Abstract
Background
Fatty acids, a considerable fraction of lipid molecules, participate in fundamental physiological processes. They undergo activation into their corresponding CoA esters for oxidation or esterification into complex lipids (e.g. triglycerides, phospholipids and cholesterol esters), a process that is carried out by acyl-CoA synthases (ACS). Here we analyze the evolution of the gene family encoding for the long-chain acyl-CoA synthetases (Acsl) in vertebrates.
Results
By means of phylogenetics and comparative genomics we show that genome duplications (2R) generated the diversity of Acsl genes in extant vertebrate lineages. In the vertebrate ancestor two separate genes originated the current Acsl1/5/6 and the Acsl3/4 gene families, and the extra gene duplicates in teleosts are a consequence of the teleost specific third round of genome duplication (3R). Moreover, the diversity of Acsl family members is broader than anticipated. Our strategy uncovered a novel uncharacterized Acsl-like gene found in teleosts, spotted gar, coelacanth and possibly lamprey, which we designate Acsl2. The detailed analysis of the Acsl2 teleost gene locus strongly supports the conclusion that it corresponds to a retained 2R paralogue, lost in tetrapods.
Conclusions
We provide here the first evolutionary analysis of the Acsl gene family in vertebrates, showing the specific contribution of 2R/3R to the diversity of this gene family. We find also that the division of ACSL enzymes into two groups predates at least the emergence of deuterostomes. Our study indicates that genome duplications significantly contributed to the elaboration of fatty acid activation metabolism in vertebrates.
Keywords: acyl-CoA long chain synthetase, Gene loss, Genome duplication, Differential paralogue retention, Acsl2
Background
Two rounds of genome duplication (1R and 2R) have now been clearly established to have occurred in early vertebrate evolution [1], with a further round taking place in teleost ancestry (3R) [2]. Extra independent genome duplications have been determined in salmonids [3], and in ray-finned fish paddlefish [4]. These events have modeled the genomes of extant vertebrate lineages in terms of gene numbers and the overall genome architecture, contributing to the appearance of numerous innovations [5]. In addition to the increase in gene counts resulting from 1R/2R/3R, the comprehension and detection of gene loss processes in combination with the differential retention of paralogues poses important challenges to enlighten vertebrate evolution [6-9].
Fatty acids (FA) are a particularly important category of lipid molecules, being a considerable source of energy and a significant component of bio-membranes. FA metabolism involves among others, processes such as hydrolysis, beta-oxidation, synthesis, esterification and activation. The later comprises a two-step, ATP dependent reaction, with the formation of an intermediate acyl-AMP which is then converted to acyl-CoA. Acyl-CoA synthetases (ACSs) are the key enzymes responsible for this fundamental initial step in lipid metabolism. They can act on non-polar hydrophobic FA substrates and convert them into water-soluble products (acyl-CoAs), which are then integrated into metabolic pathways such as oxidation of acyl-CoAs to obtain ATP, storage in the form of triglycerides (TGA) or use as building blocks for other lipid molecules. The human genome contains 26 ACS genes divided into 6 distinct families: Short-chain ACS family (ACSS); Medium-chain ACS family (ACSM); Long-chain ACS family (ACSL); Very long-chain ACS family (ACSVL), Bubblegum ACS family (ACSBG) and ACS-Family (ACSF) [10-12]. This division reflects the chain length of their preferred substrate. ACSL enzymes play a paramount role in humans, since FAs with 12 to 20 carbons (C12-C20) are highly prevalent in the diet and are preferentially converted to acyl-CoA by these enzymes [12,13]. Further, several pathological conditions have been linked to ACSL enzymes such as inadequate lipid metabolism leading to diabetes [14], X-linked 63 mental retardation (MRX63-OMIM300387) [15,16] and cancer [17]. In mammals, previous studies identified five distinct Acsl genes, which were further organized into two separate groups (Acsl1, Acsl5 and Acsl6) and (Acsl3 and Acsl4) [10,12,18]. It is worth mentioning that the current Acsl gene nomenclature omits the “Acsl2” which was dropped since shortly after its identification it was found to be identical to Acsl1 in human and additionally rodent “Acsl2” was also renamed Acsl6 since it shared more identity with human Acsl6[18]. The advent of whole genome sequencing projects allowed the identification of Acsl genes in non-mammalian species, but no approach has been made in order to unravel the evolutionary history of this family [12,19]. Additionally, recent findings have illustrated the need to consider genome duplication processes (and gene loss) in combination with extensive species analysis for proper evolutionary insights regarding lipid metabolic gene networks to be drawn [20,21]. Moreover, non-mammalian species, such as the zebrafish, have been recently proposed as alternative models to study lipid metabolism [22]. Therefore, a comparative and phylogenetic approach involving a broader number of vertebrate species should shed light into the evolutionary history of ACSL enzymes and their function. In this study we demonstrate that genome duplications in stem vertebrate ancestry and the teleost specific genome duplication were instrumental in the generation of Acsl gene diversity. Moreover, we show that the variety of Acsl family members is broader than anticipated. Our strategy uncovered a novel uncharacterized Acsl-like gene found in teleosts and coelacanth, which we designate Acsl2. The detailed analysis of the Acsl2 teleost gene locus strongly supports the suggestion that it corresponds to a retained paralogue, lost in other vertebrates classes (“an ohnolog gone missing”). Finally, we provide the first comparative transcription analysis between the human and zebrafish Acsl gene repertoire.
Results
ACSL gene repertoire in vertebrates
Human ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6 sequences were used to perform Blastp searches and collect Acsl-like sequences from various available genomes. We analyzed a total of 21 species in order to include all major vertebrate lineages. Our database search determined the presence of five ACSL genes in humans, mouse, opossum, chicken, anole lizard, western clawed frog, and the coelacanth. In the spotted gar, an out-group of the teleost specific genome duplication [23], we found 5 sequences though 2 were partial (Additional file 1). Blast searches in teleost fish genomes hinted at a larger Acsl gene set, with nine hits in zebrafish, pufferfish, green spotted puffer and medaka and seven in stickleback. However, detailed sequence analysis suggested a number of inconsistent annotations in the Ensembl database. For example, we found three Acsl1 gene annotations in medaka, (1-ENSORLG00000019563, 2-ENSORLG00000018806 and 3-ENSORLG00000008655), however, when aligning the DNA and amino acid sequence of the first two sequences we observe that they are identical (not shown). Given that the annotated Acsl1 copy ENSORLG00000018806 is located within a contig that presents extensive regions that are poorly resolved we consider that this species presents 2 gene copies of Acsl1 and select ENSORLG00000019563 and ENSORLG00000008655 for further studies. The green spotted pufferfish again shows three annotated copies of Acsl1 with two of these (ENSTNIG00000000345 and ENSTNIG00000010115) located in the same scaffold with the same orientation and contiguously (Additional file 2). These annotations are partial sequences, one corresponding to the N-terminal and the other corresponding to the C-terminal of the protein. Here we assume that these annotations correspond to a single gene poorly assembled. Therefore we consider that the green spotted puffer presents two Acsl1 genes and we use only the correctly annotated gene (ENSTNIG00000018054) for further analysis. Finally we find two annotated Acsl1 genes in pufferfish (1-ENSTRUG00000017576 and 2-ENSTRUG00000001450) were the second gene corresponds to a partial sequence which was not used for further analysis. We investigated also the genomes of three Chondrichthyans, the elephant shark, catshark, and little skate. Our investigation identified 4 full sequences and several partial (Additional file 1).
Finally, the search in the lamprey genome resulted in four Acsl-like gene hits (1-ENSPMAG00000008135, 2-ENSPMAG00000004625, 3-ENSPMAG00000005099 and 4-ENSPMAG00000005133). Three of these correspond to partial sequences (449 residues) and were not used for further analysis. Finally, in the investigated invertebrate species, acorn worm and amphioxus, we recovered 3 Acsl sequences from acorn worm and 4 Acsl sequences from amphioxus. After clarifying all inconsistent gene annotations a set of ACSL sequences from various species were collected to perform phylogenetics (Additional file 3).
Phylogenetics indicates vertebrate specific Acsl gene expansions
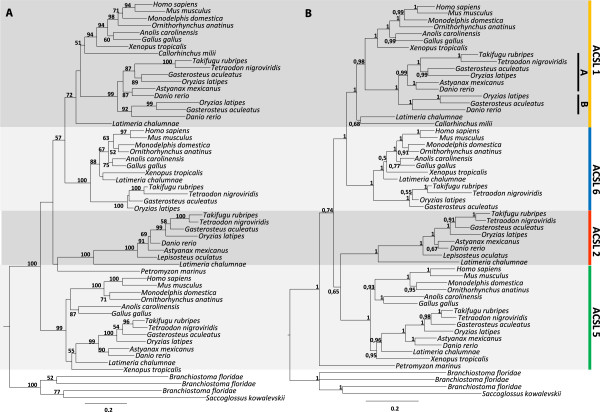
Preliminary phylogenetic analysis confirmed that Acsl3 and Acsl4 form a distinct group from Acsl1, Acsl5 and Acsl6 as previously reported (data not shown) [10,12,18]. Thus, we have reconstructed each phylogeny separately (Figure 1 and Figure 2). In the Maximum likelihood analysis Figure 1A it is possible to observe that invertebrate sequences out-group four statistical well supported clades comprising Acsl1, Acsl5, Acsl6 and an unidentified Acsl group. However, the exact phylogenetic relationships between each isoform are not statistically supported with the bootstrap analysis. In the Bayesian analysis (Figure 1B) we find again the invertebrate sequences out-grouping four statistically well supported vertebrate clades. The unidentified Acsl group is composed of teleost, rayfin fish and coelacanth sequences. In the Maximum likelihood analysis a lamprey sequence also groups with this novel clade (though weakly supported). We name this new gene lineage Acsl2. The overall tree branching pattern in Maximum likelihood and Bayesian analysis is indicative that the expansion of Acsl1/5/6/novel clade took place after the radiation of the vertebrate lineage approximately 500 million years ago, although independent gene expansions have taken place in amphioxus and the acorn worm (Figure 1A and B). We find representatives of Acsl1/5/6 in all of the examined vertebrate species, with the exception of lamprey and chondrichthyans where the presence of partial sequences impedes a final conclusion regarding the full Acsl gene repertoire in these lineages (see Additional file 1). Nevertheless, this cannot be taken as an indication of gene loss due to the poor genome sequence coverage. The phylogenetic trees also indicate that Acsl1 has specifically duplicated in the teleost lineage. Even though only medaka, zebrafish and stickleback present these duplicates, we antecipate that pufferfish and the green spotted pufferfish probably retain these two copies.
Figure 1.
Phylogenetic analysis of Acsl1, Acsl2, Acsl5, Acsl6 sequences. (A) Maximum likelihood tree, numbers at nodes represent percentage of bootstrap values (only values above 50% are shown); (B) Bayesian tree, number at the branch nodes refer to posterior probabilities.
Figure 2.
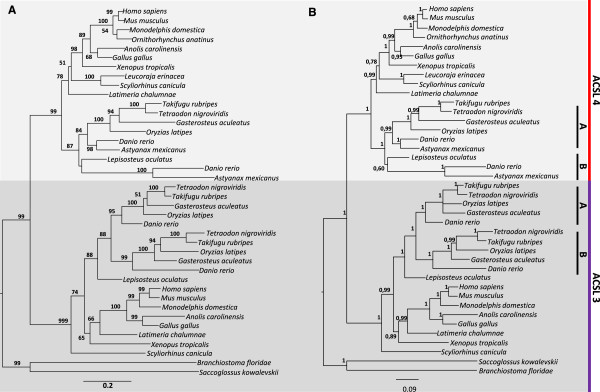
Phylogenetic analysis of Acsl3 and Acsl4 sequences. (A) Maximum likelihood tree, numbers at nodes represent percentage bootstrap values (only values above 50% are shown); (B) Bayesian tree, number at branch nodes refer to posterior probabilities.
Regarding the Acsl3 and Acsl4 trees (Figure 2), both in the Maximum likelihood and Bayesian analysis we observe that the invertebrate Acsl-like sequences again out-group two well supported groups containing vertebrates sequences. Also, it is possible to recognize that all teleost species here analyzed present a lineage specific duplication of Acsl3 (Acsl3a and Acsl3b). In zebrafish and cave fish we find an Acsl4 duplicate; microsynteny analysis of this locus in zebrafish suggests that this extra gene copy is also a teleost specific 3R duplicate (Additional file 4). However, despite extensive database search, we did not retrieve other Acsl4-like sequences in other teleost species. Acsl3 and Acsl4 gene copies were also found in the cat shark and little skate (Figure 2).
The phylogenetic analysis also resolves a further inaccurate annotation in the western clawed frog. In the Ensembl database two ORFs are annotated as Acsl4 genes (1-NP_001090679.1-ENSXETG00000033126 and 2-ENSXETG00000012429). After observing the localization of these two sequences in the phylogenetic tree, we find that one of the annotated “Acsl4” groups within the Acsl3 clade, which is consistent with a synteny analysis. Therefore we consider that the western clawed frog presents one Acsl4 gene (ENSXETG00000012429) and one inaccurate annotation of an Acsl3 gene (ENSXETG00000033126) (Additional file 5).
In summary, the phylogenetic data indicates that the Acsl1, Acsl2, Acsl5 and Acsl6 and Acsl3 and Acsl4 have all duplicated before vertebrate radiation, with episodes of lineage specific expansion observed in studied invertebrate deuterostomes. In addition, teleost fish underwent specific duplications in Acsl1 and Acsl3, and possibly Acsl4 in zebrafish and in cave fish.
Genome duplications contributed to the diversity of Acsl genes in vertebrates
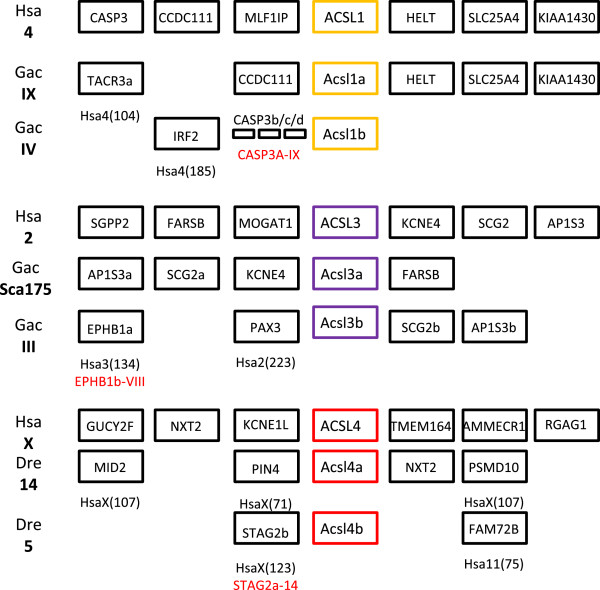
Our phylogenetic analysis clearly indicates that despite the existence of several Acsl gene copies in the studied invertebrate deuterostome species, the expansion of the Acsl1/Acsl2/Acsl5/Acsl6 and of the Acsl3/Acsl4 clades took place in the vertebrate ancestor. Thus, we next analyzed the contribution of 2R and 3R in the generation of Acsl gene diversity. We started by examining the genomic location of each human ACSL gene and respective flanking gene families (Figure 3). Human ACSL1, ACSL5 and ACSL6 localize respectively to Chr4q35, Ch10q25.2 and Ch5q31 (Figure 3A), regions which are part of the 2R NK-paralogon [24-26]. The analysis of the flanking gene families revealed that those which are multi-copy and whose duplication timing coincides with vertebrate emergence, typically have their members localizing to Hsa4, Hsa5, Hsa8, Hsa10 and/or Hsa2. For example, TCF7L2 gene which flanks ACSL5 has two other duplicates mapping to Hsa4 (LEF1) and Hsa5 (TCF7); CASP3 which maps close to ACSL1 has a paralogue, CASP7, mapping close to ACSL5; PDLIM4 mapping downstream of ACSL6 has two paralogues, PDLIM1 and PDLIM3, localizing to Hsa10 and Hsa4 respectively. Overall, the majority of genes flanking human ACSL1, ACSL5 and ACSL6 revealed conserved macrosynteny and therefore support the hypothesis that these regions are related, with the duplication timing coinciding with 2R. Furthermore, using the proposed vertebrate ancestral genome reconstruction [27], we find that the Hsa4, Hsa10 and Hsa5 belong to the same ancestral group, group C (Figure 3C). In summary, from a single ancestral chromosome C in the vertebrate ancestor, derived four chromosomes (C0, C1, C2 and C3) [27] as a result of 1R/2R. Each human ACSL locus maps to a distinct ancestral C chromosome: ACSL1-C1, ACSL6-C0 and ACSL5-C2 (Figure 3A and C). We would expect to find a fourth ACSL gene which should map to the ancestral chromosome C3 distributed in present day human genome at Hsa2/7/8 (see next section).
Figure 3.
Microsyteny analysis of the Acsl human loci and their mapping location in the ancestral vertebrate chromosomes. (A) Location of the Acsl1, Acsl5, Acsl6 and neighboring genes in the human genome and corresponding paralogues; (B) location of Acsl3, Acsl4 and neighboring genes in the human genome and corresponding paralogues; (C and D) schematic representation of the duplication history of the ancestral vertebrate chromosomes C and F.
Regarding human ACSL3 and ACSL4 genes we find that they map to chromosomes Chr2q34 and ChrXq22 respectively. Neighboring gene families have paralogues in the following set of chromosomes HsaX, Hsa2, Hsa7, Hsa11, Hsa13, Hsa14 and Hsa17 (Figure 3B), with no apparent conserved macrosynteny. However, ACSL3 and ACSL4 map to chromosome regions derived from the 2R duplication of the proto-chromosome F, at F0 and F4 respectively (Figure 3D). Accordingly, after the first round of genome duplication one F proto-chromosome underwent an additional fission event, resulting in three proto-chromosomes. Two of these proto-chromosomes underwent the second WGD, giving rise to four ancestral chromosomes (F1, F2, F3 and F4) and the third chromosome gave rise to the F0. The gene families flanking ACSL3/ACSL4 have in most cases duplicates in regions assigned to F chromosomes [27], thus providing strong support to the hypothesis that both were 2R generated.
Extra gene copies of Acsl1, Acsl3, and Acsl4 (in zebrafish) were found in our survey. The analysis of the loci of Acsl1, Acsl3 and Acsl4 (Figure 4) in stickleback and zebrafish provides solid support to the conclusion that 3R contributed for the increase of the Acsl gene set in teleosts. We find that 3R specific duplicates can be observed outflanking each pair of Acsl duplicates (Casp3, Ephb1 and Stag2) (Figure 4).
Figure 4.
Conserved synteny of Acsl1 , Acsl3 and Acsl4 between human and teleost ( G. aculeatus and D. rerio ) indicate the contribution of 3R in the generation of extra gene copies.
Acsl2 is a potential 2R paralogue gone missing in tetrapods
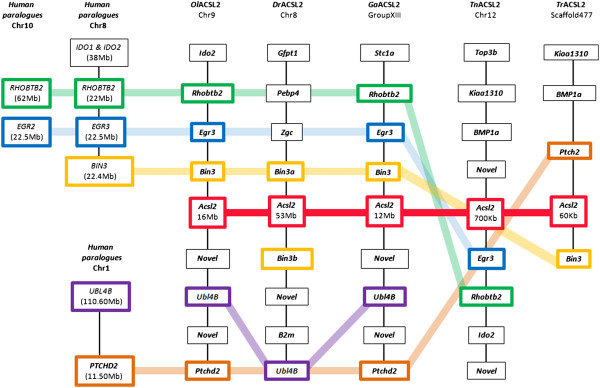
The phylogenetic analysis showed the presence of a previously unidentified Acsl gene, Acsl2, paralogous to Acsl1, Acsl5 and Acsl6. To enlighten the evolutionary origin of the Acsl2 gene we analyzed the genomic locus of this novel gene in teleost species (Figure 5). We show that the Acsl2 gene is confined to a largely conserved locus in teleost fish. A large set of neighboring gene families have their human orthologues mapping to Hsa8. The following genes EGR3, BIN3, RHOBTB2, BMP1, PEBP4, STC1, and IDO2 are close together at Hsa8 and constitute a conserved block, with UBL4b and PTCHD2 localizing in Hsa1 (Figure 5; data not shown). Most importantly, the conserved syntenic block in Hsa8 maps back to the ancestral chromosome C3 which corresponds to the expected localization of a fourth copy of the ACSL gene after 2R, absent in the human genome. Further, gene families which are multicopy have their paralogues localizing to Hsa10/5/4 as expected. These finds together with the phylogenetic analysis suggest that the Acsl2 gene corresponds to a retained paralogue conserved in teleosts and lost in the tetrapod lineage.
Figure 5.
Synteny map of Acsl2 loci in teleost fish, and the location of human orthologues.
Gene expression data indicates functional partitioning and diversification
Given that the teleosts have additional Acsl gene copies, we proceeded to analyze the gene expression of Acsl genes in zebrafish and performed a comparative analysis with the human ACSL gene repertoire. In zebrafish a high Acsl1a mRNA content is observed in all analyzed tissues with the exception of the eye (Figure 6A), while Acsl1b is only marginally expressed in testis, ovary, kidney and heart (Figure 6A). The human ACSL1 has a similar expression pattern with high expression in brain, heart, spleen, kidney, ovary and testis and medium to low expression in liver, lung and eye (Figure 5C). These findings are in agreement with previous studies in Rattus norvegicus in which it was found that Acsl1 is highly expressed in major energy metabolizing tissues namely heart, liver and adipose tissues [28]. Regarding ACSL5, in human we find that this enzyme is highly expressed in all tissues here analyzed with the exception of the ovary (Figure 6C). When observing the data obtained for zebrafish we find a distinct expression pattern. High Acsl5 mRNA transcription is observed in the testis, ovary, kidney, gut and liver, while spleen, gill, heart, eye and brain have a low/absent gene transcription (Figure 6A).
Figure 6.
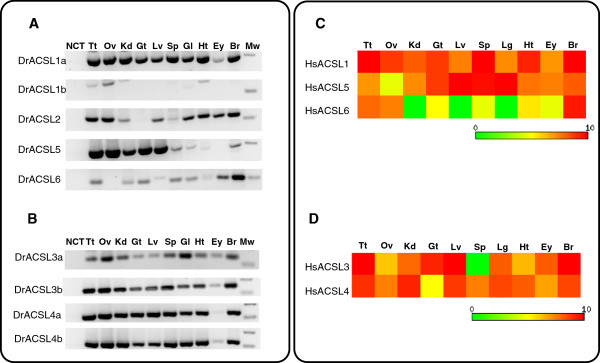
Comparative expression analysis of Acsl genes in human and zebrafish tissues.(A and B) Tissue expression profile of zebrafish Acsl genes; (C and D) Tissue expression heatmap of Human Acsl genes; NCT negative template control, Tt: testis, Ov: ovary, Kd: kidney, Gt: gut, Lv: Liver, Sp: spleen, Gl: gill, Ht: heart, Ey: eye; Br: brain, Lg: lung, Mw: molecular weight marker.
Concerning ACSL6, in opposition to ACSL1 and ACSL5; this enzyme presents a fairly restricted expression pattern in human being highly expressed in testis, ovary and brain. In zebrafish this restricted expression pattern is also observed, with Acsl6 being found essentially in brain (Figure 6A). The teleost Acsl2 transcription is high in testis, ovary, gill, heart, eye, and brain.
The expression pattern of Acsl3a and Acsl3b in zebrafish, reveals that Acsl3a is preferably expressed in ovary, gill and brain; nevertheless this gene is also expressed at lower levels in all other tissues (Figure 6B). Acsl3b is expressed in all tissues with comparatively higher levels to Acsl3a, with the exception of eye. The expression pattern of the Acsl4a and Acsl4b is highly similar being highly expressed in all tissues here analyzed with a slight decrease in gut and liver and low expression in eye (Figure 6B). When observing the expression pattern of ACSL4 in human we observe that this gene is also highly expressed in all tissues with the exception of gut and eye (Figure 6D).
Discussion
ACSL are key enzymes involved in the initial steps of FA metabolism. These enzymes preferentially activate FA with C12-C20 (the most abundant in the human diet), which may then intervene in a variety of metabolic pathways. Although the ACSL family has been the focus of various studies, their evolutionary history has not been investigated before. Here we combine extensive database search with phylogenetics and comparative genomics to provide a reliable depiction of the evolution of Acsl in vertebrates (Figure 7). Initial analyses revealed several inaccurate gene annotations. After an exhaustive analysis we were able to clarify several of these and perceive that the diversity of Acsl genes is broader than anticipated. The gene repertoire varies significantly between mammals (5), teleosts (7/8), and the invertebrate studied species (3/4). Through phylogenetics we were able to reconstruct the Acsl gene duplication timings in relation to the divergence of major vertebrate groups. The five mammalian Acsl genes have been organized into two separate groups: Acsl1, Acsl5, Acsl6 and Acsl3, Acsl4, on the basis of sequence homology and gene organization [10,12,18]. We now propose that this division is evolutionarily significant and dates back to at least deuterostome ancestry since clear co-orthologues of both gene groups exist in hemichordates and cephalochordates. Although, various Acsl1/2/5/6 genes were found in amphioxus and the acorn worm, these represent independent lineage duplications. The exact duplication timing of a proto-Acsl gene to originate the ancestor of Acsl1/2/5/6 and Acsl3/4 is at present unknown but probably dates as far back as the origin of the Bilateria (not shown; LFCC unpublished results). In the agnathan lamprey we were only able to retrieve one complete Acsl sequence, although several partial sequences were also evident. Thus, at this stage a final conclusion concerning the full repertoire of Acsl genes in lamprey is premature. The findings that the Acsl gene family expanded significantly in the time window coincident with the emergence of vertebrates lead us to test the contribution of genome duplications. Using the proposed ancestral vertebrate genome reconstruction [27] we were able to trace back the duplication history of Acsl genes in the gnathostome ancestor. We find that human Acsl1, Acsl5 and Acsl6, map to chromosomes C1, C2 and C0 respectively which originated from duplication of the ancestral proto-chromosome C (Figure 7). This observation is also supported by the duplication history analysis of the flanking gene families. In teleosts we found extra gene copies within the Acsl1/5/6 clade. These were partially explained by the contribution of the teleost specific genome duplication (3R) (Acsl1a and Acsl1b), but not entirely. A novel gene with no clear orthologues in tetrapods was found in the analyzed teleost species, the spotted gar, coelacanth and possibly lamprey. The phylogenetic analysis clearly indicated that this represents a distinct gene lineage which we name Acsl2. To enlighten the origin of Acsl2, we investigated the genomic locus in teleosts and cross-compared it with the human genome. We find that the most parsimonious explanation for the retrieved data is that Acsl2 is a 2R paralogue retained in teleosts and lost in tetrapods, similar to what was found in a distinct ACS gene family [21]. Thus, the novel uncharacterized Acsl2 gene corresponds to a quadruplicate Acsl paraloguous to Acsl1, Acsl5 and Acsl6.
Figure 7.
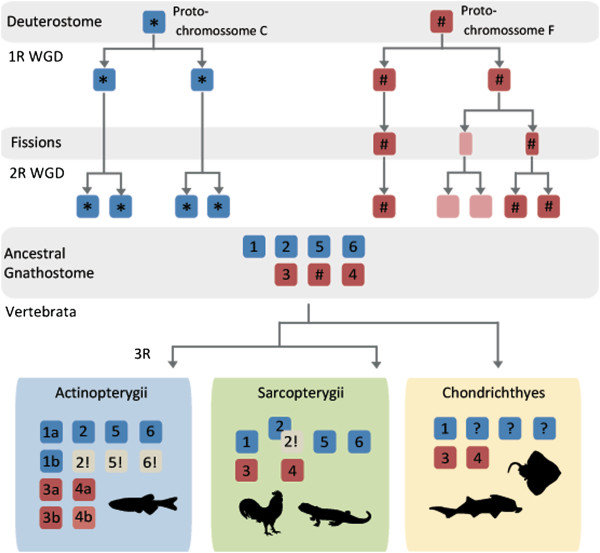
Proposed evolutionary history and duplication timing of the Acsl gene family in vertebrates. Questions marks indicates unknown data, and exclamation signals gene loss.
Similarly, we found also that the human orthologues of Acsl3 and Acsl4 map to chromosome regions related by duplication. Both genes map to genomic regions remnants of 2R resulting from the duplication of the ancestral proto-chromosome F. The most plausible explanation for the unequal distribution of Acsl gene copies within F0, F1, F2, F3 and F4, is a chromosome fission event occurred after 1R of WGD which resulted in the splitting of the genetic information into two distinct chromosomes, which then underwent the second genome duplication (2R) and originated F1, F2, F3 and F4 (Figure 7). The detailed comparative genomic and phylogenetic analysis again highlighted the contribution of 3R to the gene number increment observed in teleosts (Figure 7).
It has been previously suggested that major vertebrate innovations occurred after genome duplication events [5]. Whole genome duplications lead to the expansion of gene numbers, facilitating gene diversification, sub-functionalization along with the rise of novel functions and gene loss. These ultimately enable evolutionary radiation. FA composition and metabolism is known to be different in some vertebrate groups, for example in teleosts [20]. Similar to our findings in Acsl, recent studies have also revealed that various gene families involved in lipid metabolic pathways have evolved distinct gene repertoires in vertebrate lineages, including fish, with clear functional and regulatory impacts [20,21,29,30]. The retention of such a larger Acsl gene set after 2R/3R, with simultaneous processes of differential loss, could be indicative that novel Acsl gene functions have emerged in vertebrate ancestry. In effect, the variety of ACSLs in mammals is apparently associated with distinct substrate preferences. ACSL1 uses FA with C16 to C18 both saturated and mono unsaturated, ACSL3 displays a high activity with C12:0 (laurate), C14:0 (myristatate), C20:4 (arachidonate) and C20:5 (eicosapentaenoic acid) [31]. In contrast, ACSL4 presents a clear preference for polyunsaturated FA with C2O:4 and C20:5 [10,32]. ACSL5 shows substrate specificity similar to ACSL1, favorably utilizing palmitic (C16:0), palmitoleic (C16:1), oleic (C18:1) and linoleic (C18:2) acids [33]. Finally, ACSL6 preferentially uses FAs with C16 to C20 saturated and polyunsaturated, although alternative splicing generates isoforms with distinct substrate specificities [10,34]. We propose that teleost orthologues have probably retained similar FA substrate preferences with respect to Acsls. However, novel Acsl family members were also discovered in this study. Thus, we performed a comparative tissue transcription analysis between zebrafish and human. We selected zebrafish as a model, given that this species has been previously suggested as a model organism for the study of lipid metabolism [22], and coincidentally this species also presents the largest set of Acsl genes. The comparison of the expression data between human and zebrafish revealed a similar expression profile, with the exception of Acsl2 and Acsl5, and the fish specific duplicates (Acsl3 and Acsl4). We observe that the zebrafish Acsl5 is expressed in fewer tissues when compared to the human orthologue, with the teleost Acsl2 apparently compensating the lack of Acsl5 transcription in various tissues. This setting suggests that in teleost fish functions are shared between Acsl2 and Acsl5. Regarding the 3R duplicated members Acsl3a and Acsl3b, and Acsl4a and Acsl4b we find similar tissue expression distributions in opposition to Acsl1a and Acsl1b. In the latter Acsl1b is co-expressed with Acsl1a in a small set of specific tissues (testis, ovary and heart). We hypothesize that Acsl1b plays a specific role in these tissues, distinct from the role played by Acsl1a, hinting towards a sub-functionalization after duplication. Although, we have not tested the FA specificity of the novel Acsl repertoire described in this work, we cannot ignore that the retention of a larger Acsl gene number in teleosts could also be related with the specific acquisition of novel substrate preferences, which future studies should address.
Conclusion
In summary, we demonstrate the importance of genome duplications, 2R and 3R, in the generation of the Acsl diversity in vertebrate species.
Methods
Database mining and identification of Acsl sequences
ACSL family members were identified in the Ensembl, GenBank and JGI (Joint genome institute) databases through Blastp searches using as reference annotated human ACSL sequences. In order to include all major vertebrate lineages we analysed eutherian metatherian and prototherian mammals: Homo sapiens (human), Mus musculus (mouse); Monodelphis domestica (opossum); Ornithorhynchus anatinus (platypus); birds: Gallus gallus (chicken); reptiles: Anolis carolinensis (anole lizard); amphibians: Xenopus tropicalis (western clawed frog); Latimeria chalumnae (Coelacanth); Lepisosteus oculatus (spotted gar); teleosts: Danio rerio (zebrafish), Astyanax mexicanus (blind cave fish), Takifugu rubripes (pufferfish), Tetraodon nigroviridis (green spotted puffer) Oryzias latipes (medaka) and Gasterosteus aculeatus (stickleback); chondrichthyans: Leucoraja erinacea (little skate), Scyliorhinus canicula (small-spotted catshark) and Callorhinchus milii elephant shark,and jawless fish hyperoartia: Petromyzon marinus (sea lamprey). Sequences searches were also made in an invertebrate chordate Branchiostoma floridae (amphioxus) and the hemichordate Saccoglossus kowalevskii (acorn worm).
Sequence alignment and phylogenetic analysis
All ACSL amino acid sequences retrieved in the database mining were initially aligned in MAFFT alignment software using default parameters [35] and manually curated with the exclusion of regions of uncertain homology, gaps, and of partial sequences. Revised sequence alignments were then submitted to Protest online server version 2.4 [36] available at http://darwin.uvigo.es/software/prottest_server.html, in order to select the appropriate protein evolution model according to our dataset. Here we found that ACSL3 and ACSL4 group follows a JTT + I + G model, while the LG + I + G + F model suits best the ACSL1, ACSL2 ACSL5 and ACSL6 group. Phylogenetic analyses were performed and a Maximum likelihood (PhyML) tree with 1000 bootstrap replicates was constructed using the online platform of the PhyML 3.0 avaliable at http://www.atgc-montpellier.fr/phyml/. Bayesian inference of phylogeney was performed with MrBayes version 3.2.2 [37] on CIPRES Science Gateway [38]. Analysis for both Acsl3/4 and Acsl1/2/5/6 amino acid sequences were performed under a mixed substitution model, with two parallel runs with 1 million generations, each with four chains one cooled and 3 heated. Trees were sampled every 100 generations; final consensus tree was calculated with the fifty percent majority rule and from the remaining trees after a 0.25 burin.
Comparative genomics
All ACSL genes were mapped into the human chromosomes, the location of each gene and the neighboring genes were collected from Ensembl and GenBank databases. ACSL loci in human were used as a model for comparison. The Ensembl paralogue and orthologue prediction tools were used to infer duplication history patterns of flanking ACSL genes. For some flanking gene families we run phylogenetics to confirm relationships with the methodology described above (not shown).
Gene transcription analysis
Adult wild-type zebrafish obtained from our own breeding stock were used for gene expression analysis. Animals were anesthetized and killed by cervical transection in accordance with the Portuguese Animals and Welfare Law (Decreto-Lei nº 197/96) approved by the Portuguese Parliament in 1996. Institutional animal approval by CIIMAR/UP and DGV (Ministry of Agriculture) was granted for this study. After collection all tissues were preserved in RNAlater and stored at -20°C. Total RNA was isolated using an Illustra RNAspin Mini RNA Isolation Kit (GE Healthcare, UK) according to the manufactures recommendations, including the on-column treatment of isolated RNA with RNase-free DNase I. RNA concentration was calculated using Qubit fluorometer instrument (Invitrogen, Carlsbad CA), integrity confirmed by electrophoresis and the RNA stored at -80°C until further use. The cDNA was synthesized from 250 ng of total RNA with the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufactures protocol. Forward and reverse primers were designed using sequences available in Ensembl with Primer3 software [39]. All of primer sets match exon sequence and flank an intron consequently avoiding genomic DNA amplification. Primers sets were created for the following genes ACSL1a; (Forward-5′ CAGGATGGGCAAAGAATAGAG 3′, Reverse-5′ TTTCAGTGTTGGTGTGAGGAG 3′, annealing at 55°C) ACSL1b; (Forward-5′ GCACAGCGAGATGTTCAC 3′, Reverse-5′ AAGTCCAATCCAAATGTCAGG 3′, annealing at 54°C) ASCL2 gene; (Forward-5′ GTAGTTCCAGATCCAGAAGTGTTC 3′ reverse-5′ CGCCGTCATGTCCTCCAG 3′, annealing at 56°C) ACSL5; (Forward-5′ CGCAGAGAAACTGGGATTGAAAGG 3′, Reverse-5′ TGGCTTTGAGTGTTGGAGTGAGG 3′, annealing at 58°C) and ASCL6 (Forward-5′ CCTCGTGGGCTCAGAAGAAAG 3′ Reverse-5′CGCACCATGTCCTCCAGAATA 3′, annealing at 58°C). PCR was performed using 2 μl of zebrafish cDNA and Phusion® Flash high-fidelity Master Mix (FINNZYMES). PCR parameters were as follows: initial denaturation at 98°C for 10 s, followed by 35 cycles of denaturation at 98°C for 1 s, annealing for 5 s and elongation at 72°C for 10s and a final step of elongation at 72°C for 1 min. PCR products were then loaded onto 2% agarose gel stained with GelRed and run in TBE buffer at 80 V. In silico expression analysis, for ACSL gene in Human, was performed using ESTs available from Unigene [40] as count per million transcripts, all values are displayed as Log2 transcripts per million. Heat map was created using the collected EST data and matrix2png web interface v1.2 [41].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
The original idea for this study was conceived by LFCC. LFCC and MLM performed all the experimental analysis; MMS, MARH, and IC participated in the discussion regarding lipid metabolism and physiology. The manuscript was written by LFCC and MLM, and edited by all other co-authors. All authors have read and approved the final manuscript.
Supplementary Material
tBlastn search of ACSLlike sequences in Transcriptomic contigs.
Partial Acsl gene annotations in green spotted pufferfish.
NCBI accession numbers and Ensembl gene ID.
Synteny maps of Zebrafish ACSL 3R duplicates.
Xenopus tropicalis Acsl4 and Acsl3 corresponding location in human.
Contributor Information
Mónica Lopes-Marques, Email: monicaslm@hotmail.com.
Isabel Cunha, Email: isabel.cunha@ciimar.up.pt.
Maria Armanda Reis-Henriques, Email: mahenriq@icbas.up.pt.
Miguel M Santos, Email: santosmafm@gmail.com.
L Filipe C Castro, Email: filipe.castro@ciimar.up.pt.
Acknowledgements
This work was supported by Fundação para a Ciência e a Tecnologia (Portugal), through grants PTDC/MAR/68106/2006 and PTDC/MAR/68885/2006, the European Regional Development Fund (ERDF) through the COMPETE - Operational Competitiveness Programme and national funds through FCT – Fundação para a Ciência e a Tecnologia, under the project “PEst-C/MAR/LA0015/2013, and SFRH/BD/84238/2012 awarded to M Lopes-Marques. We thank three anonymous reviewers for their insightfull comments.
References
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;13(7198):1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;13(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Moghadam HK, Ferguson MM, Danzmann RG. Whole genome duplication: challenges and considerations associated with sequence orthology assignment in Salmoninae. J Fish Biol. 2011;13(3):561–574. doi: 10.1111/j.1095-8649.2011.03030.x. [DOI] [PubMed] [Google Scholar]
- Crow KD, Smith CD, Cheng J-F, Wagner GP, Amemiya CT. An independent genome duplication inferred from Hox paralogs in the American paddlefish-a representative basal ray-finned fish and important comparative reference. Genome Biol Evol. 2012. [DOI] [PMC free article] [PubMed]
- Shimeld SM, Holland PWH. Vertebrate innovations. Proc Natl Acad Sci. 2000;13(9):4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;13(5):563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Mulley JF, Holland PWH. Parallel retention of Pdx2 genes in cartilaginous fish and coelacanths. Mol Biol Evol. 2010;13(10):2386–2391. doi: 10.1093/molbev/msq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Kuratani S. Genome-Wide Detection of Gene Extinction in Early Mammalian Evolution. Evolution: Genome Biology and; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmark J, Sundström G, Ocampo Daza D, Larhammar D. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;13(1):859–871. doi: 10.1093/molbev/msq257. [DOI] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med. 2008;13(5):507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Ellis JM. Peroxisomal acyl-CoA synthetases. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2012;13(9):1420–1411. doi: 10.1016/j.bbadis.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 2007;13(12):2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- Li LO, Klett EL, Coleman RA. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochimica et Biophysica Acta (BBA) - Mol Cell Biol Lipids. 2009;13(3):246–251. doi: 10.1016/j.bbalip.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;13(Supplement 2):S9–S15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo I, Frints SGM, Fryns JP, Meloni I, Pescucci C, Ariani F, Borghgraef M, Raynaud M, Marynen P, Schwartz C. et al. A third MRX family (MRX68) is the result of mutation in the long chain fatty acid-CoA ligase 4 (FACL4) gene: proposal of a rapid enzymatic assay for screening mentally retarded patients. J Med Genet. 2003;13(1):11–17. doi: 10.1136/jmg.40.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni I, Muscettola M, Raynaud M, Longo I, Bruttini M, Moizard M-P, Gomot M, Chelly J, des Portes V, Fryns J-P. et al. FACL4, encoding fatty acid-CoA ligase 4, is mutated in nonspecific X-linked mental retardation. Nat Genet. 2002;13(4):436–440. doi: 10.1038/ng857. [DOI] [PubMed] [Google Scholar]
- Cao Y, Pearman AT, Zimmerman GA, McIntyre TM, Prescott SM. Intracellular unesterified arachidonic acid signals apoptosis. Proc Natl Acad Sci. 2000;13(21):11280–11285. doi: 10.1073/pnas.200367597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashek DG, Bornfeldt KE, Coleman RA, Berger J, Bernlohr DA, Black P, DiRusso CC, Farber SA, Guo W, Hashimoto N. et al. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J Lipid Res. 2004;13(10):1958–1961. doi: 10.1194/jlr.E400002-JLR200. [DOI] [PubMed] [Google Scholar]
- Grove TJ, Sidell BD. Fatty acyl CoA synthetase from Antarctic notothenioid fishes may influence substrate specificity of fat oxidation. Comp Biochem Physiol B Biochem Mol Biol. 2004;13(1):53–63. doi: 10.1016/j.cbpc.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Morais S, Monroig O, Zheng X, Leaver M, Tocher D. Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of ELOVL5- and ELOVL2-like elongases. Marine Biotechnol. 2009;13(5):627–639. doi: 10.1007/s10126-009-9179-0. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Lopes-Marques M, Wilson JM, Rocha E, Reis-Henriques MA, Santos MM, Cunha I. A novel acetyl-CoA synthetase short-chain subfamily member 1 (Acss1) gene indicates a dynamic history of paralogue retention and loss in vertebrates. Gene. 2012. [DOI] [PubMed]
- Flynn EJ, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J Lipid Res. 2009;13(8):1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted Gar, an outgroup for the teleost genome duplication. Genetics. 2011;13(4):799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SL, Holland PWH. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr Biol. 2000;13(17):1059–1062. doi: 10.1016/S0960-9822(00)00676-X. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Holland PWH. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev. 2003;13(5):459–465. doi: 10.1046/j.1525-142X.2003.03052.x. [DOI] [PubMed] [Google Scholar]
- Luke GN, Castro LFC, McLay K, Bird C, Coulson A, Holland PWH. Dispersal of NK homeobox gene clusters in amphioxus and humans. Proc Natl Acad Sci. 2003;13(9):5292–5295. doi: 10.1073/pnas.0836141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;13(9):1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashek DG, Li LO, Coleman RA. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J Lipid Res. 2006;13(9):2004–2010. doi: 10.1194/jlr.M600150-JLR200. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Monroig Ó, Leaver MJ, Wilson J, Cunha I, Tocher DR. Functional Desaturase Fads1 (Δ5) and Fads2 (Δ6) Orthologues Evolved before the Origin of Jawed Vertebrates. PLoS ONE. 2012;13(2):e31950. doi: 10.1371/journal.pone.0031950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LFC, Wilson J, Goncalves O, Galante-Oliveira S, Rocha E, Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol Biol. 2011;13(1):132. doi: 10.1186/1471-2148-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Kang M-J, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of Rat acyl-CoA synthetase 3. J Biol Chem. 1996;13(28):16748–16752. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- Kang M-J, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci. 1997;13(7):2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa E, Iijima H, Suzuki T, Sasano H, Sato H, Kamataki A, Nagura H, Kang M-J, Fujino T, Suzuki H. et al. A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J Biochem. 1998;13(3):679–685. doi: 10.1093/oxfordjournals.jbchem.a022165. [DOI] [PubMed] [Google Scholar]
- Soupene E, Dinh N, Siliakus M, Kuypers F. Activity of the acyl-CoA synthetase ACSL6 isoforms: role of the fatty acid Gate-domains. BMC Biochem. 2010;13(1):18. doi: 10.1186/1471-2091-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;13(15):1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2009;13(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;13(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc Gateway Comput Environ Workshop (GCE) 2010. pp. 1–8.
- Rozen S, Skaletsky H. Primer3 On the WWW for general users and for biologist programmers. 1999. pp. 365–386. [DOI] [PubMed]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2010;13(suppl 1):D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;13(2):295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
tBlastn search of ACSLlike sequences in Transcriptomic contigs.
Partial Acsl gene annotations in green spotted pufferfish.
NCBI accession numbers and Ensembl gene ID.
Synteny maps of Zebrafish ACSL 3R duplicates.
Xenopus tropicalis Acsl4 and Acsl3 corresponding location in human.