Abstract
New technologies for DNA sequencing, coupled with advanced analytical approaches, are now providing unprecedented speed and precision in decoding human genomes. This combination of technology and analysis, when applied to the study of cancer genomes, is revealing specific and novel information about the fundamental genetic mechanisms that underlie cancer’s development and progression. This review outlines the history of the past several years of development in this realm, and discusses the current and future applications that will further elucidate cancer’s genomic causes.
Cancer is a Disease of the Genome
Theodor Boveri initially proposed in 1902 that a single cell with scrambled chromosomes and hence uncontrolled cell division was the origin of a cancerous tumor. This hypothesis was supported by the work of many biologists, culminating in the descriptions by Janet Rowley in the 1970’s[1–4]. Although controversial at the time she proposed it, her microscopic observations of leukemia chromosomes established a link between specific chromosomal translocations and different types of leukemia[5, 6]. As a result of these initial observations and many more that followed, it is entirely appropriate to describe cancer as a disease of the genome. In particular, there are not only somatic alterations that are unique to tumor cell genomes, ranging from point mutations to chromosomal translocations but also specific inherited or “germline” genomic alterations are known to confer increased susceptibility to cancer development. Since 2008, using new technologies for DNA sequencing, our ability to characterize the somatic alterations present in cancer genomes has been radically transformed, as these technologies provide a “microscope” with the highest resolution: the single nucleotide.
The aforementioned “next-generation” or “massively parallel” DNA sequencing technology is embodied in several different instrument platforms, all of which have been profiled in reviews [7, 8], and all of which have achieved remarkable advances in capacity, read length and accuracy since their initial introduction in the mid-2000’s. Our group was the first to utilize the Solexa technology (now Illumina) to sequence and analyze a complete tumor and normal genome from the same individual, an acute myeloid leukemia (AML) patient, in 2008 [9]. In this effort, we required the Human Genome Reference sequence as a template against which we aligned the 32 bp Solexa reads from the tumor and normal genomes separately. We first compared the variant calls to those obtained from a high density SNP array as a means of estimating the breadth and depth to which we had covered the genome. After this comparison, at around 28-fold coverage, we identified in excess of 3 million putative single nucleotide variants in both the tumor and normal genomes. By implementing a decision tree algorithm, a commonly implemented means to calculate conditional probabilities such as the probability of a sequence variant being somatic, we were able to identify 10 genes with point mutations or small insertion/deletion changes that were somatic, or unique to the tumor genome. This work established the basic approach to whole genome somatic mutation discovery, although the data and algorithmic approaches have changed over time, effectively broadening the comprehensiveness with which one can characterize the extent of genome alterations in cancer.
Our first effort in AML was strategic, in that leukemia cells derived from bone marrow biopsies are tumor-rich with few normal cells, and the M1 subtype we studied is characterized by diploid chromosomes (hence lack of aneuploidy and copy number alterations so common in solid tumors). It was also driven by the fact that the treatment of AML patients hadn’t changed dramatically in ~25 years, leaving the majority of patients with normal cytogenetics and hence in a so-called “intermediate risk” category (see Figure 1) that provided little to no information to them or to their oncologist regarding their potential outcome in the disease course. In this regard, our efforts to-date and those of others now have established three genes (IDH1, IDH2 and DNMT3A) that either alone or in combination with other frequently mutated genes, predict poor outcomes for those AML patients whose genomes contain the mutation [10–12]. Of these three, DNA methyltransferase 3A (DNMT3A), a de novo DNA methyltransferase, is mutated in ~34% of cytogenetically normal patients and predicts poor outcome when mutated[10, 13]. This prognostic correlation to poor outcome in the current clinical paradigm for cytogenetically normal de novo AML (e.g. induce to remission with chemotherapy and monitor for relapse) suggests that DNMT3A mutant AML patients should instead proceed directly to stem cell transplant upon achieving first remission. In addition to prognostic mutations, large-scale tumor sequencing efforts have identified new frequently mutated genes across multiple types of solid and liquid tumors. The decreasing cost of producing the next-generation sequencing data for whole genome coverage has now resulted in large multi-tumor studies that permit the genomic impact on cellular pathways to be evaluated across all types of somatic alterations [14–19].
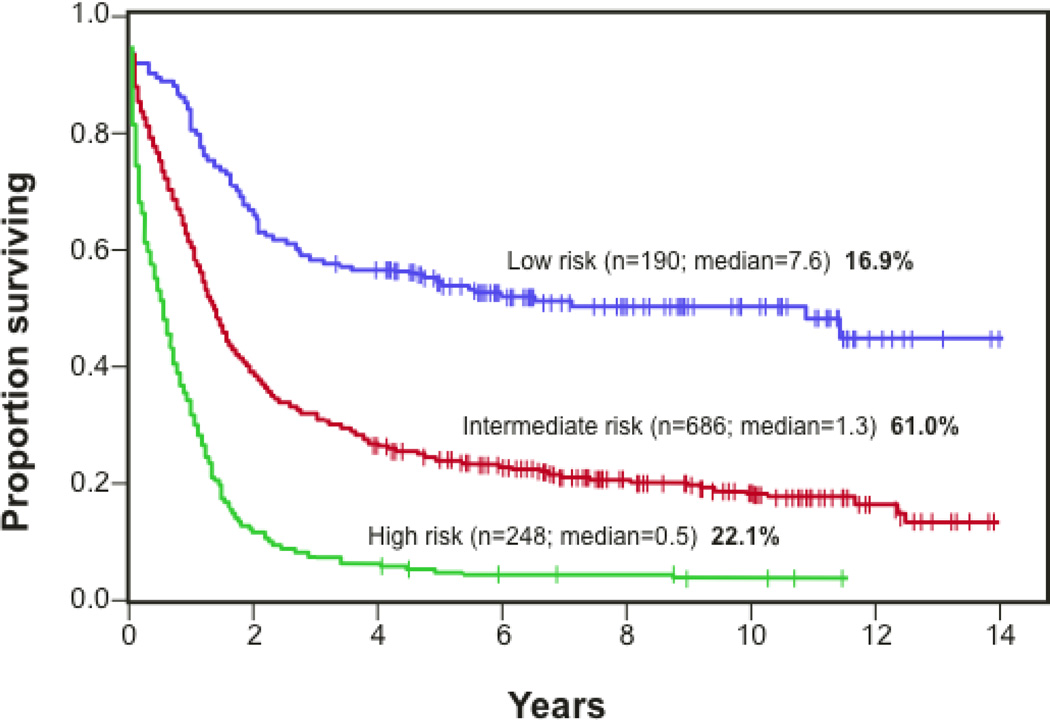
Figure 1.
A Kaplan-Meier curve illustrating the current cytogenetics and pathology-based stratification of acute myeloid leukemia patient risk. High risk cases are classified by complex cytogenetics that do not conform to known translocations, whereas low risk patients have straightforward cytogenetics that include known translocations (t15;17, t8;12, inv16) and for which well-defined therapeutics define the standard of care. Most patients are classified by normal/diploid cytogenetics as intermediate risk cases. Because the cytogenetic profiles of these patients are not prognostic for risk, genome sequencing has been pursued in an effort to identify genes that are frequently altered in the tumor and for which correlations to outcome can be made. (Figure from J.C. Byrd et al., Blood 2002. 100: 4325–36).
Genome sequencing in solid tumors provides several potential challenges, including the fact that any tumor section used for genomic DNA isolation will include normal cells such as stromal cells, blood vessels and immune cells, all of which contribute a normal genomic DNA signature to that provided by the tumor cells. Although sequencing to a sufficient coverage (defined as the –fold oversampling of the genome required to produce sufficient sequence read depth genome-wide for variant discovery) will permit somatic mutation discovery regardless of the tumor cellularity, most studies focus on tumors with >60% tumor nuclei present (based on conventional pathology estimates) so the sequencing coverage remains tractable from an economic standpoint. Genomic aneuploidy and large-scale amplification of chromosomes also impact the coverage calculation, since these regions contribute more DNA to the sequencing library than diploid or haploid regions and sequencing must compensate for this disparity until all regions are sufficiently covered by sequencing read data. Certain tumor types are more diffuse, such as pancreas or prostate, and require either block macro-dissection or laser capture microdissection (LCM) to enhance the tumor nuclei that contribute to the genomic DNA isolation. While this sounds ideal, the yield of genomic DNA from LCM is relatively low (<100 ng) and modified methods are required to generate whole genome libraries of sufficient complexity to represent the tumor genome. Another challenge is presented by the cellular heterogeneity displayed by many solid tumors, evident in differential immunohistochemistry staining and low-resolution genomic screens [20], indicating that not all genomes of all tumor cells are equivalent. A deep sampling of the collective tumor genomes in a DNA isolate by next-generation methods, coupled with advanced mathematical analysis of the data can provide a structure for modeling the tumor cell populations, their relative proportions, and their associated mutational profiles.
NGS Studies of Cancer Progression
This approach was published recently in a study designed to compare the tumor genomes of patients with de novo AML to their relapse genomes[21]. After sequencing each genome (de novo tumor and relapse tumor) and the matched normal from skin for each patient, somatic mutations and structural variants were identified. Some of these appeared to be unique to the relapse sample in each case. We then obtained high sequencing read depth at each somatic mutation site in the de novo and relapse tumors, and characterized the reads that contained the mutated base(s) at each site to calculate an allele frequency of that variant in the tumor cell population. Using kernel density estimation, we then identified groups of mutations present at the same allele frequencies, indicative of their prevalence in the tumor cell population. This comparison of allele frequency groups between de novo and relapse disease allowed us to model the relative numbers of tumor subclones at each disease presentation, and defined AML progression as a clonal process, as illustrated in Figure 2. Namely, all subclones originate from a founder clone that shares all but the newest mutations, and relapse disease shares mutations with the founder clone as well as new mutations that portend its proliferative advantage in the relapse presentation.
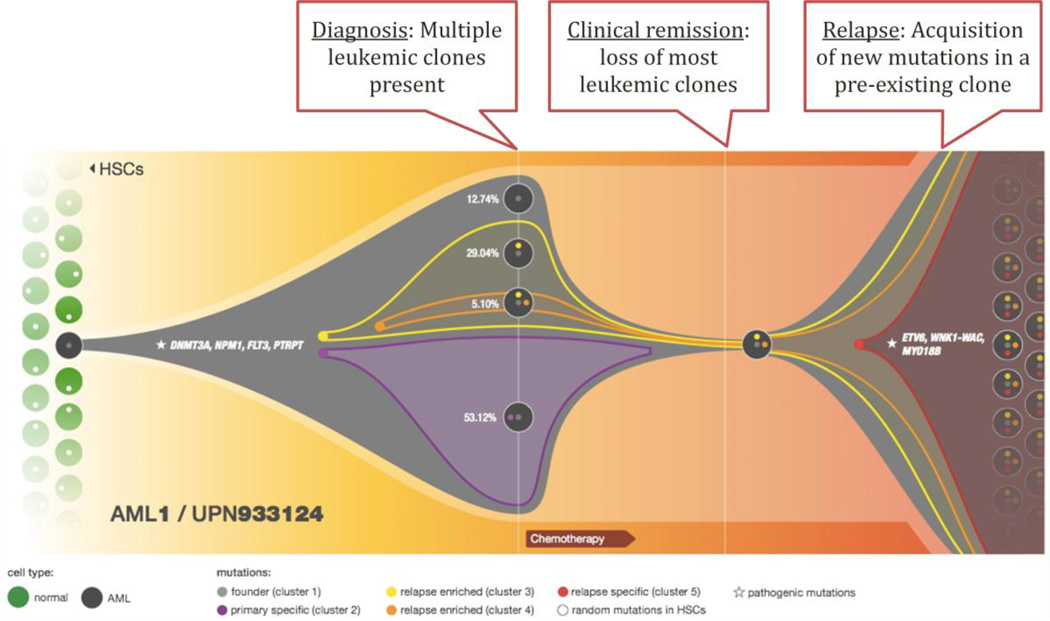
Figure 2.
Model of the clonal progression process that occurs between the initial (de novo) and relapse presentation in AML patients. At diagnosis, this patient has an oligoclonal disease characterized by four different subclones, each present at a specific proportion in the tumor cell population and with a specific mutational profile. Chemotherapy used to induce the patient into remission decreases clonal heterogeneity but a single subclone persists, acquires new mutations, and again proliferates in the bone marrow as a relapse-specific subclone.
In a similar study, with a slightly different experimental design, we recently explored the differences between myelodysplastic syndrome (MDS) genomes and the genomes found in those patients’ secondary AML (sAML) tumors. MDS identifies a heterogeneous group of syndromes characterized by dysplasia and ineffective hematopoesis. Since about 1/3 of these patients progress to sAML for reasons that are not well understood at the genomic level, we characterized these genomes to understand novel somatic variants in the sAML cells. In our study, the results were quite different than the de novo to relapse AML study outlined above. Namely, we found that the sAML genomes were all oligoclonal (comprised of several related tumor cell subclones, each with unique sets of mutations), each containing a pre-existing MDS founder clone that was out-competed in the sAML tumor cell population in some cases. We hypothesized that the oligoclonal nature of the sAML presentation may contribute to the very poor response rates of these patients to conventional chemotherapies that often induce remission in de novo AML treatment (Graubert et al., accepted for publication).
Akin to de novo leukemia and relapse is metastatic tumor occurrence in patients with a primary solid tumor presentation. Similarly, the question of genetic relatedness between primary tumor cells and metastatic tumor cells is of interest, although as before, solid tumors present challenges in that typically the metastatic tumor is not surgically removed and/or banked, once diagnosed. There are, however, exceptions and two published reports to-date have studied this genetic relatedness in primary breast tumors and subsequent metastases. The first study involved a patient with lobular breast cancer that was followed 9 years later by a recurrent tumor in the breast[22]. The second manuscript described a “trio” of tumors from one patient, including a primary basal-like ductal breast tumor, a brain metastasis that developed 8 months after the primary tumor was diagnosed, and a xenograft-propagated tumor derived from the primary tumor after its surgical removal [23]. Both studies established a genetic relatedness between the primary and the metastatic tumors, albeit one that becomes more distant with time between the primary and metastatic disease diagnoses. In the second example, the metastatic tumor appeared to be enriched for a specific subclone within the primary disease that was characterized by certain low allele frequency mutations in the primary tumor genome rising to much higher allele frequencies in the metastatic tumor genome. More studies of this type are needed to fully understand the potential for metastasis and the roles of specific mutations in the tendency for certain tumors to metastasize.
The use of different initial preparatory methods and post-sequencing computational data analyses has expanded the scope of cancer genomics inquiry to include expressed and non-coding RNA (“RNA-seq”), and DNA methylation (“methyl-seq”) comparisons of tumor and matched non-malignant tissues from the same patient. If anything, the wealth of genomic information that can be collected from each tumor case proves two things; our relatively primitive ability to integrate data from different “omes” and our inability to quickly characterize the impact of different types of genomic alterations on tumor biology. Nevertheless, these cataloguing efforts will undoubtedly be valuable when coupled with downstream efforts to investigate the impact of genomic alterations on protein and pathway function in cell-based systems. Data integration, similarly, provides a challenge for computational and systems biologists—and one set of efforts will inform the other, ultimately advancing our understanding of tumor biology.
NGS-Based Diagnosis in Cancer Care
As analytical abilities that interpret next-generation sequencing data using mathematical or statistical methods become more integrated, and are coupled with secondary validation assays that verify the predicted mutations or alterations correctly identified by the analysis, the remarkable pace of the cancer genomics discovery process already evidenced in just the past three years will continue. While these efforts are valuable and worthwhile, one ultimate goal is to improve patient care, including the precision of diagnosis. A clear ramification of this capability is the translation of next-generation sequencing to clinical diagnosis, especially as it relates to the identification of mutated genes that can be “targeted” using either small molecule inhibitors or specific antibodies. The first example of such an approach was published by a group in Vancouver at the British Columbia Genome Sequencing Centre, and entailed a patient with metastatic lung tumors who originally had presented with a papillary adenocarcinoma of the tongue[24]. Through a brilliant combination of genome and RNA sequencing, coupled with KEGG pathway analysis and DrugBase exploration of targeted therapies available to treat the variant genes they identified, the patient experienced a dramatic recovery with the drug Sunitinib™ that addressed a RET over-expression identified from RNA sequencing. After four months, a CT scan diagnosed disease recurrence and Sorafenib™ and Sulindac™, also indicated from the initial genomic analysis, replaced the Sunitinib™ treatment. The patient again responded by tumor regression for an additional three months, followed by metastatic progression, whereupon a third genome sequence was conducted with analysis indicating extreme resistance to Sunitinib™ and Sorafenib™ had developed, based on up-regulation of MAPK/ERK and PI3K/AKT pathways. This important work establishes a paradigm that NGS analysis of DNA and RNA from tumors can effectively be interpreted in light of the available targeted therapies and that relief from tumor burden can be obtained. However, until we better understand the processes by which tumors can be successfully drugged with targeted therapies and not result in the presentation of new, drug-resistant subclones, multiple genome sequencing assays may be required to achieve disease regression and stabilization.
Another diagnostic impact of next-generation sequencing is in the resolution of atypical genomic presentations of cancers where the clinical diagnostic paradigm uses defined reagents that address known cytogenetic abnormalities. One example of the latter application of NGS is described in our manuscript regarding the genome sequencing-based diagnosis of acute promyelocytic leukemia (APL) in a patient [25]. APL was characterized in the 1970’s by cytogenetics because of its canonical translocation between chromosomes 15 and 17, often reciprocal, whereby the genes PML (promoter and first three exons) and RARα (exons 3–9 and 3’ UTR) are juxtaposed and the resulting fusion transcript contributes to APL development [26–29]. In this study, the patient presented with classical pathologic hallmarks of APL but upon cytogenetic evaluation to identify the t15;17, was found to be negative for this and for the reciprocal translocation. Further complicating her treatment was that cytogenetic evaluation genome-wide indicates multiple rearrangements, classified as “complex cytogenetics”. Typically, the latter diagnosis indicates stem cell transplant (SCT) as the treatment standard of care, since these patients are categorized as “high risk”. Because of the associated morbidity and mortality associated with SCT, and because the cellular pathology was indicative of APL, we sequenced the patient’s tumor genome from bone marrow and a comparator normal from skin, once the patient was in remission. Within seven weeks that mirrors the time required for FISH cytogenetic diagnosis of APL’s t15;17, we determined by sequence read pair analysis focused initially on chromosomes 15 and 17, evaluating anomalously mapping read pairs that identify structurally variant regions of the genome (relative to the human reference genome and the patient’s normal genome), that the PML-RARα fusion had indeed occurred in this patient. However, this fusion was arrived at by a completely novel mechanism of cryptic insertion between chromosome 15 (77 kb containing the first three exons of PML) and chromosome 17, effectively juxtaposing the two genes and producing the anticipated fusion transcript of PML-RARα as is arrived at by t15;17, as shown in Figure 3. This information was verified in a CLIA environment, using PCR of the assembled junction sequences identified by NGS data assembly, and then provided to the patient’s oncologist for consideration in her treatment. As such, this patient was consolidated with all-trans retinoic acid (ATRA) and is leukemia-free now two years following her treatment.
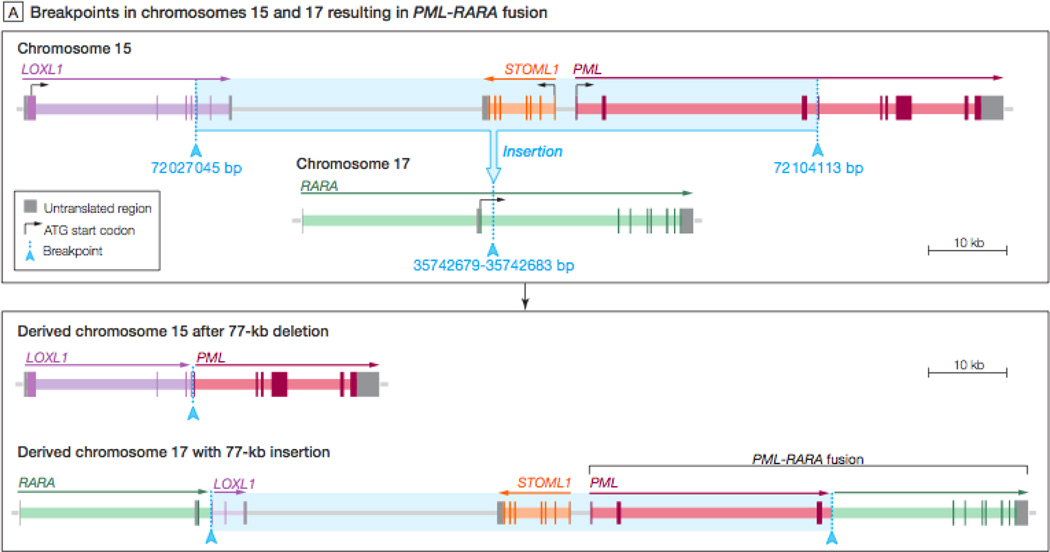
Figure 3.
Diagrammatic representation of the cryptic insertion identified in an acute promyelocytic patient genome. In this event, a 77kb portion of chromosome 15 containing the PML first three exons and a portion of the LoxL1 gene was inserted into chromosome 17, between exons 3 and 4 of the RARa gene. The resulting sequences of chromosomes 15 and 17 are shown at the bottom of the figure.
In conclusion, the development over three years of ultra high-throughput sequencing technologies known as “next-generation” or “massively parallel” has dramatically changed the landscape of cancer genomics. This trajectory is advancing rapidly and is beginning to impact the diagnosis and treatment of cancer. Certainly, our understanding of cancer as a disease of the genome already has, and will continue to be impacted in a dramatic and lasting way.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann Genet. 1973;16(2):109–112. [PubMed] [Google Scholar]
- 2.Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet. 1977;1(8010):549–550. doi: 10.1016/s0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- 3.Rowley JD, et al. Further evidence for a non-random chromosomal abnormality in acute promyelocytic leukemia. Int J Cancer. 1977;20(6):869–872. doi: 10.1002/ijc.2910200608. [DOI] [PubMed] [Google Scholar]
- 4.Golomb HM, et al. Correlation of clinical findings with quinacrine-banded chromosomes in 90 adults with acute nonlymphocytic leukemia: an eight-year study (1970–1977) N Engl J Med. 1978;299(12):613–619. doi: 10.1056/NEJM197809212991201. [DOI] [PubMed] [Google Scholar]
- 5.Rowley JD. The clinical usefulness of chromosome studies in patients with leukemia. Compr Ther. 1980;6(7):57–64. [PubMed] [Google Scholar]
- 6.Rowley JD. Identification of the constant chromosome regions involved in human hematologic malignant disease. Science. 1982;216(4547):749–751. doi: 10.1126/science.7079737. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 8.Mardis ER. New strategies and emerging technologies for massively parallel sequencing: applications in medical research. Genome Med. 2009;1(4):40. doi: 10.1186/gm40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley TJ, et al. DNMT3A Mutations in Acute Myeloid Leukemia. N Engl J Med. 2010 doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 14. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. This manuscript is from The Cancer Genome Atlas project and provides one of the first examples of comprehensive data integration across DNA and RNA data from a very large sample set.
- 15.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah SP, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360(26):2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 20.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 23.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones SJ, et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010;11(8):R82. doi: 10.1186/gb-2010-11-8-r82. Using next-generation sequencing of DNA and RNA from clinical samples obtained from a metastatic patient initially diagnosed with adenocarcinoma of the tongues, Jones et al. describe the first example of medical interpretation of sequencing data. This description includes key considerations regarding determining best therapeutic options for an individual patient.
- 25.Welch JS, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305(15):1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grisolano JL, et al. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89(2):376–387. [PubMed] [Google Scholar]
- 27.Brown D, et al. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 1997;94(6):2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly LM, et al. PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proc Natl Acad Sci U S A. 2002;99(12):8283–8288. doi: 10.1073/pnas.122233699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wartman LD, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121(4):1445–1455. doi: 10.1172/JCI45284. This manuscript describes the first whole genome sequencing of a mouse model of human cancer, and correlates the genomic alterations identified in the mouse model to those already studied in the human equivalent of the disease.