Abstract
The processes that direct skull remodeling are of interest to both human-oriented studies of cranial dysplasia and evolutionary studies of skull divergence. There is increasing awareness that these two fields can be mutually informative when natural variation mimics pathology. Here we describe a zebrafish mutant line, devoid of blastema(dob), which does not have a functional fgf20a protein, and which also presents cranial defects similar to both adaptive and clinical variation. We used geometric morphometric methods to provide quantitative descriptions of the effects of the dob mutation on skull morphogenesis. In combination with whole-mount in situ hybridization labeling of normal fgf20a expression and assays for osteoblast and osteoclast activity, the results of these analyses indicate that cranial dysmorphologies in dob zebrafish are generated by aberrations in post-embryonic skull remodeling via decreased osteoblasotgenesis and increased osteoclastogenesis. Mutational effects include altered skull vault geometries and midfacial hypoplasia that are consistent with key diagnostic signs for multiple human craniofacial syndromes. These phenotypic shifts also mimic changes in the functional morphology of fish skulls that have arisen repeatedly in several highly successful radiations (e.g., damselfishes and East-African rift-lake cichlids). Our results offer the dob/fgf20a mutant as an experimentally tractable model with which to examine post-embryonic skull development as it relates to human disease and evolution.
Introduction
The developmental genetic processes that promote both disease and evolutionary divergence are in some cases identical, since similar shifts in ontogeny may promote either fitness or failure in different species or in different ecological circumstances (Albertson et al., 2009). Developmental investigations therefore have relevance to both human health and studies of adaptive divergence. Most developmental work has focused on patterning that occurs in early ontogeny (Albertson and Yelick 2004), but the developmental shaping that occurs during anatomical remodeling is a major component of morphogenesis, and many tissues are reshaped throughout life (e.g., bone; Suda et al. 1992, Slemenda et al. 1997, Hadjidakis and Androulakis 2006). Structures that are remodeled over long periods of time may generate either adaptive or maladaptive morphologies over wide age ranges. In evolutionary terms, anatomical remodeling therefore represents a broad target for selective forces, but the developmental controls of remodeling have been relatively understudied.
Understanding osteological remodeling is of particular importance to studies of skull form, function and disease. In many species the bones of the skull undergo dramatic morphological changes over the course of development, and significant remodeling can occur many months or even years subsequent to the completion of embryonic stages (Brodie 1941, Maes and Kronenberg 2011). This remodeling can lead to dramatic shifts in shape, and developmental changes in cranial shape often reflect ontogenetic changes in foraging niche (Luczkovich et al. 1995, Mullaney and Gale 1996, Erickson et al. 2003, Denoel et al. 2006, Tibbetts et al. 2008, Hellig et al. 2010). Since changes in the functional morphology of cranial structure have been identified as a major factor in important and well-described adaptive radiations (e.g., African cichlid fishes, Darwin's finches, Damselfishes, and Mammals; Cooper and Westneat 2009, Cooper et al. 2010, Goswami et al. 2011, Mallarino et al. 2011), studies of the controls of skull morphogenesis are critical for establishing the ontogenetic underpinnings of evolutionary divergence.
The skulls of fishes, perhaps more than any other vertebrate group, have been extensively studied in terms of the functional morphology of feeding (e.g.; Westneat 1995, Ferry-Graham et al. 2002, Hernández et al. 2002, Westneat 2003, Carroll et al. 2004, Rice and Westneat 2005, Bellwood et al. 2006, Konow et al. 2008, Rice et al. 2008, Cooper and Westneat 2009, Cooper et al. 2010). Moreover, there is a rapidly expanding body of research focused on using the zebrafish as a model organism for understanding the genetic controls of skull formation and the mutations responsible for anatomical cranial disorders in humans (Schilling and Kimmel 1997, Kimmel et al. 2001a, Kimmel et al. 2001b, Schilling et al. 2010, Parsons et al. 2011). This developmental work augments the comparative studies of fish trophic anatomy in a way that makes fish skulls a uniquely promising subject for understanding the changes in skull development that are responsible for both disease and adaptive changes in feeding morphology.
There is, however, a disjunction between these two lines of inquiry; studies of fish functional morphology have focused mainly on the adult skull, whereas developmental genetic studies of zebrafish have focused almost exclusively on embryonic stages. Before these two fields can be fully complementary a common ground must be reached. To this end, we present a study in which we discuss the effects of fgf20a deficiency on zebrafish post-embryonic skull growth and remodeling in terms of both human craniofacial disorders and the evolutionary diversification of the functional morphology of fish feeding.
Fibroblast growth factors (Fgfs) function as ligands in a signaling system that is ancient in origin, and which is involved in a wide variety of developmental events that include the regulation of cellular proliferation, differentiation, migration, and survival (Itoh and Ornitz 2004, Katoh and Katoh 2006). Multiple Fgf family genes play important roles in skeletal growth and development (Iseki et al. 1997, Hurley et al. 1999, Sabbieti et al. 1999, Liu et al. 2002, Ohbayashi et al. 2002, Cowan et al. 2003, Hajihosseini et al. 2004, Albertson and Yelick 2007, Hung et al. 2007, Liu et al. 2007), and most of the 23 known Fgfs are expressed in the skull (Hajihosseini and Heath 2002, Katoh and Katoh 2006, Degnin et al. 2010). Normal skull growth and development requires a balance between cell proliferation, differentiation and destruction, and this balance can be destabilized in multiple ways by the disruption of Fgf signaling (Morriss-Kay and Wilkie 2005).
Mutagenesis screening in the zebrafish (Danio rerio; Hamilton, 1882) was used to create the mutant line devoid of blastema(dob), which possesses a null mutation in fgf20a (Whitehead et al. 2005). Fgf20 is an evolutionarily conserved target of the canonical WNT signaling pathway (Chamorro et al. 2005, Katoh and Katoh 2006, Katoh 2007). It is one of the canonical Fgfs that function as ligands that activate membrane-bound Fgfrs in a paracrine manner (Degnin et al. 2010). Fgf20 is known to function in the brain, epidermis and the gastrointestinal tract, it has been identified as being necessary for normal heart development, and in humans FGF20 has been implicated in Parkinson's disease and some types of carcinogenesis (van der Walt et al. 2004, Chamorro et al. 2005, Lavine et al. 2005, Katoh and Katoh 2006, Cohen et al. 2007, Fan et al. 2007, Satake et al. 2007, Beenken and Mohammadi 2009, Wells et al. 2012). The zebrafish fgf20a protein has 76.9% total amino-acid identity with human FGF20 (Katoh and Katoh 2005).
The similarity of zebrafish calvarial (i.e., skull vault) anatomy to that of humans has prompted their nomination as an appropriate model for studying human skull disorders (Quarto and Longaker 2005). There is considerable variation in the normal development of skull shape and this can complicate the task of determining mutational effects (Rosas and Bastir 2002, Zollikofer and de Leon 2002, Gonzalez-Jose et al. 2005, Bastir et al. 2006, Sardi et al. 2007) Quantitative descriptions of morphology can therefore improve the precision with which mutational effects are identified and described and such analytical techniques present a valuable set of tools for investigating the physiological mechanisms that determine the genotype to phenotype relationship (Cooper and Albertson 2008, Oates et al. 2009).
We examined the effects of the dob/fgf20a mutation at two developmental stages (1 cm standard length and adult) using a morphometric approach to quantify and compare cranial shape variation among dob and AB strain wild-type (ABWT) fish, which is the strain into which the dob mutation was induced (Whitehead et al. 2005). We combined this approach with whole-mount in situ hybridization (WISH) analysis of fgf20a expression in ABWT fish, and a comparison of calvarial suture patterns in dob and ABWT specimens. In addition, we surveyed dob mutants for more general mineralized tissue defects. Finally, we used assays for osteoclast and osteoblast activity to compare bone remodeling in both groups.
Materials and Methods
Specimens
Zebrafish were maintained under standard conditions at 28 °C using a 14-h light/10-h dark cycle. Homozygous recessive dob mutants and ABWT fish were produced by natural matings and reared until they reached a standard length of 1 cm (junvenile specimens), or until 150-180 dpf (adult specimens). All specimens were euthanized using an overdose of Tricaine Methanesulfonate (Western Chemical, Inc.). Digital images were obtained by photographing specimens using a Zeiss Axiocam digital imaging system mounted to an M2 Bio stereomicroscope (Carl Zeiss, Inc.). All specimens were photographed while submerged in 80% glycerol.
Head shape comparisons
Specimens were cleared and stained according to Pothoff (1983) with slight modifications, stored in 80% glycerol, and then photographed in both dorsal and lateral views. Geometric morphometric methods were used to analyze the skull shapes of dob and ABWT specimens (see below). As our initial results identified the existence of upper jaw mutational effects, we removed the maxillary and premaxillary bones from adult ABWT and dob specimens and photographed them in dorsal view. The shapes of these bones were then analyzed using an outline-based, semilandmark approach.
All skull bones had formed by the time the fish reached 1 cm standard length. In order to determine if Fgf20a affected skull patterning or skull remodeling we compared the skull shape anatomies of mutant and wild-type fish at the 1 cm and adult stages. Dorsal images were collected first using the following sample sizes: 1 cm ABWT (N=28); 1 cm dob (N=16); adult ABWT (N=12); adult dob (N=15). An initial attempt was made to close the jaws of cleared and stained specimens in order to achieve homologous positioning, but this proved difficult and resulted in the damage of some specimens that could not be used in lateral shape analyses. Lower jaw LMs were therefore excluded from the lateral data. Lateral sample sizes: 1 cm ABWT (N=24); 1 cm dob (N=13); adult ABWT (N=12); adult dob (N=15).
The program tpsDig2 (Rohlf 2006) was used to establish the positions of homologous anatomical landmarks (LM) and semilandmarks on specimen images. For both the juvenile and adult fish, the positions of 11 lateral LM and 9 dorsal LM (4 midline LM and 5 pairs of bilaterally homologous LM) were determined (Figure 1). Adult left and right premaxillae and maxaillae were photographed in the dorsal view and 75 semilandmarks were evenly spaced around the periphery of each bone image using tpsdig. The programs tpsUtil and tpsrelW were then used to superimpose these semilandmarks using a chord-distance (Procrustes distance) based “sliders” method. Maxillary and premaxillary shapes were analyzed separately. For each specimen the semilandmark positions from the left and right bones were averaged by first reflecting the right-side semilandmarks in tpsDig2 and then calculating a consensus form using tpsRelW. Procrustes sumperimpositions of LM data from multiple specimens were used to remove variation in position that was unrelated to shape (scale and orientation) from the dorsal, lateral, maxillary and premaxillary data. The program CoordGen was used to perform Procrustes transformations.
Figure 1.
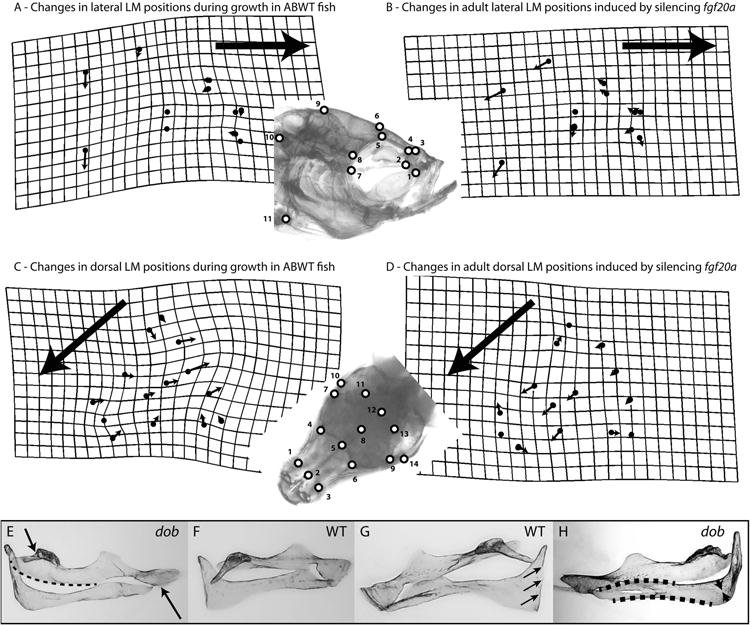
Depictions of skull and upper jaw bone shape transformations that occurred as a result of normal development or which were induced by silencing fgf20a signaling. The lateral and dorsal LM analyzed in this study are depicted in the inserts. The grids and vectors in panels A-D describe the differences between the mean LM configurations (i.e., consensus configurations calculated after Procrustes superimposition of the LM data) of 2 datasets. Deformation grids depict the minimal amount of “bending” needed to transform one mean LM configuration to another and were calculated using a thin-plate spline method. Vectors denote the direction and magnitude of positional differences between homologous LM in the 2 mean LM configurations being compared. Deformation grids and vectors were calculated using TwoGroupMac7. Large arrows indicate the anterior direction in panels A-D. A. Developmental changes in lateral ABWT skull shape that occur between 1 cm and adult (1 cm ABWT to adult ABWT transformation of the LM configuration). B. Changes in lateral adult skull shape induced by the loss of fgf20a signaling (adult ABWT to adult dob transformation of the LM configuration). C. Developmental changes in dorsal WT skull shape that occur between 1 cm and adult (1 cm ABWT to adult ABWT transformation of the LM configuration). D. Changes in dorsal adult skull shape induced by the loss of fgf20a signaling (adult ABWT to adult dob transformation of the LM configuration). E-H. The maxillae and premaxillae in these panels are shown in dorsal view. Maxillae and premaxillae were left articulated during their removal from the specimens. E and F. The maxillary shape differences depicted in in these panels are indicative of the shape changes induced by the loss of fgf20a expression. E. Maxilla from a dob specimen. Loss of fgf20a expression induces shortening and contortion of the premaxillad wing (upper arrow), a pronounced posterior-medial expansion of articular head (dotted line), and a reduction and contortion of the shank (lower arrow). F. ABWT maxilla. G and H. The premaxillary shape differences depicted in in these panels are indicative of the shape changes induced by the loss of fgf20a expression. G. ABWT premaxilla with a straight medial edge on the ascending arm (arrows). H. Premaxilla from a dob specimen. Loss of fgf20a expression induces a reduction in the anterior and posterior curvature of the dentigerous arm (dotted lines), a pronounced reduction of the area of bone in the angle between the dentigerous and ascending arms (arrows) and irregularity in the shape of the medial edge on the ascending arm.
Single factor MANOVAs that employed resampling to determine the significance of the F-ratio (2000 permutations) were used to test both the dorsal and lateral data for shape differences between the following groups: 1 cm ABWT vs. 1 cm dob; adult ABWT vs. adult dob; 1 cm ABWT vs. adult ABWT; and 1 cm dob vs. adult dob. MANOVA was also used to test for maxillary and premaxillary shape differences between adult ABWT and adult dob fish. A Goodall's F-test that employed resampling to determine the significance of the F-ratio (4900 bootstraps) was also used to test for differences in mean skull shape between the same groups examined using MANOVA. To determine if there were similar mutational effects (if any) on skull shape at the two developmental stages the Procrustes distances between the means of juvenile LM configurations (1 cm ABWT vs. 1 cm dob) were compared to the Procrustes distances between means of adult LM configurations (adult ABWT vs. adult dob) using a resampling test (4900 bootstraps). The program CVAGenMac7b was used to perform MANOVA tests. Goodall's F-tests and tests for differences in Procrustes distances were both performed using the program TwoGroupMac7.
In order to determine if the ABWT and dob skull shape datasets had similar orientations in “shape space” we used a bootstrapping procedure (4900 sets) to define the 95% confidence interval (CI) for the angle that described the difference in the orientations of the shape spaces. These comparisons examined the orientations of lateral and dorsal datasets that contained both 1 cm and adult LM data. If the observed angle fell within one of the 95% CIs derived from either dataset, then the orientations of the two original shape spaces were considered to be not significantly different. The orientations of the first PC axes derived from each of the data sets were compared directly, but axes subsequent to PC1 could not be examined individually. All analyses that involved multiple axes determined whether the alignments of planes (when only 2 PC axes were examined) or multi-dimensional hyperplanes (“flat” surfaces of >2 dimensions embedded in larger dimensional spaces) were significantly different. When bootstrapping any two of the original sets of data, the sample sizes of the bootstrap sets produced from the larger of the two were the same as the sample sizes of both original data sets. Re-sampling the smaller of the original two data sets created two bootstrap sets of the same size as the original. These comparisons were performed using the program SpaceAngle. For further methodological details in regard to comparing shape spaces see Parsons et al. (2009).
Comparisons of Developmental Trajectories
The developmental skull shape trajectories of ABWT and dob fish (dorsal and lateral data) were compared using “trajectory analysis” in the geomorph package for the R programing language (Adams and Otárola-Castillo 2013). This analysis was used to estimate of the angle between the developmental trajectories for the two genotypes and to test whether the trajectories were significantly different. The null hypothesis is that the residuals from the trajectories are interchangeable between the two groups and this was tested by permuting the residuals (9999 iterations). A similar analysis was performed using the program VecCompare7. MANCOVA was used to compare ABWT and dob skull shapes (dorsal and lateral data) by comparing datasets that included both 1 cm and adult fish. This permitted testing for significant effects of genotype, age and the interaction between genotype and age on skull shape. MANCOVA tests were performed using geomorph (Adams and Otárola-Castillo 2013).
Comparisons of morphological variation
The morphological variation (i.e., shape diversity) exhibited by the juvenile and adult skulls of both genotypes (both lateral and dorsal data) was quantified by calculating the “Foote disparity” (D) of each set of LM data using the following formula:
where di is the distance from the centroid of the entire group (the centroid of the mean shape of all the specimens in a data set) to the centroid of the Procustes transformed LMs derived from each individual specimen in that group, and N is the total number of specimens in that dataset (Foote 1993). A permutation procedure (2000 runs) was used to calculate the 95% CI of the difference between datasets. If the observed shape variation difference was greater than the upper bound of the 95% CI, then the difference was considered to be significant. The program DisparityBox6 was used to perform these calculations.
The programs CoordGen, DisparityBox6, SpaceAngle TwoGroupMac7, and VecCompare7 are part of the Integrated Morphometrics Programs (IMP), and compiled stand-alone versions are freely available at http://ww3.canisius.edu/∼sheets/morphsoft.html.
Comparisons of adult ABWT and dob upper jaw length
Error is introduced into coordinate-based shape analyses if moveable elements (e.g. limbs, jaws) are in non-homologous positions in different specimens. These differences in orientation will be mistakenly perceived to be differences in shape. Since there were differences in the degree of gape among specimens, lateral shape analyses did not include LM data from the jaws. Non-destructive manipulation of fish jaws did not prove feasible. Although there exist analytical procedures that can be used to correct for jaw angle differences (e.g the geomorph package in R; Adams and Otárola-Castillo 2013), zebrafish jaws are more biomechanically complex than those of tetrapods. The motion of lower jaw abduction is transferred to the premaxillary bones via highly kinetic maxillas in a manner that results in premaxillary protrusion (Hernández et al. 2002). An appropriate transformation that would allow homologous positioning of jaw LM would therefore require the simultaneous correction of multiple angles and is not currently feasible.
Since many of the dob specimens had shortened upper jaws (Figure 2) that did not occlude properly with their lower jaws, we tested for differences in relative upper jaw length (upper jaw length standardized by lower jaw length) in dob and ABWT fish. We used tpsDig2 to record the length of both the upper jaw (measured as the distance from the lower, i.e. posterio-ventral, tip of the shank of the maxilla to the upper, i.e., anterio-dorsal, tip of the articulation head of the maxilla) and the lower jaw (measured as the distance from the point of the articular-quadrate articulation, i.e. the lower jaw joint, to the anterior tip of the dentary). Standardized upper jaw lengths of dob and ABWT fish were compared using ANOVA. Anatomy after Barel et al. (1976). The comparison of ratios (e.g., standardized upper jaw length) is statistically problematic and may violate the assumptions of ANOVA, but we include the results here in order to highlight an interesting pattern that we observed in our data. This analysis should be considered exploratory.
Figure 2.
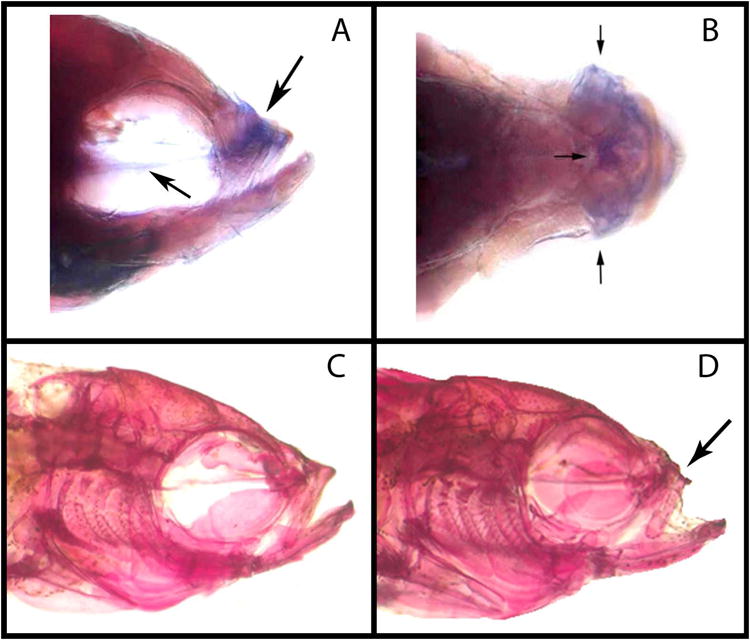
Expression labeling of fgf20a in a ABWT zebrafish, and head morphology of ABWT and dob fish. Lateral (A) and dorsal (B) views of fgf20a expression labeling (indicated by arrows) in the midface (upper arrow in panel A, all arrows in panel B) and the parasphenoid bone (lower arrow in panel A) of a ABWT adult zebrafish. C. The head of an adult ABWT zebrafish. D. The head of an adult dob zebrafish.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed on adult ABWT zebrafish as described previously (Albertson and Yelick 2007). Anti-sense digoxigenin-labeled riboprobe was generated from full-length fgf20a (Whitehead et al. 2005), and fractionated to ∼300 nucleotides to facilitate probe penetration according to Elizondo et al. (2005).
Assays for osteoblast and osteoclast activity
Since bone remodeling relies on a balance between deposition and resorption, the activity of osteoblasts and osteoclasts was quantified using assays for alkaline and acid phosphatase, respectively. The eyes and viscera of adult specimens were removed prior to their fixation on a rocker for 1 hour at room temperature. The fixative solution was prepared by adding the following compounds in their respective ratios: 25 ml citrate solution (citric acid, 18 mmol/l, sodium citrate, 9 mmol/l, sodium chloride, 12 mmol/l with surfactant, buffered at pH 3.6), 65 ml acetone, and 8 ml 37% formaldehyde. All compounds were brought to room temperature prior to preparation of the fixative solution. Fixation was performed in 50 ml test tubes, and enough fixative was made to allow for generous submersion of all specimens (∼15 fish/tube).
Subsequent to fixation, all specimens were rinsed in diH20 2 × 10 minutes on a rocker. To insure accurate comparisons of alkaline and acid phosphatase staining, all dob and ABWT fish were stained for equal periods of time in their respective solutions, and care was taken to ensure approximately equal densities of fish and staining solutions in all of the test tubes in which the staining procedures were performed.
Alkaline phosphatase (AP) staining for osteoblast activity was performed using a leukocyte alkaline phosphatase kit (Sigma-Aldrich, 86C-1KT). Immediately after fixation and rinsing, specimens were submerged in an alkaline dye solution for 2 hours in the dark. Using 15 ml test tubes, we added 5 fish per tube and enough alkaline dye solution to complete 15 ml total.
The alkaline dye was prepared by adding the following compounds in their respective ratios: 2 ml of a diazonium salt solution was added to 45 ml of room temperature diH20; and then adding 1 ml naphthol AS-BI alkaline solution (naphthol AS-BI phosphate, 4 mg/ml, in AMPD buffer, 2 mol/l, pH 9.5.). The diazonium salt solution was prepared by mixing 1 ml of fast blue alkaline solution (Fast blue BB base, 5 mg/ml, in hydrochloric acid, 0.4 mol/l, with stabilizer) with 1ml of sodium nitrite solution (0.1 mol/l) via gentle inversion. All staining solutions were prepared immediately before use.
Tartrate resistant acid phosphatase (TRAP) staining for osteoclast activity was performed using a leukocyte acid phosphatase kit (Sigma-Aldrich, 387A-IKT). Initial specimen preparation, fixation and rinsing followed the same protocol as the AP staining. After rinsing in diH20 the specimens were transferred to 15 ml tubes (5 fish/tube), and the tubes were filled to a volume of 15 ml with a pre-prepared TRAP staining solution and incubated at 37° C in the dark for 4.5 hours.
The TRAP solution was prepared by adding the following compounds in their respective ratios: A diazotized fast garnet GBC solution was first prepared by mixing 0.5 ml of fast garnet GBC base solution (fast garnet GBC base, 7.0 mb/ml, in 0.4 mol/l hydrochloric acid with stabilizer) with 0.5 ml of a sodium nitrite solution (0.1 mol/l) via gentle inversion, and this solution was then allowed to stand for 2 minutes. The diazotized fast garnet solution (1.0 ml) was then added to 45 ml of diH20 that had been pre-warmed to 37° C, and this solution was then mixed with 0.5 ml of naphthol AS-BI phosphate solution (naphthol AS-BI phosphoric acid, 12.5 mg/ml), 2.0 ml of acetate solution (acetate buffer, 2.5 mol/l, pH 5.2 ± 0.1), and 1.0 ml of tartrate solution (L(+)- tartrate buffer, 0.335 mol/l, pH 4.9 ± 0.1).
After staining, all AP and TRAP specimens were immediately rinsed in H20 3 × 10 minutes on a rocker. AP specimens were rinsed in tap water, and TRAP specimens were rinsed in diH20. The fish were then cleared at 37° C using trypsin (1 g trypsin powder in 30 ml H2O+30 ml sodium borate saturated H2O), and then bleached in 10% H202. All specimens were then placed in 25% glycerol for 24 hours, transferred to 50% glycerol for 24 hours, and then stored in 80% glycerol in the dark.
Specimens were photographed in both dorsal and lateral (right side only) views, and pixel density analyses (PDA) were performed by quantifying the amount of AP or TRAP staining in the head of each fish, and then comparing the levels of staining in the dob and ABWT specimens using ANOVA. The degree of staining was measured by examining the digital photograph of each specimen using Adobe Photoshop CS5 (version 12.0.1; Adobe Systems Inc.). The eyedropper tool was used to select the color displayed in stained areas of the head (either AP or TRAP) in a representative specimen, and this color was then used to determine the number of stained pixels in all specimens for that type of assay.
The number of pixels within the head of each specimen was determined by outlining the region using the Photoshop polygonal lasso tool, and the pixel number displayed in the histogram window was recorded. The number of stained pixels within the outline was then determined by selecting a color range based on the representative stain color and using a fuzziness setting of 100. The number of pixels within the outlined area that matched these color parameters was then recorded from the histogram window. The ratio of stained to unstained pixels was calculated for each specimen using Excel (Excel for Mac, version 14.0.2; Microsoft Inc.) and this data was analyzed using ANOVA.
Results
Shape, shape variation and growth
Loss of fgf20a signaling had a significant effect on zebrafish skull shape, but this effect did not manifest until after the initial formation of all skull bones (i.e, subsequent to the fish achieving lengths of 1 cm). Loss of signaling strongly affected the midface and upper jaws while also shifting calvarial morphology and other aspects of skull anatomy (Figures 1,2). The results of both MANOVA and Goodall's F tests indicated that adult ABWT and mutant zebrafish had significantly different skull shapes (Table 1). This was true for both the lateral and dorsal LM datasets. The shapes of the adult maxillae and premaxillae of the two genotypes were also significantly different (Table 1; Figure 1). There was no significant difference in skull shape between the ABWT and dob fish at the 1 cm stage, but within each genotype 1 cm fish had lateral and dorsal LM configurations that were significantly different from those of adults (Table 1). Analysis of the lateral LM data using MANCOVA indicated that genotype, age and the genotype × age interaction all significantly affected skull shape (Table 1). MANCOVA did not return a significant effect for genotype alone when the dorsal LM data were examined, but there were significant effects for both age and the genotype × age interaction (Table 1).
Table 1. Shape comparisons.
| Comparison | MANOVA F ratio (df) | MANOVA p-value | Goodall's F-test F ratio | Goodall's F-test p-value |
|---|---|---|---|---|
| Lateral 1 cm ABWT vs. 1 cm dob | 1.22 (1, 35) | 0.2739 | 1.22 | 0.2655 |
|
| ||||
| Lateral adult ABWT vs. adult dob | 2.33 (1, 25) | 0.0225* | 2.34 | 0.0416* |
|
| ||||
| Lateral 1 cm ABWT vs. adult ABWT | 15.19 (1, 34) | 0.0005* | 15.21 | 0.0002* |
|
| ||||
| Lateral 1 cm dob vs. adult dob | 13.86 (1, 26) | 0.0028* | 13.87 | 0.0002* |
|
| ||||
| Dorsal 1 cm ABWT vs. 1 cm dob | 1.10 (1, 42) | 0.3348 | 1.11 | 0.3200 |
|
| ||||
| Dorsal adult ABWT vs. adult dob | 3.19 (1, 25) | 0.0185* | 3.20 | 0.0288* |
|
| ||||
| Dorsal 1 cm ABWT vs. adult ABWT | 20.66 (1, 38) | 0.0076* | 20.67 | 0.0002* |
|
| ||||
| Dorsal 1 cm dob vs. adult dob | 18.40 (1, 29) | 0.0061* | 18.41 | 0.0002* |
|
| ||||
| Adult maxilla shape, ABWT vs. dob | 3.95 (1, 25) | 0.0045* | 4.07 | 0.0041* |
|
| ||||
| Adult premaxilla shape, ABWT vs. dob | 3.21 (1, 25) | 0.0255* | 3.25 | 0.214* |
| MANCOVA results | Genotype: F ratio, p-value | Age: F ratio, p-value | Genotype × Age: F ratio, p-value | Residuals df |
|---|---|---|---|---|
| Lateral data (both ages, ABWT & dob) | 2.66, 0.015* | 26.32, 0.001* | 2.80, 0.017* | 60 |
|
| ||||
| Dorsal data (both ages, ABWT & dob) | 1.79, 0.132 | 36.34, 0.001* | 2.31, 0.040* | 67 |
comparisons that were statistically significant at α=0.05
Within age groups there was no significant effect of genotype on skull shape variation (both lateral and dorsal data; Table 2). There was also no significant effect of age on skull shape variation within genotypes (both lateral and dorsal data; Table 2). Genotype also had no significant effect on the variation of adult maxillary and premaxillary shape (Table 2).
Table 2. Shape variation comparisons.
| Comparisons | Foote disparity | Difference in shape variation | 95% upper bound of the permutation generated difference | Number of permutations (out of 2000) in which the magnitude of the observed difference was exceeded |
|---|---|---|---|---|
| Lateral LM 1 cm ABWT | 0.00261 | |||
| vs. | 0.00015 | 0.00112 | 1572 | |
| Lateral LM 1 cm dob | 0.00245 | |||
|
| ||||
| Lateral LM adult ABWT | 0.00258 | |||
| vs. | 0.00054 | 0.00167 | 1093 | |
| Lateral LM adult dob | 0.00312 | |||
|
| ||||
| Dorsal LM 1 cm ABWT | 0.00827 | |||
| vs. | 0.00108 | 0.00384 | 1201 | |
| Dorsal LM 1 cm dob | 0.00718 | |||
|
| ||||
| Dorsal LM adult ABWT | 0.00594 | |||
| vs. | 0.00086 | 0.00202 | 938 | |
| Dorsal LM adult dob | 0.00508 | |||
|
| ||||
| Adult maxilla ABWT | 0.00581 | |||
| vs. | 0.00136 | 0.00250 | 639 | |
| Adult maxilla dob | 0.00444 | |||
|
| ||||
| Adult premaxilla ABWT | 0.00319 | |||
| vs. | 0.00070 | 0.00195 | 1000 | |
| Adult premaxilla dob | 0.00389 | |||
When the upper bound of the 95% confidence interval was greater than the observed difference, then the two groups being compared were considered to not have significantly different disparities. This was the case for all comparisons.
The Procrustes distances between the mean shapes of adult ABWT and dob fish were greater than the distances between the mean shapes of 1 cm ABWT and dob fish (both lateral and dorsal data), but there was overlap of the 95% estimates of mean shape for both the lateral and dorsal comparisons (Figure 3). The developmental trajectories of the ABWT and dob fish were significantly different, with an angular difference of 36.01 degrees for the lateral data (p = 0.0016) and 27.62 degrees for the dorsal data (p = 0.0199). The same angles were returned by both the geomorph analyses and VecCompare. The length of the developmental trajectories did not differ significantly between genotypes for either the lateral or dorsal data (Figure 3). The ABWT and dob LM datasets (1 cm and adult combined) did not have significantly different orientations in morphospace. This was true for the first 5 principal component axes (at least) for both the lateral and dorsal datasets.
Figure 3.
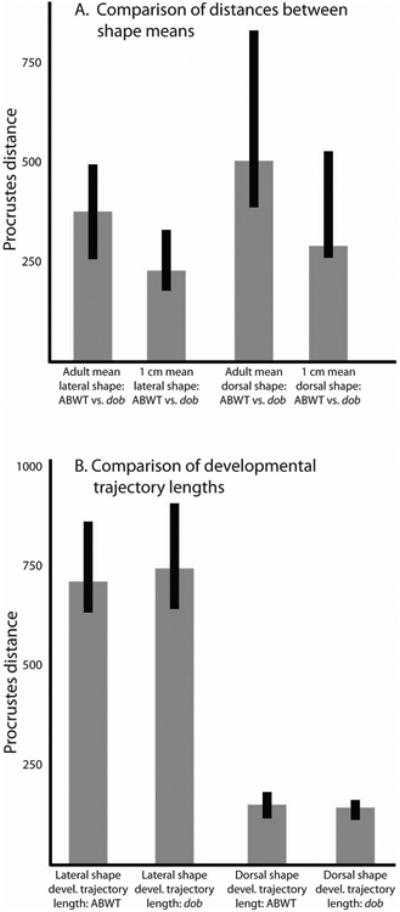
Comparisons of Procrustes distances between shape means and along developmental trajectories. A. Comparisons of the Procrustes distances between the mean shapes of ABWT and dob at two developmental stages for both the lateral and dorsal LM data. B. Comparisons of developmental trajectory lengths (measured as the Procrustes distances between mean 1 cm skull shapes and adult skull shapes) between ABWT and dob fish for both the lateral and dorsal LM data.
The laterally observable anatomical differences induced by the lost of fgf20a (Figures 1,2) included a general reduction of the upper jaw and mid-facial region of the skull, ventral extension of the pre-orbital process (LM 1), a posterio-ventral displacement of the anterior edge of the parasphenoid (LM 2), a posterior shift of the lateral edges of the prefrontal region of the calvaria (LM 3-6), posterio-ventral shift in the posterior edge of the parasphenoid (LM 7), strong posterior-ventral shifts in the lateral edge of the posterior margin of the parietal bone (LM 9) and the articulation of the skull with the vertebral column (LM 10), and a strong posterior-ventral shift in the articulation of the pectoral girdle with the pectoral fin (LM 11; Figure 1) and an increase in the distance between the posterior-dorsal margin of the parietal bone and the joint between the skull and the vertebral column (landmarks 10, 11). ANOVA results also indicated that upper jaw lengths were significantly shorter in adult dob fish (F= 12.43; p=0.002).
The dorsally observable anatomical differences induced by the loss of fgf20a (Figure1) included a widening of the anterior edge of the prefrontal region (LM 1,3), reduction of the prefrontal region via a relatively strong anterior shift of its posterior edge (LM 4-6), posterior-lateral movement of the anterior edges of the parietal bones (LM 7,9), an anterior shift of the anterio-medial edges of the parietal bones along the sagittal suture (LM 8), and a weak anterior shift of the posterior edge of the parietal bones (LM 11-13). The dorsal surface of ABWT fish skulls underwent more extensive anatomical changes between the 1 cm and adult stages than did the lateral surfaces and these changes were strongly associated with skull elongation (Figure 1).
Loss of fgf20a signaling induced significant changes in the shapes of the maxillae and premaxillae of dob fish (Table 1). These bones lie within a region where fgf20a expression is normally high in post-larval zebrafish (Figure 2). In the maxillae, the loss of fgf20a induced a shortening and contortion of the premaxillad wing (upper arrow, panel E, Figure 1), a pronounced posterior-medial expansion of articular head (dotted line, panel E, Figure 1), and a reduction and contortion of the shank (lower arrow, panel E, Figure 1). In the premaxillae, loss of fgf20a signaling induced a reduction in the anterior and posterior curvature (dotted lines, panel H, Figure 1) of the dentigerous arm (the anatomical terms employed here were developed for toothed fishes), a pronounced reduction of the area of bone in the angle between the dentigerous and ascending arms (arrows, panel H, Figure 1) and irregularity in the shape of the normally straight medial edge of the ascending arm (large arrow, panels G and H, Figure 1). In ABWT fish the medial edges of the ascending arms of the left and right premaxillae were joined along their entire anterior-posterior length by a suture, but in the dob fish this connection often had an incomplete posterior extension.
Scale and lateral line defects
In dob animals, scales were malformed with irregular and often fissured posterior edges (Figure 5E). These results are consistent with the silencing of FGF20 resulting in the loss of scales, spurs and feathers in the chicken (Wells et al. 2012). Cranial lateral line canals were often incompletely formed in dob animals (Figure 5A-C). In zebrafish, cranial lateral line canal development commences at post-embryonic stages (i.e., > 21dpf), and involves bony walls extending upward from the underlying bone and fusing above the neuromast (Webb and Shirey 2003). Fusion of bone extends away from neuromasts resulting in the formation of hollow canals, with openings/pores between each neuromast (Webb and Shirey 2003). Bony walls are clearly seen growing up from the underlying bone in dob fish, but growth is either significantly delayed or arrested before these lips of bone fuse (e.g., preorbital canals, Figure 5A-C). These qualitative observations are consistent with the quantitative shape data presented above, and suggest that dob mutants suffer from aberrant bone growth.
Figure 5.
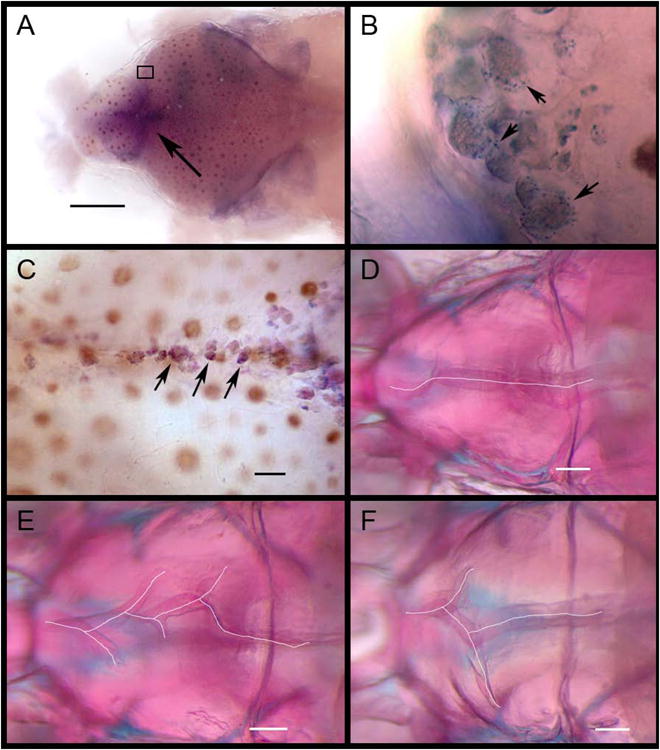
Labeling of ABWT fgf20a expression and cranial suture morphology in ABWT and dob zebrafish. A. Dorsal view of labeled fgf20a expression in the calvaria of a ABWT fish. Arrow indicates the frontal fontanel. Scale bar = 500 μm. B. Magnified view of the boxed area in A showing labeled fgf20a expression in putative osteoblasts (some indicated by arrows) along the anterior growing edge of the right frontal bone. C. Labeled fgf20a expression in putative osteoblasts within the developing interfrontal suture of a ABWT fish. Some indicated by arrows. Scale bar = 100 μm. D. Normal interfrontal suture anatomy in a ABWT zebrafish. E and F. Aberrant branched suturing in dob zebrafish. Scale bar = 200 μm in D-F.
Expression of fgf20a and suture defects
The shape changes associated with fgf20a deficiency were located in anatomical regions that matched those in which fgf20a expression were detected via WISH labeling in ABWT fish (Figure 2). These include the maxilla and premaxilla (downward pointing arrow in panel A, Figure 1; all arrows in panel B, Figure 1) and the parasphenoid bone (upward pointing arrow in panel A, Figure 2). These bones should receive a considerable portion of the forces exerted by the jaw adductor muscles when the upper and lower jaws occlude during biting (Cooper et al. 2011), and are therefore likely to experience substantial remodeling throughout life (Bentolila et al. 1998, Bonewald 2011). The expression of fgf20a was also detected in groups of cells along the edges of calvarial sutures and dob mutants exhibited malformations of these sutures (Figure 6). The interfrontal sutures of ABWT zebrafish were intact and relatively straight with respect the midline (Figure 6D), whereas mutants possessed conspicuously branched sutures (Figure 6E-F). This defect was variable in expression, but was limited to the interfrontal suture.
Figure 6.
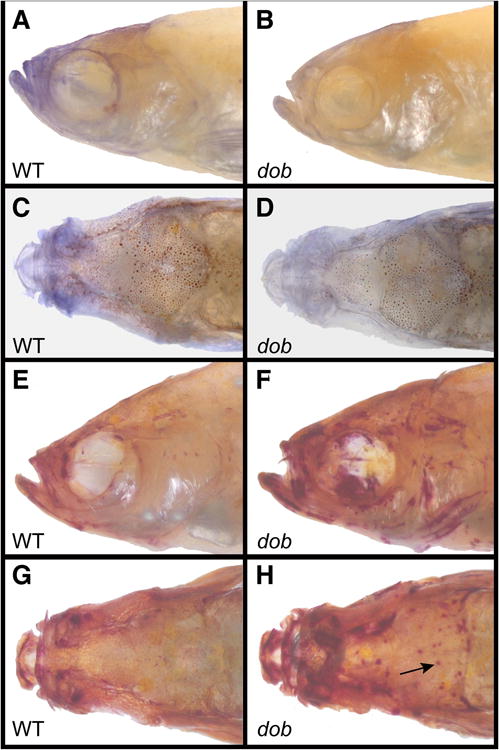
Osteoblast and osteoclast activity in representative ABWT fish (left column) and dob fish (right column). A. Lateral view of ABWT osteoblast activity labeled with alkaline phosphatase (AP). B. Lateral view of osteoblast activity labeled with AP in a dob fish. C. Dorsal view of ABWT osteoblast activity labeled with AP. D. Dorsal view of osteoblast activity labeled with AP in a dob fish. E. Lateral view of ABWT osteoclast activity labeled with tartrate resistant acid phosphatase (TRAP). F. Lateral view of osteoclast activity labeled with TRAP in a dob fish. G. Dorsal view of ABWT osteoclast activity labeled with TRAP. H. Dorsal view of osteoclast activity labeled with TRAP in a dob fish. Arrow indicates a possible bone absorption pit.
Aberrant bone metabolism in dob/fgf20a mutants
Adult dob and ABWT zebrafish exhibited significant differences in osteoblast and osteoclast activity (Table 3, Figure 7). Osteoblast activity was higher in ABWT fish, while osteoclast activity was higher in dob fish. Those anatomical regions that showed different osteoblast and osteoclast activity in mutant and ABWT fish roughly corresponded to regions that exhibited normal fgf20a expression in ABWT fish (e.g., maxillae, premaxillae and parasphenoid bones) and localized craniofacial shape distortions in dob fish, with differences in AP staining concentrated in the mid-facial region (Figure 7A-D), and differences in TRAP staining showing some localization to the mid-facial area and along the calvarial sutures (Table 3, Figure 7E-H). The dob specimens also frequently exhibited a large number of localized points of TRAP expression on their calveria (arrow, Figure 6H), which may represent absorption pits in which there is a high level of osteoclast activity.
Table 3. ANOVA results for AP and TRAP comparisons.
| ABWT mean | dob mean | F-ratio | p-value | |
|---|---|---|---|---|
| AP staining (lateral) | 0.468 | 0.278 | 11.890 | 0.001* |
| AP staining (dorsal) | 0.187 | 0.121 | 4.469 | 0.040* |
|
| ||||
| TRAP staining (lateral) | 0.146 | 0.378 | 34.312 | <0.001* |
| TRAP staining (dorsal) | 0.171 | 0.358 | 14.858 | <0.001* |
All comparisons were statistically significant at α=0.05
Figure 7.
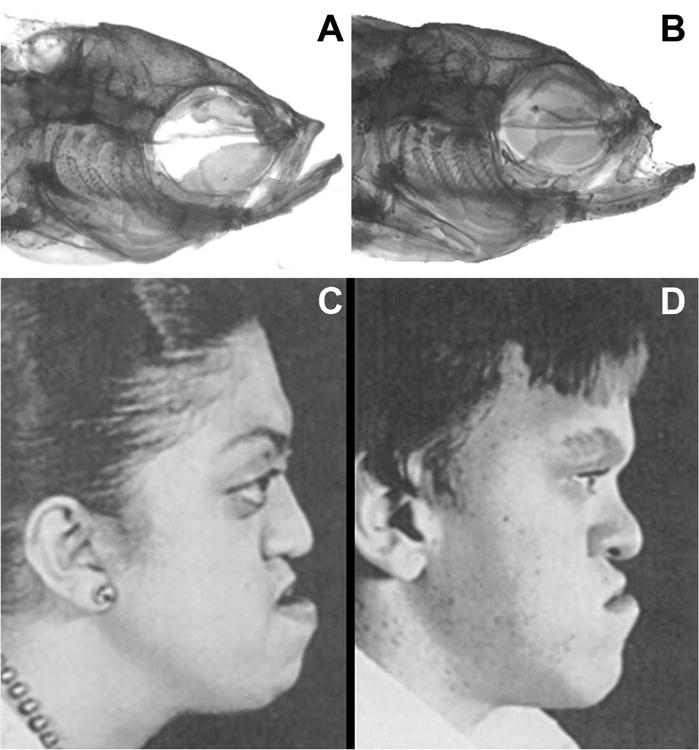
A. Normal head morphology in a ABWT zebrafish. B. Aberrant head morphology in a dob zebrafish with midfacial hypoplasia (arrow) similar to that seen in C and D. C. Adolescent female with Crouzon's syndrome. D. Adolescent male with Apert's syndrome. The images in C and D are from Plastic Surgery: Pediatric Plastic Surgery, 1st Edition, Volume 4, pp. 3022 and 3044, with permission (McCarthy 1990).
Discussion
Skulls are remodeled throughout life (Maes and Kronenberg 2011), and mutations in the genes that direct this process can result in cranial anatomies that depart from the ancestral form. Significant deviations from the normal anatomical variation present within a species are frequently deleterious, and are therefore described as diseases, but on those occasions when new morphologies confer selective advantages they can promote the process of evolutionary divergence. Searching for the genetic underpinnings of adaptive diversification has typically involved genetic mapping studies directed at finding the genes associated with specific phenotypes. We suggest that sufficient data has been accumulated from laboratory mutant strains to allow the matching of mutational effects to specific adaptive changes. Such data, especially when combined with genetic mapping, can facilitate the search for genes and/or signaling transduction pathways of adaptive importance in natural radiations.
Studying variation in anatomical form has been approached from multiple directions and prominent among them are investigations focused on the etiology of genetic disease and studies of the genetic factors that underlie adaptive diversification. Notably, however, recent efforts to link genotype and phenotype have yielded results that illuminate both of these fields, and the use of “evolutionary mutant model” species shows great potential for augmenting medically oriented studies of model organisms when naturally evolved diversity arises in characters associated with human disease (Albertson et al. 2009, Detrich and Amemiya 2010). As discussed below, insights gleaned from developmental investigations in the zebrafish dob mutant illustrate these points.
The zebrafish dob/fgf20a mutant as a model for post-embryonic craniofacial development and disease
The evidence presented here indicates that fgf20a is important for normal mid-facial and calvarial growth and homeostasis in zebrafish, and that a key aspect of its influence on skull morphogenesis is the regulation of osteoblast and osteoclast activity during craniofacial remodeling. Notably, the skull shape differences that significantly distinguished dob and ABWT zebrafish skulls correspond to known skull deformations in humans with syndromic craniosynostoses, and particularly resemble those present in some of the most common syndromes where midfacial hypoplasia is present (Figure 8). While the manifestation of craniosynostotic syndromes is often associated with gain-of-function mutations (Morriss-Kay and Wilkie, 2005), the cellular response to such mutations may still effect bone metabolism, which could explain the similarity of effects. Examining similar gain-of-function mutations in zebrafish Fgfr's during skull development would be a fruitful line of further inquiry. Moreover, the phenotypic divergence connected with craniosynostotic disorders can be highly variable (Ito et al. 2005), and the full list of genes whose dysfunction can produce or contribute to craniosynostosis is as yet incomplete (Morriss-Kay and Wilkie 2005). We therefore suggest that FGF20 may be a promising candidate for further study in regard to human cranial disease.
Figure 8.
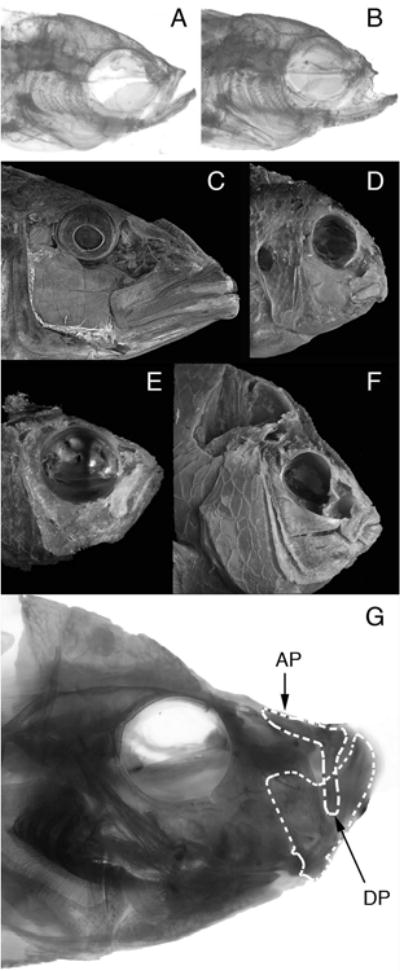
Comparative differences in the functional morphology of fish feed. A. Adult ABWT zebrafish. B. Adult dob zebrafish. C. Tyrannochromis macrostoma, a highly piscivorous cichlid from Lake Malawi in Eastern Africa. This fish uses large jaws and fast bite speeds to feed in the water column (pelagic feeding). D. Labeotropheus fuelleborni, a highly herbivorous cichlid from Lake Malawi in Eastern Africa. This fish uses small jaws and strong bites to scrape tough algae from rocks (benthic feeding). C and D depict fishes that represent the extremes of skull morphology that have evolved among the Lake Malawi cichlids. The anatomical differences that differentiate these skulls define the largest aspect of skull shape variation that has evolved among these fishes. E. Teixeirichthys jordani, a highly planktivorous (pelagic feeding) damselfish from the Indo-West Pacific. F. Hypsypops rubicundus, a hard-biting, benthic-feeding damselfish from the Eastern Pacific. E and F depict fishes that represent the extremes of skull morphology that have evolved among the damselfishes. The anatomical differences that differentiate these skulls define the largest aspect of skull shape variation that has evolved in this lineage. G. Caprichromis orthognathus, a specialized predator of the eggs and fry of mouthbrooding cichlids endemic to Lake Malawi. This fish has a highly reduced upper jaw relative to the lower jaw. Note the similarity of the reduced upper jaws in C. orthognathus and the dob zebrafish (panel B). AP – Ascending process of the premaxillary bone in the upper jaw. DP – dentigerous process of the premaxillary bone in the upper jaw. The dentigerous process articulates directly with the lower jaw via a ligamentous connection.
Suture malformations in dob mutants were similar to those observed in aceti282a/+/fgf8a deficient zebrafish in that both affected the midline sutures (Albertson and Yelick 2007). The specific defects in dob zebrafish were, however, slightly different from those in aceti282a/+ animals. In dob mutants the sagittal sutures were broken and branched, whereas in fgf8 haploinsufficent zebrafish these sutures were curved and asymmetrically positioned with respect to the midline, but they were intact (Albertson and Yelick, 2007). This difference may indicate slightly different roles for fgf8 and fgf20a in suture development. Another explanation is that, since sutures could only be examined in aceti282a heterozygous animals, differences in suture defects reflect a dosage effect. Alternatively, it is possible that both suture “defects” are a secondary response to defective skull geometries, and have nothing to do with suture formation per se.
The expression of fgf20a was observed in anatomical regions that are affected by the loss of fgf20a function (Figures 1, 2, 6 and 7), which is consistent with a primary role for this molecule in regulating the development of these traits. Expression was seen along cranial sutures (Figure 6 A,C), within putative osteoblasts along the growing edge of the calvarial bones (Figure 6 A,B), and around the anterior skull and upper jaw elements of ABWT zebrafish (Figure 2 A,B). The zebrafish upper jaw and anterior skull is a functionally dynamic region that likely undergoes continuous remodeling (Albertson and Yelick 2007). The results of the AP and TRAP assays support the assertion that fgf20a is necessary for normal skull remodeling in both this region and the calvaria (Table 3, Figure 7). The FGF20 ligand is known to bind to the receptor protein FGFR2c (Katoh 2009), and since mutation of FGFR2 has been associated with osteoblast dysfunction in Apert craniosynostosis (Miraoui et al. 2010), our data are consistent with the hypothesis that FGF20 mutations may contribute to (i.e., modify) certain craniosynostotic syndromes.
The importance of anatomical quantification in developmental studies
Morphometric techniques aid in the visualization, description and depiction of morphogenic effects. We suggest that the quantification of anatomical form can augment any developmental study of morphogenesis in which there is variation in the trait of interest, and that the application of these analytical methods will prove especially useful in analyses of complex phenotypes (e.g., the skull). The integration of quantitative methods and experimental embryology is becoming an increasingly important tool for developmental biologists (Albertson and Yelick 2004, 2007, Cooper and Albertson 2008, Oates et al. 2009, Goody et al. 2010, Parsons et al. 2010, Young et al. 2010, Sanger et al. 2011), and this approach has particular utility when we seek to better understand the etiology of human diseases with variable phenotypes.
Fgf20a as a candidate gene for factors underlying major trends in fish skull evolution
Evolutionary changes in jaw size have been identified as a major factor in the diversification of fish feeding morphology (see Figure 8; Schluter and McPhail 1992, Robinson et al. 1993, Lu and Bernatchez 1999, Rundle et al. 2000, Kristjansson et al. 2002, Gillespie and Fox 2003, Wainwright et al. 2004, Wund et al. 2008, Cooper and Westneat 2009, Cooper et al. 2010), and variation in this trait is strongly associated with divergence along a bentho-pelagic ecological axis, such that fishes with larger jaws tend to feed higher in the water column using rapid bites that are relatively weak (e.g., piscivores, planktivores), while those with smaller jaws tend to feed on benthic organisms using stronger bites (e.g., herbivores and durophagous fishes; Cooper and Westneat 2009, Cooper et al. 2010). Variation in relative jaw size is the most extensive aspect of skull evolution that has arisen over millions of years in such fishes as the damselfishes (386 currently described species; Cooper and Westneat 2009) and several highly successful lineages of East-African rift-lake cichlids (>1,500 species; Cooper et al. 2010), while rapid and much more recent divergence in jaw size is also well documented in many distantly related fish clades (Malmquist 1992, Walker 1997, Adams et al. 1998, Fraser et al. 1998, Ruzzante et al. 1998, Walker and Bell 2000, Kristjansson et al. 2002, Robinson and Parsons 2002, Ostbye et al. 2005). Bone remodeling is a likely mechanism that has promoted/facilitated these divergences, especially at lower taxonomic levels.
The identification of the genetic factors that determine jaw length is clearly of importance for evolutionary-developmental studies of adaptive diversification in fishes. In the two examples of extensive adaptive divergence in jaw size mentioned above (damselfishes and cichlids; Figure 9), coordinated changes in both upper and lower jaw size exemplified the most important aspects of skull evolution (Cooper and Westneat 2009, Cooper et al. 2010). Mutations that produce changes in upper jaw size alone are therefore only part of this divergence, but Fgf/Fgfr signaling has been determined to be a covariance-generating process in skull development that can modulate morphological integration (Martinez-Abadias et al. 2011). While the integrated upper and lower jaws and their supporting structures were identified as a potential anatomical, biomechanical and evolutionary module that has influenced East African cichlid divergence (Cooper et al. 2010), there is also evidence that the upper jaws constitute an integrated sub-module within this larger structure among cichlids (Parsons et al., 2011), which indicates that certain genetic changes can allow the upper and lower jaws to evolve independently.
Reduced (hypoplastic) upper jaws appear convergently in multiple cichlid fishes from Lake Malawi in East Africa (Figure 9, panel G), many piranhas and the closely related pacus (Serrasalminae), and the “halfbeaks” (Hemiramphidae), which exhibit the most extreme upper jaw reduction, just to name a few examples. The opposing trend is best exemplified by the billfishes (Xiphioidei), all of which possess a dramatically extended upper jaws that protrude far beyond the lower jaw (Collette et al. 2006). These trends are consistent with the idea that while the upper and lower jaws represent an integrated unit among teleosts, a further degree of modularity must also exist such that certain developmental pathways influence each character complex alone. Fgf20a offers an inroad into the signaling pathways that modulate the size of the upper jaw complex and might be an important target for future evolutionary genetic studies. Whether the phenotypic manifestation of silencing Fgf20a/Fgfr signaling in fishes is more properly described as a disruption of the integration between upper and lower jaw development, or strictly as an alteration of upper jaw development alone requires further investigation.
Conclusions
That divergence in jaw size has been a major factor in vertebrate evolution is made clear by even a cursory examination of cranial diversity in almost any vertebrate clade. Whether comparing the morphology of a sparrow with a heron, a bluegill sunfish with a swordfish, a giant anteater with a sloth, or an adult human with an adult chimpanzee (or Homo sapiens with some of the extinct prognathous hominids), changes in the size of the jaws relative to the rest of the skull have been an important part of adaptive divergence. Variation in jaw prominence is also part of the normal quantitative cranial variation currently exhibited by humans, such that multiple lineages have different means for this trait, while each lineage also displays a relatively large degree of jaw size variation (Hanihara 2000). That such a variable character would also exist in debilitating extreme forms that are classified as diseases is not entirely surprising, and studies of the genetic factors that contribute to the phenotypic variation exhibited by relative jaw size may have strong importance for medical studies of micrognathism, debilitating proganthism, and some of the jaw and facial hypoplasias that frequently accompany many craniosynostotic disorders.
Depending on the organism and the environment in which they manifest, phenotypic changes will be either adaptive, mal-adaptive, or of no consequence to selection. In different species, or even populations, the same genetic mutation may therefore either cause a disease or promote adaptive divergence, and it is becoming increasingly recognized that there is an inherent link between evo-devo studies of adaptive radiation and genetic developmental studies of disease. The genotype to phenotype connection of interest is frequently the same, while the positive or negative nature of the mutational consequences may be entirely a product of circumstance. Studies of developmental variation in model organisms focus on the genetic factors that produce phenotypic variation within populations maintained as laboratory lines, while evolutionary developmental studies focus on the genetic factors that promote adaptive divergence in natural populations. Cross-fertilization of knowledge between the multiple, rapidly-expanding disciplines concerned with understanding developmental variation are therefore likely to allow us to understand macroevolutionary patterns in terms of microevolutionary shifts, while simultaneously enhancing our understanding of the genetics of disease in individual species, such as humans.
Figure 4.
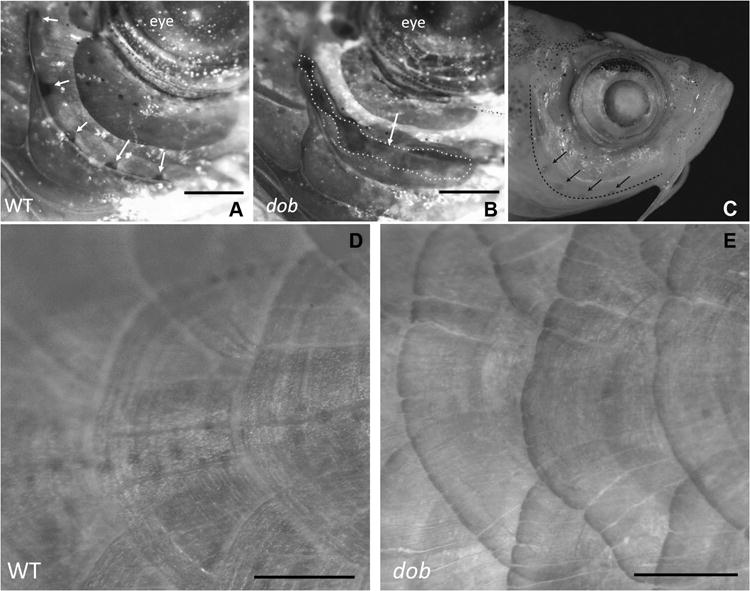
The effect of the loss of fgf20a expression on zebrafish mineralized tissue anatomy. A. Lateral view of a ABWT specimen with normal lateral line formation in the preopercle bone. Lateral line pores are indicated by arrows. B. Lateral line canals are incompletely fused in dob fish, resulting in gaping clefts in the canals (dotted line/arrow). For A and B dorsal is up and anterior is to the right. C. ABWT zebrafish head showing the edge of the preopercle bone (dashed line) and lateral line pores in this bone (arrows). D. Normal body scales in a ABWT fish. E. Distorted body scales with irregular posterior margins in a dob fish. The scales depicted in D and E are located along the posterior flank of the fish. Dorsal is up and anterior is to the right. Scale bars equal 500μm.
Acknowledgments
We would like to thank an anonymous reviewer for their highly detailed suggestions for improving our analytical approach, we thank Miriam Zelditch for assistance with shape analyses, and we thank Sarah Collins and Julie Adams for technical and husbandry assistance. This work was supported by grant R21DE019223 (NIH/NIDCR) awarded to R.C.A. and by start-up funding provided by WSU to W.J.C.
References
- Adams CE, Fraser D, Huntingford FA, Greer RB, Askew CM, Walker AF. Trophic polymorphism amongst Arctic charr from Loch Rannoch, Scotland. Journal of Fish Biology. 1998;52:1259–1271. [Google Scholar]
- Adams DC, Otárola-Castillo E. geomorph: an r package for the collection and analysis of geometric morphometric shape data. Methods in Ecology and Evolution. 2013;4:393–399. [Google Scholar]
- Albertson RC, Cresko W, Detrich HW, Postlethwait JH. Evolutionary mutant models for human disease. Trends in Genetics. 2009;25:74–81. doi: 10.1016/j.tig.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Yelick PC. Zebrafish: 2nd Edition Cellular and Developmental Biology. Elsevier Academic Press Inc; San Diego: 2004. Morphogenesis of the jaw: Development beyond the embryo; pp. 437–454. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Yelick PC. Fgf8 haploinsufficiency results in distinct cranlofacial defects in adult zebrafish. Developmental Biology. 2007;306:505–515. doi: 10.1016/j.ydbio.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel CDN, Witte F, van Oijen MJP. The shape of the skeletal elements in the head of a generalized Haplochromis species: H. elegans Trewavas 1933 (Pisces: Cichlidae) Netherlands Journal of Zoology. 1976;26:163–265. [Google Scholar]
- Bastir M, Rosas A, O'Higgins P. Craniofacial levels and the morphological maturation of the human skull. Journal of Anatomy. 2006;209:637–654. doi: 10.1111/j.1469-7580.2006.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nature Reviews Drug Discovery. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood DR, Wainwright PC, Fulton CJ, Hoey AS. Functional versatility supports coral reef biodiversity. Proceedings of the Royal Society B-Biological Sciences. 2006;273:101–107. doi: 10.1098/rspb.2005.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila V, Boyce TM, Fyhrie DP, Drumb R, Skerry TM, Schaffler MB. Intracortical remodeling in adult rat long bones after fatigue loading. Bone. 1998;23:275–281. doi: 10.1016/s8756-3282(98)00104-5. [DOI] [PubMed] [Google Scholar]
- Bonewald LF. The Amazing Osteocyte Journal of Bone and Mineral Research. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie AG. On the growth pattern of the human head. From the third month to the eighth year of life. American Journal of Anatomy. 1941;68:209–262. [Google Scholar]
- Carroll AM, Wainwright PC, Huskey SH, Collar DC, Turingan RG. Morphology predicts suction feeding performance in centrarchid fishes. Journal of Experimental Biology. 2004;207:3873–3881. doi: 10.1242/jeb.01227. [DOI] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. Embo Journal. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Wang ZS, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1 - positive cardiac progenitor cells through regulation of FGF signaling. Journal of Clinical Investigation. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette BB, McDowell JR, Graves JE. Phylogeny of recent billfishes (Xiphioidei) Bulletin of Marine Science. 2006;79:455–468. [Google Scholar]
- Cooper WJ, Albertson RC. Quantification and variation in experimental studies of morphogenesis. Developmental Biology. 2008;321:295–302. doi: 10.1016/j.ydbio.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Cooper WJ, Parsons K, McIntyre A, Kern B, McGee-Moore A, Albertson RC. Bentho-Pelagic Divergence of Cichlid Feeding Architecture Was Prodigious and Consistent during Multiple Adaptive Radiations within African Rift-Lakes. PLoS ONE. 2010;5:A38–A50. doi: 10.1371/journal.pone.0009551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Wernle J, Mann KA, Albertson RC. Functional and Genetic Integration in the Skulls of Lake Malawi Cichlids. Evolutionary Biology. 2011;38:316–334. [Google Scholar]
- Cooper WJ, Westneat MW. Form and function of damselfish skulls: rapid and repeated evolution into a limited number of trophic niches. BMC Evolutionary Biology. 2009;9 doi: 10.1186/1471-2148-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CM, Quarto N, Warren SM, Salim A, Longaker MT. Age-related changes in the biomolecular mechanisms of calvarial osteoblast biology affect fibroblast growth factor-2 signaling and osteogenesis. Journal of Biological Chemistry. 2003;278:45040–45040. doi: 10.1074/jbc.M304698200. vol 278, pg 32005, 2003. [DOI] [PubMed] [Google Scholar]
- Degnin CR, Laederich MB, Horton WA. FGFs in Endochondral Skeletal Development. Journal of Cellular Biochemistry. 2010;110:1046–1057. doi: 10.1002/jcb.22629. [DOI] [PubMed] [Google Scholar]
- Denoel M, Whiteman HH, Wissinger SA. Temporal shift of diet in alternative cannibalistic morphs of the tiger salamander. Biological Journal of the Linnean Society. 2006;89:373–382. [Google Scholar]
- Detrich HW, Amemiya CT. Antarctic Notothenioid Fishes: Genomic Resources and Strategies for Analyzing an Adaptive Radiation. Integrative and Comparative Biology. 2010;50:1009–1017. doi: 10.1093/icb/icq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizondo MR, MacDonald EL, Parichy DM. Dwarf zebrafish, altered bone development, and kidney stone formation in trpm7 mutants. Developmental Biology. 2005;295:322–323. [Google Scholar]
- Erickson GM, Lappin AK, Vliet KA. The ontogeny of bite-force performance in American alligator (Alligator mississippiensis) Journal of Zoology. 2003;260:317–327. [Google Scholar]
- Fan HH, Vitharana SN, Chen T, O'Keefe D, Middaugh CR. Effects of pH and polyanions on the thermal stability of fibroblast growth factor 20. Molecular Pharmaceutics. 2007;4:232–240. doi: 10.1021/mp060097h. [DOI] [PubMed] [Google Scholar]
- Ferry-Graham LA, Bolnick DI, Wainwright PC. Using functional morphology to examine the ecology and evolution of specialization. Integrative and Comparative Biology. 2002;42:265–277. doi: 10.1093/icb/42.2.265. [DOI] [PubMed] [Google Scholar]
- Foote M. Contributions of Individual Taxa to Overall Morphological Disparity. Paleobiology. 1993;19:403–419. [Google Scholar]
- Fraser D, Adams CE, Huntingford FA. Trophic polymorphism among Arctic charr Salvelinus alpinus L, from Loch Ericht, Scotland. Ecology of Freshwater Fish. 1998;7:184–191. [Google Scholar]
- Gillespie GJ, Fox MG. Morphological and life-history differentiation between littoral and pelagic forms of pumpkinseed. Journal of Fish Biology. 2003;62:1099–1115. [Google Scholar]
- Gonzalez-Jose RG, Ramirez-Rozzi F, Sardi M, Martinez-Abadias N, Hernandez M, Pucciarelli HM. Functional-cranial approach to the influence of economic strategy on skull morphology. American Journal of Physical Anthropology. 2005;128:757–771. doi: 10.1002/ajpa.20161. [DOI] [PubMed] [Google Scholar]
- Goody MF, Kelly MW, Lessard KN, Khalil A, Henry CA. Nrk2b-mediated NAD+ production regulates cell adhesion and is required for muscle morphogenesis in vivo: Nrk2b and NAD+ in muscle morphogenesis. Developmental Biology. 2010;344:809–826. doi: 10.1016/j.ydbio.2010.05.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Milne N, Wroe S. Biting through constraints: cranial morphology, disparity and convergence across living and fossil carnivorous mammals. Proceedings of the Royal Society B: Biological Sciences. 2011;278:1831–1839. doi: 10.1098/rspb.2010.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidakis DJ, Androulakis II. Bone Remodeling. Annals of the New York Academy of Sciences. 2006;1092:385–396. doi: 10.1196/annals.1365.035. [DOI] [PubMed] [Google Scholar]
- Hajihosseini MK, Heath JK. Expression patterns of fibroblast growth factors-18 and-20 in mouse embryos is suggestive of novel roles in calvarial and limb development. Mechanisms of Development. 2002;113:79–83. doi: 10.1016/s0925-4773(01)00656-6. [DOI] [PubMed] [Google Scholar]
- Hajihosseini MK, Lalioti MD, Arthaud S, Burgar HR, Brown JM, Twigg SRF, Wilkie AOM, Heath JK. Skeletal development is regulated by fibroblast growth factor receptor 1 signalling dynamics. Development. 2004;131:325–335. doi: 10.1242/dev.00940. [DOI] [PubMed] [Google Scholar]
- Hamilton F. An account of the fishes found in the River Ganges and its branches. Archibald Constable; Edinburgh: 1822. [Google Scholar]
- Hanihara T. Frontal and facial flatness of major human populations. American Journal of Physical Anthropology. 2000;111:105–134. doi: 10.1002/(SICI)1096-8644(200001)111:1<105::AID-AJPA7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hellig CJ, Kerschbaumer M, Sefc KM, Koblmuller S. Allometric shape change of the lower pharyngeal jaw correlates with a dietary shift to piscivory in a cichlid fish. Naturwissenschaften. 2010;97:663–672. doi: 10.1007/s00114-010-0682-y. [DOI] [PubMed] [Google Scholar]
- Hernández LP, Barresi MJF, Devoto SH. Functional morphology and developmental biology of zebrafish: Reciprocal illumination from an unlikely couple. Integrative and Comparative Biology. 2002;42:222–231. doi: 10.1093/icb/42.2.222. [DOI] [PubMed] [Google Scholar]
- Hung IH, Yu K, Lavine KJ, Ornitz DM. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Developmental Biology. 2007;307:300–313. doi: 10.1016/j.ydbio.2007.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley MM, Tetradis S, Huang YF, Hock J, Kream BE, Raisz LG, Sabbieti MG. Parathyroid hormone regulates the expression of fibroblast growth factor-2 mRNA and fibroblast growth factor receptor mRNA in osteoblastic cells. Journal of Bone and Mineral Research. 1999;14:776–783. doi: 10.1359/jbmr.1999.14.5.776. [DOI] [PubMed] [Google Scholar]
- Iseki S, Wilkie AOM, Heath JK, Ishimaru T, Eto K, MorrissKay GM. Fgfr2 and osteopontin domains in the developing skull vault are mutually exclusive and can be altered by locally applied FGF2. Development. 1997;124:3375–3384. doi: 10.1242/dev.124.17.3375. [DOI] [PubMed] [Google Scholar]
- Ito S, Sekido K, Kanno H, Sato H, Tanaka M, Yamaguchi K, Yamamoto I. Phenotypic diversity in patients with craniosynostoses unrelated to Apert syndrome: the role of fibroblast growth factor receptor gene mutations. Journal of Neurosurgery. 2005;102:23–30. doi: 10.3171/ped.2005.102.1.0023. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends in Genetics. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Katoh M. Networking of WNT, FGF, notch, BMP, and hedgehog signaling pathways during carcinogenesis. Stem Cell Reviews. 2007;3:30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- Katoh M. FGFR2 Abnormalities Underlie a Spectrum of Bone, Skin, and Cancer Pathologies. J Invest Dermatol. 2009;129:1861–1867. doi: 10.1038/jid.2009.97. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Comparative genomics on FGF20 orthologs. Oncology Reports. 2005;14:287–290. [PubMed] [Google Scholar]
- Katoh M, Katoh M. FGF signaling network in the gastrointestinal tract (Review) International Journal of Oncology. 2006;29:163–168. [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Keynes RJ. Neural crest patterning and the evolution of the jaw. 2001a:105–120. doi: 10.1046/j.1469-7580.2001.19910105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Developmental Biology. 2001b;233:239–257. doi: 10.1006/dbio.2001.0201. [DOI] [PubMed] [Google Scholar]
- Konow N, Bellwood DR, Wainwright PC, Kerr AM. Evolution of novel jaw joints promote trophic diversity in coral reef fishes. Biological Journal of the Linnean Society. 2008;93:545–555. [Google Scholar]
- Kristjansson BK, Skulason S, Noakes DLG. Morphological segregation of Icelandic threespine stickleback (Gasterosteus aculeatus L) Biological Journal of the Linnean Society. 2002;76:247–257. [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang XQ, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Developmental Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Developmental Biology. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Xu JS, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes & Development. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu GQ, Bernatchez L. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): Support for the ecological speciation hypothesis. Evolution. 1999;53:1491–1505. doi: 10.1111/j.1558-5646.1999.tb05413.x. [DOI] [PubMed] [Google Scholar]
- Luczkovich JJ, Norton SF, Gilmore RG. The Influence Of Oral Anatomy On Prey Selection During The Ontogeny Of 2 Percoid Fishes, Lagodon-Rhomboides And Centropomus-Undecimalis. Environmental Biology of Fishes. 1995;44:79–95. [Google Scholar]
- Maes C, Kronenberg HM. Postnatal bone growth: growth plate biology, bone formation, and remodeling. In: Glorieux FH, Pettifor J, Juppner H, editors. Pediatric Bone: Biology & Diseases. Academic Press; Boston: 2011. p. 868. [Google Scholar]
- Mallarino R, Grant PR, Grant BR, Herrel A, Kuo WP, Abzhanov A. Two developmental modules establish 3D beak-shape variation in Darwin's finches. Proceedings of the National Academy of Sciences. 2011;108:4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmquist HJ. Phenotype-Specific Feeding-Behavior of 2 Arctic Charr Salvelinus-Alpinus Morphs. Oecologia. 1992;92:354–361. doi: 10.1007/BF00317461. [DOI] [PubMed] [Google Scholar]
- Martinez-Abadias N, Heuze Y, Wang YL, Jabs EW, Aldridge K, Richtsmeier JT. FGF/FGFR Signaling Coordinates Skull Development by Modulating Magnitude of Morphological Integration: Evidence from Apert Syndrome Mouse Models. PLoS ONE. 2011;6:9. doi: 10.1371/journal.pone.0026425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JG, editor. Plastic surgery. 1. W. B. Saunders; Philadelphia: 1990. [Google Scholar]
- Miraoui H, Ringe J, Häupl T, Marie PJ. Increased EFG- and PDGFα- receptor signaling by mutant FGF-receptor 2 contributes to osteoblast dysfunction in Apert craniosynostosis. Human Molecular Genetics. 2010;19:1678–1689. doi: 10.1093/hmg/ddq045. [DOI] [PubMed] [Google Scholar]
- Morriss-Kay GM, Wilkie AOM. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. Journal of Anatomy. 2005;207:637–653. doi: 10.1111/j.1469-7580.2005.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney MD, Gale LD. Ecomorphological relationships in ontogeny: Anatomy and diet in gag, Mycteroperca microlepis (Pisces: Serranidae) Copeia. 1996:167–180. [Google Scholar]
- Oates AC, Gorfinkiel N, Gonzalez-Gaitan M, Heisenberg CP. Quantitative approaches in developmental biology. Nat Rev Genet. 2009;10:517–530. doi: 10.1038/nrg2548. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes & Development. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostbye K, Naesje TF, Bernatchez L, Sandlund OT, Hindar K. Morphological divergence and origin of sympatric populations of European whitefish (Coregonus lavaretus L.) in Lake Femund, Norway. Journal of Evolutionary Biology. 2005;18:683–702. doi: 10.1111/j.1420-9101.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- Parsons K, Andreeva V, Cooper WJ, Yelick PC, Albertson RC. Morphogenesis of the zebrafish jaw: Development beyond the embryo. In: Westerfield M, Detrich HW, Zon LI, editors. Methods in Cell Biology: The Zebrafish. Elsevier Academic Press Inc; San Diego: 2010. [Google Scholar]
- Parsons KJ, Andreeva V, Cooper WJ, Yelick PC, Albertson RC. Morphogenesis of the Zebrafish Jaw: Development Beyond the Embryo. In: Detrich HW, Westerfield M, Zon LI, editors. Methods in Cell Biology, Vol 101: Zebrafish: Cellular and Developmental Biology, Pt B. Third. 2011. pp. 225–248. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Cooper WJ, Albertson RC. Limits of Principal Components Analysis for Producing a Common Trait Space: Implications for Inferring Selection, Contingency, and Chance in Evolution. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothoff T. Ontogeny and Systematics of Fishes. American Society of Ichthyologists and Herpetologists; Austin, TX: 1983. Clearing and staining technique. [Google Scholar]
- Quarto N, Longaker MT. The zebrafish (Danio rerio): a model system for cranial suture Patterning. Cells Tissues Organs. 2005;181:109–118. doi: 10.1159/000091100. [DOI] [PubMed] [Google Scholar]
- Rice AN, Cooper WJ, Westneat MW. Diversification of coordination patterns during feeding behaviour in cheiline wrasses. Biological Journal of the Linnean Society. 2008;93:289–308. [Google Scholar]
- Rice AN, Westneat MW. Coordination of feeding, locomotor and visual systems in parrottishes (Teleostei : Labridae) Journal of Experimental Biology. 2005;208:3503–3518. doi: 10.1242/jeb.01779. [DOI] [PubMed] [Google Scholar]
- Robinson BW, Parsons KJ. Changing times, spaces, and faces: tests and implications of adaptive morphological plasticity in the fishes of northern postglacial lakes. Canadian Journal of Fisheries and Aquatic Sciences. 2002;59:1819–1833. [Google Scholar]
- Robinson BW, Wilson DS, Margosian AS, Lotito PT. Ecological and Morphological-Differentiation of Pumpkinseed Sunfish in Lakes without Bluegill Sunfish. Evolutionary Ecology. 1993;7:451–464. [Google Scholar]
- Rohlf FJ. 2006 tpsDig2 http://life.bio.sunysb.edu/morph/%5D.
- Rosas A, Bastir M. Thin-plate spline analysis of allometry and sexual dimorphism in the human craniofacial complex. American Journal of Physical Anthropology. 2002;117:236–245. doi: 10.1002/ajpa.10023. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nagel L, Boughman JW, Schluter D. Natural selection and parallel speciation in sympatric sticklebacks. Science. 2000;287:306–308. doi: 10.1126/science.287.5451.306. [DOI] [PubMed] [Google Scholar]
- Ruzzante DE, Walde SJ, Cussac VE, Macchi PJ, Alonso MF. Trophic polymorphism, habitat and diet segregation in Percichthys trucha (Pisces : Percichthyidae) in the Andes. Biological Journal of the Linnean Society. 1998;65:191–214. [Google Scholar]
- Sabbieti MG, Marchetti L, Abreu C, Montero A, Hand AR, Raisz LG, Hurley MM. Prostaglandins regulate the expression of fibroblast growth factor-2 in bone. Endocrinology. 1999;140:434–444. doi: 10.1210/endo.140.1.6442. [DOI] [PubMed] [Google Scholar]
- Sanger TJ, Norgard EA, Pletscher LS, Bevilacqua M, Brooks VR, Sandell LJ, Cheverud JM. Developmental and genetic origins of murine long bone length variation. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2011;316B:146–161. doi: 10.1002/jez.b.21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi ML, Ventrice F, Rozzi FR. Allometries throughout the late prenatal and early postnatal human craniofacial ontogeny. Anatomical Record-Advances in Integrative Anatomy and Evolutionary Biology. 2007;290:1112–1120. doi: 10.1002/ar.20581. [DOI] [PubMed] [Google Scholar]
- Satake W, Mizuta I, Suzuki S, Nakabayashi Y, Ito C, Watanabe M, Takeda A, Hasegawa K, Sakoda S, Yamamoto M, Hattori N, Murata M, Toda T. Fibroblast growth factor 20 gene and Parkinson's disease in the Japanese population. Neuroreport. 2007;18:937–940. doi: 10.1097/WNR.0b013e328133265b. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Le Pabic P, Hoffman TL. Using transgenic zebrafish (Danio rerio) to study development of the craniofacial skeleton. Journal of Applied Ichthyology. 2010;26:183–186. [Google Scholar]
- Schluter D, McPhail JD. Ecological Character Displacement and Speciation in Sticklebacks. American Naturalist. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- Slemenda CW, Peacock M, Hui S, Zhou L, Johnston CC. Reduced Rates of Skeletal Remodeling Are Associated with Increased Bone Mineral Density During the Development of Peak Skeletal Mass. Journal of Bone and Mineral Research. 1997;12:676–682. doi: 10.1359/jbmr.1997.12.4.676. [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Martin TJ. Modulation of Osteoclast Differentiation. Endocrine Reviews. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- Tibbetts IR, Day RD, Carseldine L. Development of the pharyngeal dentition of two herbivorous halfbeaks (Teleostei : Hemiramphidae) and implications for the hemiramphid ontogenetic trophic shift. Marine and Freshwater Research. 2008;59:117–124. [Google Scholar]
- van der Walt JM, Noureddine MA, Kittappa R, Hauser MA, Scott WK, McKay R, Zhang FY, Stajich JM, Fujiwara K, Scott BL, Pericak-Vance MA, Vance JM, Martin ER. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. American Journal of Human Genetics. 2004;74:1121–1127. doi: 10.1086/421052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright PC, Bellwood DR, Westneat MW, Grubich JR, Hoey AS. A functional morphospace for the skull of labrid fishes: patterns of diversity in a complex biomechanical system. Biological Journal of the Linnean Society. 2004;82:1–25. [Google Scholar]
- Walker JA. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L (Gasterosteidae) body shape. Biological Journal of the Linnean Society. 1997;61:3–50. [Google Scholar]
- Walker JA, Bell MA. Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus) Journal of Zoology. 2000;252:293–302. [Google Scholar]
- Webb JF, Shirey JE. Postembryonic development of the cranial lateral line canals and neuromasts in zebrafish. Developmental Dynamics. 2003;228:370–385. doi: 10.1002/dvdy.10385. [DOI] [PubMed] [Google Scholar]
- Wells K, Hadad Y, Ben-Avraham D, Hillel J, Cahaner A, Headon D. Genome-wide SNP scan of pooled DNA reveals nonsense mutation in FGF20 in the scaleless line of featherless chickens. BMC Genomics. 2012;13:257. doi: 10.1186/1471-2164-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westneat MW. Phylogenetic Systematics and Biomechanics in Ecomorphology. Environmental Biology of Fishes. 1995;44:263–283. [Google Scholar]
- Westneat MW. A biomechanical model for analysis of muscle force, power output and lower jaw motion in fishes. Journal of Theoretical Biology. 2003;223:269–281. doi: 10.1016/s0022-5193(03)00058-4. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wund MA, Baker JA, Clancy B, Golub JL, Fosterk SA. A test of the “Flexible stem” model of evolution: Ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. American Naturalist. 2008;172:449–462. doi: 10.1086/590966. [DOI] [PubMed] [Google Scholar]
- Young NM, Chong HJ, Hu D, Hallgrímsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010;137:3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollikofer CPE, de Leon MSP. Visualizing patterns of craniofacial shape variation in Homo sapiens. Proceedings Of The Royal Society Of London Series B-Biological Sciences. 2002;269:801–807. doi: 10.1098/rspb.2002.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]


