reducing mammalian availability of choline was deduced using a number of biomarkers such as choline, phosphatidylcholine and methylaminies [118]. 1H NMR-based metabolomics was applied to assess diabetes induced nephropathy and the results indicate high positive as well as negative predictive values (89% and 83.6%, respectively) which are comparable to those derived from clinical biochemistry data, 95.5% and 79.2%, respectively [119]. NMR of other nuclei such as 13C and 31P has been extensively used to understand the pathogenesis of diabetes [120–125]. For example, 13C NMR was used to investigate metabolic responses to a dextrose challenge [120] and to understand the role of reduced glycogen synthesis on muscle insulin resistance [121]. Measurements of glycogen synthesis was made using 13C NMR to study the effect of insulin resistance on both type 1 and type 2 diabetes [122, 123]. 31P NMR was employed to measure the glucose transport/phosphorylation activity in situ. The results correlated well with the glycogen synthesis rate as measured by 13C NMR [124,125].
Table 3.
NMR- and MS-based metabolomics studies of biological samples from diabetes.
| Diabetes type |
Sample type |
Analytical method |
Information derived | Reference |
|---|---|---|---|---|
| Urine | 1H NMR | High alanine, lactate, acetate and citrate, and disturbance in the glucose metabolism and TCA cycle, and a contribution from gut microbial metabolism | 113, 116 | |
| Diabetes Type 1 | Serum | 1H NMR | High positive (89%) and negative (83.6%) predictive values obtained | 119 |
| tissue | 13C NMR | Defective glucose transport/phosphorylation responsible for the lower rate of muscle glycogen synthesis | 122 | |
| Urine | 1H NMR | High lactate, citrate, glycine, alanine, hippurate, trimethylamine- Noxide, dimethylaminecreatine, acetate, betaine,and ketone bodies in diabetes | 114 | |
| Humans, rats and mice have shown similar trends in the levels of glucose, TCA cycle intermediates, polyols, amines, and amino acids | 115 | |||
| Urine and plasma | 1H NMR | Gut microbial contribution to fatty liver phenotype: trimethylamine-N-oxide | 118 | |
| Plasma | a1H NMR | High lipids, lactate, pyruvate, glucose, fucose, phosphatidylcholine, trimethylamine N-oxide and other methylamines in the adaptation to high-fat diet | 117 | |
| Diabetes Type 2 | LC-MS | Detection of phosphatidylethanolamine and lysophosphatidylcholine | 126 | |
| GC-MS, UPLC-MS | Distinguishing metabolic features between the Zucker (fa/fa) obese, Zucker lean and the lean/(fa) obese rats | 127, 128 | ||
| GC-MS | Fatty acids detected as biomarkers | 129 | ||
| NMR, UPLC-MS and GC/MS | Showed metabolic differences between normal and obese rats | 131 | ||
| Tissue | 13C NMR | Muscle glycogen synthesis is the principal pathway of glucose disposal in both normal and diabetic subjects, and defects in muscle glycogen synthesis have a dominant role in the insulin resistance | 121 | |
| 13C, 31P NMR | Increased plasma free fatty acids inhibit insulin stimulated glucose uptake at the level of glucose transport/phosphorylation rather than in the glycogen synthesis | 125 | ||
| HRMAS GC-MS, LC-MS | Low amino acids and high lactate levels detected | 132 | ||
Several investigations have demonstrated the application of metabolomics to diabetes using a combination of MS and multivariate statistics [126–129]. Plasma was analysed using LC-MS and the data subjected to PCA and PLS-DA, focusing on phospholipid metabolites. Type 2 diabetes could be distinguished from controls based on the differences in the spectral features [126]. Utilizing the advanced UPLC-MS approach, nearly 10,000 ions in rat plasma have been detected, and the ability of such metabolic data to distinguish among three rat strains, obese, lean and the lean/obese was demonstrated using multivariate analysis [127]. Nevertheless, the detection of so many features does give pause as to the required level of validation that is necessary. Targeted metabolic profiling using GC-MS has enabled detection of plasma fatty acids including non-esterified fatty acids (NEFA) and esterified fatty acids [129]. This study detected a number of additional putative biomarkers, and allowed a comprehensive understanding of the role of NEFA and the effect of treatment with thiazolidinediones.
Finally, the interest in combining NMR and MS methods for the study of diabetes is growing [130–132]. The metabolic regulatory mechanisms in diabetes was investigated by obtaining metabolic profiles in plasma from normal Wistar-derived and Zucker (fa/fa) obese rats from multiple analytical platforms including NMR, UPLC-MS and GC-MS. PCA analysis of the data readily detected the differences in the metabolite profiles between the two rat strains [131]. For example, a number of biomarkers including cholesterol, arachidonic acid, oleic acid, hexadecanoic acid, monooleoylglycerol and low and very low density lipoproteins were higher in the Zucker rats. Another recent study combining 1H HRMAS NMR, GC-MS and LC-MS examined the metabolic perturbations in type 2 diabetes and obesity. Both PCA and PLS analysis showed dramatic alterations in the levels of several metabolites such as glucose, glutamine, alanine and lactate and indicated perturbations in glycolysis, the TCA cycle, and gluconeogenesis [132].
Inborn errors of metabolism (IEM)
The IEM form a large and diverse group of diseases. A majority of these involve single genetic defects that affect a specific enzyme [133]. Individually, IEM are rare, however, their collective incidence is relatively high and lie in the range of 1 per 1,400 to 5,000 live births in the United States. In most of the disorders, problems arise due to the accumulation of metabolites that are toxic or interfere with normal function. Often, IEM are difficult to diagnose since clinical signs and symptoms overlap among the different diseases. Biochemical tests are often nonspecific and gene analysis is not always conclusive. Moreover, for several metabolic disorders, comprehensive analytical techniques have not been established making it difficult to diagnose such diseases. For example, N-acetylated metabolites in urine that are involved in several IEM are not easily detectable [134]. Currently GC/MS or tandem MS is used to detect up to 80 different IEM, although these analyses are typically based on the detection of single metabolite biomarkers. Metabolomics may have a role to play in changing the detection of at least some of these IEM. NMR spectroscopy is very useful for the diagnosis of a number of IEMs [134–138], and has been successfully combined with MS using a metabolomics approach [139]. Metabolomics appears to be particularly promising for identifying additional potential IEM biomarkers, and may prove useful for detecting borderline cases, or for sub-classifying the diseases [140].
Coronary heart disease
Coronary heart diseases (CHD) provide another excellent target for metabolomics diagnostic development. Angiography, a current diagnostic modality for CHD is both expensive and invasive. Hence, advancement in metabolomics-based assessment of CHD is highly desirable. In view of this opportunity, lipid metabolites have been assessed for some time using NMR spectroscopy to understand the risk of CHD [141,142]. The diagnostic utility of the NMR-based metabolomics approach was assessed using serum from individuals with no evidence of stenosis (normal coronary arteries, NCA), or severe CHD defined as at least 50% of stenosis (triple vessel disease, TVD) employing multivariate statistical analysis [143]. Disease and control subjects were distinctly separated from the analysis using PLS-DA and OSC. Classification was achieved mainly from subtle differences in the lipid signals of the NMR spectra between the two groups. These findings were in conformity with the results of independent studies made on individuals with and without CHD. Subsequently, NMR spectroscopy was evaluated as a diagnostic method on patients with hypertension, and the results clearly distinguished low/normal systolic blood pressure (SBP) serum samples from borderline and high SBP samples [144]. NMR-based diagnosis of CHD depends mainly on the major lipid regions of the 1H NMR spectra and many variables such as diet, gender, lifestyle and drugs affect lipid composition: failing to take such confounding variable into account can lead to a false conclusion. A study was performed to determine the predictive power of the NMR-based method in groups of male patients. This study achieved a mere 36.2% and 6.2% predictive accuracy at the 99% confidence level for untreated and treated groups, respectively, indicating that the NMR-based metabolomics method still lacks sufficient diagnostic accuracy [145]. A very recent review presents an evaluation of the metabolomics approach with emphasis on the CHD risk assessment and diagnosis using 1H NMR of plasma. It highlights the potential utility of combining in vitro 1H NMR based metabolomics and in vivo multicontrast magnetic resonance imaging for early diagnosis and multiphase risk assessment of atherothrombosis [146].
Expert Commentary
In metabolomics multi-component measurements are primarily focused on the information rich analytical techniques of MS and NMR spectroscopy. Analysis of the complex data from these methods using advanced multivariate statistics provides a powerful platform for diagnostic applications, as well as for translational and clinical research. During the past several years numerous developments have taken place in the field including a variety of advances in the analytical methods for high throughput measurements, improved statistical approaches for classifying samples based on subtle changes, and applications in the area of disease diagnosis and toxicity assessment.
Today, MS methods provide high sensitivity, however reproducibility is still a concern, as is the unique identification of unknown and interesting metabolites. NMR methods allow the identification and quantification of metabolites down to µM limits facilitated by cryogenic probes, micro-coil NMR and isotope labeling, however, spectral complexity is still a problem due to the high degree of signal overlap. Numerous multivariate statistical methods are readily available due to their incorporation into user-friendly software packages. These advancements combined with the recently developed databases of human metabolites and metabolic information, and the vast body of metabolic pathway information currently available greatly benefit the metabolomics field.
A majority of metabolomics studies have focused on using a single analytical method, NMR spectroscopy or MS. Given the complexity of the biological systems, it will be more prudent to exploit both methods in parallel, at least in the developmental stages of the field, to derive more meaningful information on metabolic variations in health and disease. The high reproducibility of NMR and high sensitivity of MS provide both supplementary and complementary data important for biomarker identification and validation. Combined multivariate analysis of data from NMR and MS will provide information which is more useful and important than using a single approach.
Five year view
There has been an explosive growth in both NMR- and MS-based metabolomics studies and applications, with a number of these being applied to assess important diseases. Metabolomics-based methods will likely have significant clinical diagnostic utility for numerous inborn errors of metabolism owing to the fact that, generally, such diseases exhibit massive metabolic disturbances. Risk assessment of cardiovascular diseases using NMR-based metabolomics is potentially highly promising because the abundant lipoproteins can be easily detected by NMR thus avoiding the often tedious separation procedures. In fact, NMR spectroscopy is already being applied for individual risk assessments. The addition of MS-based metabolomics approaches might well improve the diagnostic accuracy and utility.
Currently, metabolomics applications to diseases such as cancer and diabetes have provided better insights in to the altered metabolic pathways and the disease pathogenesis. However, for applications in early diagnosis the technology is still in its evolutionary stages. Factors such as diet, age, gender, lifestyle, drugs and environment contribute immensely to human bio-complexity, and identification of subtle metabolic variations associated with early cancer or diabetes pathogenesis is a great challenge. Further studies focused on the deconvolution of such confounding effects are required. In general, NMR detects relatively highly concentrated metabolites, and it is generally thought to lack sufficient sensitivity to detect low concentration, more specific early biomarkers. Although the latest technological advancements have demonstrated a dramatic reduction in the detection limit of NMR using pure substances, such methods are still not considered high throughput for routine biological samples analysis. Combining the latest advancements in NMR methods and targeted metabolic profiling using sensitivity enhanced approaches such as isotope labeling or others may achieve a breakthrough in biomarker detection.
While MS is intrinsically highly sensitive capable of detecting early biomarkers, problems with reproducibility arising from the chromatography of biofluids and factors such as matrix effects and/or ion suppression still present considerable challenges. However, in light of recent methodological improvements in both NMR and MS, as well as multivariate statistical methods, it can be envisaged that metabolomics will emerge as a sensitive and convenient approach for early disease diagnoses. The five year outlook for metabolomics is very strong given the rapid developments in the field, as well as the important lessons learned from early studies in the genomics and proteomics fields.
Key issues.
Advancements in 1H NMR spectroscopy and mass spectrometry (and their combination), as well as multivariate statistical analysis promise high utility for early disease diagnosis, identification of affected metabolic pathways and thereby drug discovery.
Future developments will need to concentrate on detecting subtle metabolic changes more reliably, and especially to take into account such confounding effects as diet, drug metabolites, age, gender, ethnicity and environment.
Deconvoluton of the confounding effects of diet, age, gender, etc., is currently the major challenge, and a number of studies focused on each confounding effect separately and in various combinations are required.
There is a need to improve the quantitative results of many of the biomarker studies.
Biomarker validation studies using independent samples are greatly needed.
While NMR spectroscopy is highly reproducible, its low sensitivity and resolution have impeded the growth of the field. Recent developments in NMR sensitivity enhancement may significantly improve the detection limit.
Reproducibility issues still plague MS studies, although the resolution has greatly improved lately. The potential for high-throughput MS methods that do not require chromatography needs to be validated.
Overall, in light of recent methodological improvements, it can be envisaged that metabolomics will emerge as an important tool in the field of diseases diagnosis.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Nicholson JK, Wilson ID. High-resolution proton magnetic resonance spectroscopy of biological-fluids. Prog. NMR Spectrosc. 1989;21:449–501. [Google Scholar]
- 2. Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. •• This is a pioneering paper defining the concept of the term metabonomics.
- 3.Shockcor JP, Holmes E. Metabonomic applications in toxicity screening and disease diagnosis. Curr. Top. Med. Chem. 2002;2:35–51. doi: 10.2174/1568026023394498. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson JK, Wilson ID. Understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug. Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 5. Lindon JC, Holmes E, Nicholson JK. Metabonomics and its role in drug development and disease diagnosis. Expert. Rev. Mol. Diagn. 2004;4:189–199. doi: 10.1586/14737159.4.2.189. • Provides a good description of the capabilities and applications of NMR-based metabolomics.
- 6. Robertson DG. Metabonomics in Toxicology: A Review. Toxicol. Sci. 2005;85:809–822. doi: 10.1093/toxsci/kfi102. •• Recent metabonomics technology is reviewed with particular emphasis on toxicologic applications in preclinical drug development.
- 7. Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol. Systm. Biol. 2006:1–6. doi: 10.1038/msb4100095. • This editorial highlights the importance of metabolic profiling in systems biology and human health care.
- 8. Wishart DS, Tzur D, Knox C, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. •• The most complete and comprehensive description on more than 2180 endogenous metabolites.
- 9.Griffin JL. The Cinderella story of metabolic profiling: does metabolomics get to go to the functional genomics ball? Philos. Trans.R. Soc. Lond. B. Biol. Sci. 2006;361:147–161. doi: 10.1098/rstb.2005.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villar-Garea A, Griese M, Imhof A. Biomarker discovery from body fluids using mass spectrometry. J Chromatogr B. 2007;849(1–2):105–114. doi: 10.1016/j.jchromb.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 11. Wishart DS. Metabolomics: the principles and potential applications to transplantation. Am. J. Transplant. 2005;5:2814–2820. doi: 10.1111/j.1600-6143.2005.01119.x. • This review provides excellent overview of the potential applications of metabolite profiling to monitoring organ transplants.
- 12.Pelczer I. High-resolution NMR for metabomics. Curr. Opin. Drug Discov. Devel. 2005;8:127–133. [PubMed] [Google Scholar]
- 13.Keun HC. Metabonomic modeling of drug toxicity. Pharmacol. Ther. 2006;109:92–106. doi: 10.1016/j.pharmthera.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 14. Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H–NMR-based metabolomics. Expert. Rev. Mol. Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. •• This review covers NMR-based metabolomics and its applications to biomedical science emphasizing translational research in transplantation, oncology and drug toxicity or discovery.
- 15. Lindon JC, Holmes E, Nicholson JK. Metabonomics techniques and applications to pharmaceutical research & development. Pharm. Res. 2006;23:1075–1088. doi: 10.1007/s11095-006-0025-z. •• Review covering advanced analytical techniques and statistical analysis emphasizing metabolomics applications to pharmaceutical research.
- 16.Harrigan GG, Goodacre R. Metabolic Profiling: Its Role in Biomarker Discovery and Gene Functional Analysis. Kluwer Academic. 2003:1–8. [Google Scholar]
- 17. Fiehn O. Metabolomics-the link between genotype and phenotype. Plant. Mol. Biol. 2002;48:155–171. •• Provides an excellent introduction to the field with a focus on the plant metabolomics.
- 18.Kell DB. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 19. van der Greef J, Smilde AK. Symbiosis of chemometrics and metabolomics: past, present, and future. J. Chemomet. 2005;19:376–386. • This article makes a good connection of metabolomics with chemometrics with historical perspective.
- 20.Lacey ME, Subramanian R, Olson DL, Webb AG, Sweedler JV. High-resolution NMR spectroscopy of sample volumes from 1 nL to 10 µL. Chem. Rev. 1999;99:3133–3152. doi: 10.1021/cr980140f. [DOI] [PubMed] [Google Scholar]
- 21.Raftery D. High-throughput NMR spectroscopy. Anal. Bioanal. Chem. 2004;378:1403–1404. doi: 10.1007/s00216-003-2437-5. [DOI] [PubMed] [Google Scholar]
- 22.Lindon JC. HPLC-NMR-MS: past, present and future. Drug Discovery Today. 2003;8:1021–1022. doi: 10.1016/s1359-6446(03)02843-5. [DOI] [PubMed] [Google Scholar]
- 23.Brown SC, Kruppa G, Dasseux JL. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom. Rev. 2005;24:223–231. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 24.Granger JH, Bake A, Plumb RS, Perez JC, Wilson ID. Ultra performance liquid chromatography-MS(TOF): New separations technology for high throughput metabonomics. Drug Met. Rev. 2004;36:252–252. [Google Scholar]
- 25. Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005;24:613–646. doi: 10.1002/mas.20032. •• Excellent review of the current mass spectrometry status in metabolomics.
- 26.Wilson ID, Plumb RS, Granger J, Major H, Williams R, Lenz EA. HPLC-MS-based methods for the study of metabonomics. J. Chromatog. B. 2005;817:67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 27.Mohler RE, Dombek KM, Hoggard JC, Young ET, Synovec RE. Comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry analysis of metabolites in fermenting and respiring yeast cells. Anal. Chem. 2006;78:2700–2709. doi: 10.1021/ac052106o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Venter A, Cooks RG. Extractive electrospray ionization for direct analysis of undiluted urine, milk and other complex mixtures without sample preparation. Chem. Commun. 2006;19:2042–2044. doi: 10.1039/b602614a. [DOI] [PubMed] [Google Scholar]
- 29.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 30.Cody RB, Laramee JA, Durst HD. Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal. Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 31.Johnson RA, Wichern DW. Applied Multivariate Statistical Analysis. Prentice Hall, Upper Saddle River, New Jersey: 1999. [Google Scholar]
- 32.Krzanowski WJ. Principles of Multivariate Analysis: A User’s Perspective. Oxford: Oxford University Press; 2000. Revised Edition. [Google Scholar]
- 33. Brereton RG. Chemometrics: Data Analysis for the Laboratory and Chemical Plant. England: Wiley, West Sussex; 2003. • Provides good description on many of the statistical methods used in metabolomics field.
- 34.Keun HC, Ebbels TMD, Antti H, et al. Improved analysis of multivariate data by variable stability scaling: application to NMR-based metabolic profiling. Anal. Chim. Acta. 2003;490:265–276. [Google Scholar]
- 35. Beckwith-Hall BM, Brindle JT, Barton RH, et al. Application of orthogonal signal correction to minimise the effects of physical and biological variation in high resolution 1H NMR spectra of biofluids. Analyst. 2002;127:1283–1288. doi: 10.1039/b205128c. • First use of the OSC filter in metabolomics to reduce effect of confounding factors.
- 36.Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18:143–162. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- 37.Nagana Gowda GA, Ijare OB, Somashekar BS, Sharma A, Kapoor VK, Khetrapal CL. Single-Step analysis of individual conjugated bile acids in human bile using 1H NMR Spectroscopy. Lipids. 2006;41:591–603. doi: 10.1007/s11745-006-5008-7. [DOI] [PubMed] [Google Scholar]
- 38.Bala L, Ghoshal UC, Ghoshal U, et al. Malabsorption syndrome with and without small intestinal bacterial overgrowth: A study on upper-gut aspirate using 1H NMR spectroscopy. Magn. Reson. Med. 2006;56:738–744. doi: 10.1002/mrm.21041. [DOI] [PubMed] [Google Scholar]
- 39. Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J. Proteome Res. 2007;6:498–505. doi: 10.1021/pr060464h. • Reviews the potentials of MRS and MS techniques for animal tumor metabolomics.
- 40. Beckonert O, Keun HC, Ebbels TMD, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. •• provides an excellent description on sample preparation and NMR experiments for metabolites analysis.
- 41. Saude EJ, Slupsky CM, Sykes BD. Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics. 2006;2:113–123. • Reviews several water suppression methods.
- 42.Mo H, Raftery D. Pre-SAT180, a simple and effective faraway water suppression method. J. Magn. Reson. 2008;190:1–6. doi: 10.1016/j.jmr.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu D, Chen A, Johnson CS. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J. Magn. Reson. 1995;115:260–264. [Google Scholar]
- 44.Sandusky P, Raftery D. Use of selective TOCSY NMR experiments for quantifying minor components in complex mixtures: Application to the metabonomics of amino acids in honey. Anal. Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 45.Sandusky P, Raftery D. Use of semiselective TOCSY and the Pearson Correlation for the metabonomic analysis of biofluid mixtures: Application to urine. Anal. Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 46.Dumas M-E, Canlet C, Andre F, Vercauteren J, Paris A. Metabonomic assessment of physiological disruptions using 1H - 13C HMBC NMR spectroscopy combined with pattern recognition procedures performed on filtered variables. Anal. Chem. 2002;74:2261–2273. doi: 10.1021/ac0156870. [DOI] [PubMed] [Google Scholar]
- 47.Tang H, Wang Y, Nicholson JK, Lindon JC. Use of relaxation-edited one-dimensional and two-dimensional nuclear magnetic resonance spectroscopy to improve detection of small metabolites in blood plasma. Anal. Biochem. 2004;325:260–272. doi: 10.1016/j.ab.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 48.Dumas M-E, Canlet C, Vercauteren J, Andre F, Paris A. Homeostatic signature of anabolic steroids in cattle using 1H - 13C HMBC NMR metabonomics. J. Proteome Res. 2005;4:1493–1502. doi: 10.1021/pr0500556. [DOI] [PubMed] [Google Scholar]
- 49.Xi Y, de Ropp JS, Viant MR, Woodruff DL, Yu P. Automated screening for metabolites in complex mixtures using 2D COSY NMR spectroscopy. Metabolomics. 2006;2:221–233. [Google Scholar]
- 50.Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem. Biophys. Res. Commun. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 51.Shanaiah N, Desilva MA, Nagana Gowda GA, Raftery MA, Hainline BE, Raftery D. Class selection of amino acid metabolites in body fluids using chemical derivatization and their enhanced 13C NMR. Proc. Natl. Acad. Sci. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J. Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 53. Zhang X, Wei D, Yap Y, Li L, Guo S, Chen F. Mass spectrometry-based “omics” technologies in cancer diagnostics. Mass Spectrom. Rev. 2007;26:403–431. doi: 10.1002/mas.20132. •• Reviews advancements in mass spectrometry applications in cancer diagnostics.
- 54.Wilson ID, Nicholson JK, Castro-Perez J, et al. High resolution “ultra performance” liquid chromatography coupled to a-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J. Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 55.Villas-Boas SG, Delicado DG, Akesson M, Nielsen J. Simultaneous analysis of amino and non-amino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal. Biochem. 2003;322(1):134–138. doi: 10.1016/j.ab.2003.07.018. [DOI] [PubMed] [Google Scholar]
- 56.O'Hagan S, Dunn WB, Knowles JD, et al. Closed-loop, multiobjective optimization of two-dimensional gas chromatography/mass spectrometry for serum metabolomics. Anal Chem. 2007;79(2):464–476. doi: 10.1021/ac061443+. [DOI] [PubMed] [Google Scholar]
- 57.Kelly RT, Page JS, Zhao R, et al. Capillary-Based Multi Nanoelectrospray Emitters: Improvements in Ion Transmission Efficiency and Implementation with Capillary Reversed-Phase LC-ESI-MS. Anal. Chem. 2008;80:143–149. doi: 10.1021/ac701647s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen H, Wortmann A, Zhang W, Zenobi R. Rapid in vivo fingerprinting of nonvolatile compounds in breath by extractive electrospray ionization quadrupole time-of-flight mass spectrometry. Angew Chem Int Ed Engl. 2007;46(4):580–583. doi: 10.1002/anie.200602942. • Describes a high-throughput mass spectrometry approach for breath analysis.
- 59.Gu H, Chen H, Pan Z, et al. Monitoring diet effects via biofluids and their implications for metabolomics studies. Anal Chem. 2007;79(1):89–97. doi: 10.1021/ac060946c. [DOI] [PubMed] [Google Scholar]
- 60.Vaidyanathan S, Gaskell S, Goodacre R. Matrix-suppressed laser desorption/ionisation mass spectrometry and its suitability for metabolome analyses. Rapid Commun Mass Spectrom. 2006;20(8):1192–1198. doi: 10.1002/rcm.2434. [DOI] [PubMed] [Google Scholar]
- 61.Nordstro1m A, Want E, Northen T, Lehtio1 J, Siuzdak G. Multiple Ionization Mass Spectrometry Strategy Used To Reveal the Complexity of Metabolomics. Anal. Chem. 2008;80:421–429. doi: 10.1021/ac701982e. [DOI] [PubMed] [Google Scholar]
- 62.Stoyanova R, Nicholson JK, Lindon JC, Brown TR. Sample classification based on Bayesian spectral decomposition of metabonomic NMR data sets. Anal. Chem. 2004;76:3666–3674. doi: 10.1021/ac049849e. [DOI] [PubMed] [Google Scholar]
- 63.Stoyanova R, Nicholls AW, Nicholson JK, Lindon JC, Brown TR. Automatic alignment of individual peaks in large high-resolution spectral data sets. J. Magn. Reson. 2004;170:329–335. doi: 10.1016/j.jmr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Forshed J, Torgrip RJ, Aberg KM, Karlberg B, Lindberg J, Jacobsson SP. A comparison of methods for alignment of NMR peaks in the context of cluster analysis. J. Pharm. Biomed. Anal. 2005;38:824–832. doi: 10.1016/j.jpba.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 65.Robinson MD, De Souza DP, Keen WW, et al. A dynamic programming approach for the alignment of signal peaks in multiple gas chromatography-mass spectrometry experiments. BMC Bioinformatics. 2007;8:419–432. doi: 10.1186/1471-2105-8-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katajamaa M, Oresic M. Processing methods for differential analysis of LC/MS profile data. 2005;6:179–190. doi: 10.1186/1471-2105-6-179. BMC Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Moyec L, Valensi P, Charniot JC, Hantz E, Albertini JP. Serum 1H nuclear magnetic spectroscopy followed by principal component analysis and hierarchical cluster analysis to demonstrate effects of statins on hyperlipidemic patients. NMR Biomed. 2005;18:421–429. doi: 10.1002/nbm.974. [DOI] [PubMed] [Google Scholar]
- 68.Barker M, Rayens W. Partial least squares for discrimination. J. Chemom. 2003;17:166–173. [Google Scholar]
- 69. Cloarec O, Dumas ME, Craig A, et al. Statistical Total Correlation Spectroscopy: An Exploratory Approach for Latent Biomarker Identification from Metabolic 1H NMR Data Sets. Anal. Chem. 2005;77:1282–1289. doi: 10.1021/ac048630x. • Presents a new statistical approach for metabolite peaks detection.
- 70. Crockford DJ, Holmes E, Lindon JC, et al. Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: Application in metabonomic toxicology studies. Anal. Chem. 2006;78(2):363–371. doi: 10.1021/ac051444m. •• Provides a correlative approach to connect different spectroscopic methods.
- 71.Pan Z, Raftery D. Combining NMR Spectroscopy and Mass Spectrometry in Metabolomics. Anal. Bioanal. Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 72.Chen HW, Pan Z, Talaty N, Cooks RG, Raftery D. Combining Desorption Electrospray Ionization Mass Spectrometry and Nuclear Magnetic Resonance for Differential Metabolomics without Sample Preparation. Rapid. Commun. Mass. Spectrom. 2006;20:1577–1584. doi: 10.1002/rcm.2474. [DOI] [PubMed] [Google Scholar]
- 73.Sitter B, Lundgren S, Bathen TF, Halgunset J, Fjosne HE, Gribbestad IS. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006;19:30–40. doi: 10.1002/nbm.992. [DOI] [PubMed] [Google Scholar]
- 74. Sitter B, Sonnewald U, Spraul M, Fjösne HE, Gribbestad IS. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15:327–337. doi: 10.1002/nbm.775. • Provides the most detailed assignment of biochemical components in intact human breast tissue.
- 75.Whitehead TL, Kieber-Emmons T. Applying in vitro NMR spectroscopy and 1H NMR metabonomics to breast cancer characterization and detection. Prog. NMR Spectroc. 2005;47:165–174. [Google Scholar]
- 76.Cheng LL, Chang IW, Smith BL, Gonzalez RG. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J. Magn. Reson. 1998;135:194–202. doi: 10.1006/jmre.1998.1578. [DOI] [PubMed] [Google Scholar]
- 77.Bathen TF, Jensen LR, Sitter B, et al. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res. Treat. 2006;104:181–189. doi: 10.1007/s10549-006-9400-z. [DOI] [PubMed] [Google Scholar]
- 78.Yang C, Richardson AD, Smith JW, Osterman A. Comparative metabolomics of breast cancer. Pac. Symp. Biocomput. 2007:181–192. [PubMed] [Google Scholar]
- 79.Phillips M, Cataneo RN, Ditkoff BA, et al. Volatile markers of breast cancer in the breath. Breast J. 2003;9:184–191. doi: 10.1046/j.1524-4741.2003.09309.x. [DOI] [PubMed] [Google Scholar]
- 80. Odunsi K, Wollman RM, Ambrosone CB, et al. Detection of epithelial ovarian cancer using H-1-NMR-based metabonomics. Int. J. Cancer. 2005;113:782–788. doi: 10.1002/ijc.20651. • First metabonomics report on ovarian cancer detection.
- 81.Denkert C, Budczies J, Kind T, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 82.Beger RD, Schnackenberg LK, Holland RD, Li D, Dragan Y. Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics. 2006;2:125–134. [Google Scholar]
- 83.Fang F, He X, Deng H, et al. Discrimination of metabolic profiles of pancreatic cancer from chronic pancreatitis by high-resolution magic angle spinning 1H nuclear magnetic resonance and principal components analysis. Cancer Sci. 2007;98:1678–1682. doi: 10.1111/j.1349-7006.2007.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sitter B, Bathen T, Hagen B, Arentz C, Skjeldestad FE, Gribbestad IS. Cervical cancer tissue characterized by high-resolution magic angle spinning MR spectroscopy. MAGMA. 2004;16:174–181. doi: 10.1007/s10334-003-0025-5. [DOI] [PubMed] [Google Scholar]
- 85.Lyng H, Sitter B, Bathen TF, et al. Metabolic mapping by use of high-resolution magic angle spinning 1H MR spectroscopy for assessment of apoptosis in cervical carcinomas. BMC Cancer. 2007;11(1–12) doi: 10.1186/1471-2407-7-11. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phillips M, Gleeson K, Hughes JM, et al. Volatile organic compounds in breath as markers of lung cancer: a cross-sectional study. Lancet. 1999;353:1930–1933. doi: 10.1016/S0140-6736(98)07552-7. [DOI] [PubMed] [Google Scholar]
- 87.Deng C, Li N, Zhang X. Development of headspace solid-phase microextraction with on-fiber derivatization for determination of hexanal and heptanal in human blood. J Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004;813:47–52. doi: 10.1016/j.jchromb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 88.Chen X, Xu F, Wang Y, et al. A study of the volatile organic compounds exhaled by lung cancer cells in vitro for breath diagnosis. Cancer. 2007;110:835–844. doi: 10.1002/cncr.22844. [DOI] [PubMed] [Google Scholar]
- 89.Machado RF, Laskowski D, Deffenderfer O, et al. Detection of lung cancer by sensor array analyses of exhaled breath. Am. J. Respir. Crit. Care Med. 2005;171:1286–1291. doi: 10.1164/rccm.200409-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Phillips M, Altorki N, Austin JH, et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomark. 2007;3:95–109. doi: 10.3233/cbm-2007-3204. • This study reports prediction of lung cancer with accuracy comparable to screening CT of chest.
- 91.Poli D, Carbognani P, Corradi M, et al. Exhaled volatile organic compounds in patients with non-small cell lung cancer: cross sectional and nested short-term follow-up study. Respir. Res. 2005;6:71–80. doi: 10.1186/1465-9921-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rantalainen M, Cloarec O, Beckonert O, et al. Statistically integrated metabonomic-proteomic studies on a human prostate cancer xenograft model in mice. J Proteome Res. 2006;5(10):2642–2655. doi: 10.1021/pr060124w. [DOI] [PubMed] [Google Scholar]
- 93.Cheng LL, Wu C, Smith MR, Gonzalez RG. Non-destructive quantitation of spermine in human prostate tissue samples using HRMAS 1H NMR spectroscopy at 9.4 T. FEBS Lett. 2001;494(1–2):112–116. doi: 10.1016/s0014-5793(01)02329-8. [DOI] [PubMed] [Google Scholar]
- 94.Swanson MG, Vigneron DB, Tabatabai ZL, et al. Proton HR-MAS spectroscopy and quantitative pathologic analysis of MRI/3D-MRSI-targeted postsurgical prostate tissues. Magn. Reson. Med. 2003;50(5):944–954. doi: 10.1002/mrm.10614. [DOI] [PubMed] [Google Scholar]
- 95. Burns MA, He W, Wu CL, Cheng LL. Quantitative pathology in tissue MR spectroscopy based human prostate metabolomics. Technol Cancer Res Treat. 2004;3(6):591–598. doi: 10.1177/153303460400300609. • Early application of metabolomics to prostate cancer.
- 96.Cheng LL, Burns MA, Taylor JL, et al. Metabolic characterization of human prostate cancer with tissue magnetic resonance spectroscopy. 22. Cancer Res. 2005;65(8):3030–3034. doi: 10.1158/0008-5472.CAN-04-4106. [DOI] [PubMed] [Google Scholar]
- 97.Swanson MG, Zektzer AS, Tabatabai ZL, et al. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55(6):1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 98.Kline EE, Treat EG, Averna TA, et al. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J Urol. 2006;176(5):2274–2279. doi: 10.1016/j.juro.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 99.Serkova NJ, Gamito EJ, Jones RH, et al. The metabolites citrate, myo-inositol, and spermine are potential age-independent markers of prostate cancer in human expressed prostatic secretions. Prostate. 2008;68(6):620–628. doi: 10.1002/pros.20727. [DOI] [PubMed] [Google Scholar]
- 100. Moka D, Vorreuther R, Schicha H, et al. Biochemical classification of kidney carcinoma biopsy samples using magic-angle-spinning 1H nuclear magnetic resonance spectroscopy. J. Pharm. Biomed. Anal. 1998;17:125–132. doi: 10.1016/s0731-7085(97)00176-3. • First reported results on human tumors using HRMAS technique.
- 101.Tate AR, Foxall PJ, Holmes E, et al. Distinction between normal and renal cell carcinoma kidney cortical biopsy samples using pattern recognition of (1)H magic angle spinning (MAS) NMR spectra. NMR Biomed. 2003;13:64–71. doi: 10.1002/(sici)1099-1492(200004)13:2<64::aid-nbm612>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 102.Righi V, Mucci A, Schenetti L, et al. Ex vivo HR-MAS magnetic resonance spectroscopy of normal and malignant human renal tissues. Anticancer Res. 2007;27:3195–3204. [PubMed] [Google Scholar]
- 103.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal. Biochem. 2007;363(2):185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 104.Martínez-Bisbal MC, Martí-Bonmatí L, Piquer J, et al. 1H and 13C HR-MAS spectroscopy of intact biopsy samples ex vivo and in vivo 1H MRS study of human high grade gliomas. NMR Biomed. 2004;17:191–205. doi: 10.1002/nbm.888. [DOI] [PubMed] [Google Scholar]
- 105.Cheng L, Chang I, Louis D, Gonzalez RG. Correlation of high-resolution magic angle spinning proton magnetic resonance spectroscopy with histopathology of intact human brain tumor specimens. Cancer Res. 1998;58:1825–1832. [PubMed] [Google Scholar]
- 106.Tzika AA, Cheng LL, Goumnerova L, et al. Biochemical characterization of pediatric brain tumors by using in vivo and exvivo magnetic resonance spectroscopy. J. Neurosurg. 2002;96:1023–1031. doi: 10.3171/jns.2002.96.6.1023. [DOI] [PubMed] [Google Scholar]
- 107.Tzika AA, Astrakas L, Cao H, et al. Combination of high-resolution magic angle spinning proton magnetic resonance spectroscopy and microscale genomics to type brain tumor biopsies. Int J Mol Med. 2007;20:199–208. [PubMed] [Google Scholar]
- 108.Sjøbakk TE, Johansen R, Bathen TF, et al. Characterization of brain metastases using high-resolution magic angle spinning MRS. NMR Biomed. 2008;21(2):175–185. doi: 10.1002/nbm.1180. [DOI] [PubMed] [Google Scholar]
- 109.Valonen PK, Griffin JL, Lehtimäki KK, et al. High-resolution magic-angle-spinning 1H NMR spectroscopy reveals different responses in choline-containing metabolites upon gene therapy-induced programmed cell death in rat brain glioma. NMR Biomed. 2005;18:252–259. doi: 10.1002/nbm.955. [DOI] [PubMed] [Google Scholar]
- 110.Horsztyński D, Wawer I, Grieb P. A study of lyophylized human brain tumor tissue using MAS 1H-MRS spectroscopy. Neurol Neurochir Pol. 2003;37:123–132. [PubMed] [Google Scholar]
- 111.Barton SJ, Howe FA, Tomlins AM, et al. Comparison of in vivo 1H MRS of human brain tumours with 1H HR-MAS spectroscopy of intact biopsy samples in vitro. Magn. Reson. Mater. Phy. 1999;8:121–128. doi: 10.1007/BF02590529. [DOI] [PubMed] [Google Scholar]
- 112.Griffin JL, Kauppinen RA. A metabolomics perspective of human brain tumours. FEBS J. 2007;274:1132–1139. doi: 10.1111/j.1742-4658.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 113.Zuppi C, Messana I, Tapanainen P, et al. Proton nuclear magnetic resonance spectral profiles of urine from children and adolescents with type 1 diabetes. Clin. Chem. 2002;48:660–662. [PubMed] [Google Scholar]
- 114.Messana I, Forni F, Ferrari F, Rossi C, Giardina B, Zuppi C. Proton nuclear magnetic resonance spectral profiles of urine in type II diabetic patients. Clin. Chem. 1998;44:1529–1534. [PubMed] [Google Scholar]
- 115.Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol. Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- 116.Zhang S, Nagana Gowda GA, Asiago V, et al. Correlative and quantitative 1H NMR-based metabolomics reveals specific metabolic pathway disturbances in diabetic rats. 2007 doi: 10.1016/j.ab.2008.07.041. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Toye A, Gauguier D. Genetics and functional genomics of type 2 diabetes mellitus. Genome Biol. 2003;241:1–4. doi: 10.1186/gb-2003-4-12-241. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci.(USA) 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Makinen VP, Soininen P, Forsblom C, et al. Diagnosing diabetic nephropathy by H-1 NMR metabonomics of serum. Magn. Reson. Mater. Phy. 2006;19:281–296. doi: 10.1007/s10334-006-0054-y. [DOI] [PubMed] [Google Scholar]
- 120.Hodavance MS, Ralston SL, Pelczer I. Beyond blood sugar: the potential of NMR-based metabonomics for type 2 human diabetes, and the horse as a possible model. Anal. Bioanal. Chem. 2007;387:533–537. doi: 10.1007/s00216-006-0979-z. [DOI] [PubMed] [Google Scholar]
- 121. Shulman GI, Rothman DL, Shulman RG, Brindle K. C-13 Nmr-Studies of Glucose Disposal in Normal and Non-Insulin-Dependent Diabetic Humans. Philos. T. Roy. Soc. A. 1990;333:525–529. • 13C isotope tracers method used for detailed metabolomic studies.
- 122.Cline GW, Magnusson I, Rothman DL, Petersen KF, Laurent D, Shulman GI. Mechanism of impaired insulin-stimulated muscle glucose metabolism in subjects with insulin-dependent diabetes mellitus. J. Clin. Invest. 1997;99:2219–2224. doi: 10.1172/JCI119395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roden M, Shulman GI. Applications of NMR spectroscopy to study muscle glycogen metabolism in man. Annu. Rev. Med. 1999;50:277–290. doi: 10.1146/annurev.med.50.1.277. [DOI] [PubMed] [Google Scholar]
- 124.Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur. J. Clin. Invest. 2002;32:14–23. doi: 10.1046/j.1365-2362.32.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 125.Roden M, Price TB, Perseghin G, et al. Mechanism of free fatty acid-induced insulin resistance in humans. J. Clin. Invest. 1996;97:2859–2865. doi: 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang C, Kong HW, Guan YF, et al. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Anal. Chem. 2005;77:4108–4116. doi: 10.1021/ac0481001. [DOI] [PubMed] [Google Scholar]
- 127.Plumb RS, Johnson KA, Rainville P, et al. The detection of phenotypic differences in the metabolic plasma profile of three strains of Zucker rats at 20 weeks of age using ultra-performance liquid chromatography/orthogonal acceleration time-of-flight mass spectrometry. Rapid Commun. Mass Spec. 2006;20:2800–2806. doi: 10.1002/rcm.2655. [DOI] [PubMed] [Google Scholar]
- 128.Major HJ, Williams R, Wilson AJ, Wilson ID. A metabonomic analysis of plasma from Zucker rat strains using gas chromatography/mass spectrometry and pattern recognition. Rapid Commun. Mass Spec. 2006;20:3295–3302. doi: 10.1002/rcm.2732. [DOI] [PubMed] [Google Scholar]
- 129.Yi LZ, He J, Liang YZ, Yuan DL, Chau FT. Plasma fatty acid metabolic profiling and biomarkers of type 2 diabetes mellitus based on GCMS and PLS-LDA. Febs Lett. 2006;580:6837–6845. doi: 10.1016/j.febslet.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 130.Newgard CB, Stedman SW. Metabolic profiling for understanding of diabetes and obesity mechanisms. Faseb J. 2006;20:A1307–A1307. [Google Scholar]
- 131.Williams R, Lenz EM, Wilson AJ, et al. A multi-analytical platform approach to the metabonomic analysis of plasma from normal and zucker (fa/fa) obese rats. Mol. Biosyst. 2006;2:174–183. doi: 10.1039/b516356k. [DOI] [PubMed] [Google Scholar]
- 132.Atherton HJ, Bailey NJ, Zhang W, et al. A combined H-1-NMR spectroscopy- and mass spectrometry-based metabolomic study of the PPAR-alpha null mutant mouse defines profound systemic changes in metabolism linked to the metabolic syndrome. Physiol. Genomics. 2006;27:178–186. doi: 10.1152/physiolgenomics.00060.2006. [DOI] [PubMed] [Google Scholar]
- 133.Jones PM, Bennett MJ. The changing face of newborn screening: diagnosis of inborn errors of metabolism by tandem mass spectrometry. Clin. Chim. Acta. 2002;324:121–128. doi: 10.1016/s0009-8981(02)00238-3. [DOI] [PubMed] [Google Scholar]
- 134.Engelke UFH, Liebrand-van Sambeek MLF, DeJong JGN, et al. N-Acetylated Metabolites in Urine: Proton Nuclear Magnetic Resonance Spectroscopic Study on Patients with Inborn Errors of Metabolism. Clin. Chem. 2004;50:58–66. doi: 10.1373/clinchem.2003.020214. [DOI] [PubMed] [Google Scholar]
- 135.Moolenaar SH, Engelke UFH, Wevers RA. Proton nuclear magnetic resonance spectroscopy of body fluids in the field of inborn errors of metabolism. Ann. Clin. Biochem. 2003;40:16–24. doi: 10.1258/000456303321016132. [DOI] [PubMed] [Google Scholar]
- 136. Iles RA, Hind AJ, Chalmers RA. Use of proton nuclear magnetic resonance spectroscopy in detection and study of organic acidurias. Clin. Chem. 1985;31:1795–1801. • Early NMR applications to inborn errors of metabolism.
- 137.Constantinou MA, Papakonstantinou E, Benaki D, et al. Application of nuclear magnetic resonance spectroscopy combined with principal component analysis in detecting inborn errors of metabolism using blood spots: a metabonomic approach. Anal. Chim. Acta. 2004;511:303–312. [Google Scholar]
- 138.Constantinou MA, Papakonstantinou E, Spraul M, et al. 1H NMR-based metabonomics for the diagnosis of inborn errors of metabolism in urine. Anal. Chim. Acta. 2005;542:169–177. [Google Scholar]
- 139. Pan Z, Gu H, Talaty N, et al. Principal component analysis of urine metabolites detected by NMR and DESI-MS in patients with inborn errors of metabolism. Anal. Bioanal. Chem. 2007;387:539–549. doi: 10.1007/s00216-006-0546-7. • Combined NMR and MS approach for detecting inborn errors of metabolism.
- 140.Engelke UFH, Oostendorp M, Wevers RA. Lindon JC, Nicholson JK, Holmes E, editors. NMR spectroscopy of body fluids as a metabolomics approach to inborn errors of metabolism. The handbook of metabonomics and metabolomics. 2007:375–412. [Google Scholar]
- 141. Otvos JD, Jeyarajah EJ, Bennett DW. Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clin. Chem. 1991;37:377–386. • Early pattern recognition approach for the identification of cardiovascular disease risk.
- 142.Kuller L, Arnold A, Tracy R, et al. Nuclear magnetic resonance spectroscopy of lipoproteins and risk of coronary heart disease in the cardiovascular health study. Arterioscler. Thromb. Vasc. Biol. 2002;22:1175–1180. doi: 10.1161/01.atv.0000022015.97341.3a. [DOI] [PubMed] [Google Scholar]
- 143.Brindle JT, Antti H, Holmes E, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR based metabonomics. Nat. Med. 2002;8:1439–1444. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 144.Brindle JT, Nicholson JK, Schofield PM, Grainger DJ, Holmes E. Application of chemometrics to 1H NMR spectroscopic data to investigate a relationship between human serum metabolic profiles and hypertension. Analyst. 2003;128:32–36. doi: 10.1039/b209155k. [DOI] [PubMed] [Google Scholar]
- 145.Kirschenlohr HL, Griffin JL, Clarke SC, et al. Proton NMR analysis of plasma is a weak predictor of coronary artery disease. Nat. Med. 2006;12:705–710. doi: 10.1038/nm1432. [DOI] [PubMed] [Google Scholar]
- 146. Ala-Korpela M. Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med. 2008;46(1):27–42. doi: 10.1515/CCLM.2008.006. •• This review evaluates plasma/serum based metabolomics in general with emphasis on coronary heart diseases and diabetes.