Abstract
Background
Deciphering the genetic structure of Arabidopsis thaliana diversity across its geographic range provides the bases for elucidating the demographic history of this model plant. Despite the unique A. thaliana genomic resources currently available, its history in North Africa, the extreme southern limit in the biodiversity hotspot of the Mediterranean Basin, remains virtually unknown.
Results
To approach A. thaliana evolutionary history in North Africa, we have analysed the genetic diversity and structure of 151 individuals collected from 20 populations distributed across Morocco. Genotyping of 249 genome-wide SNPs indicated that Morocco contains substantially lower diversity than most analyzed world regions. However, IBD, STRUCTURE and PCA clustering analyses showed that genetic variation is strongly geographically structured. We also determined the genetic relationships between Morocco and the closest European region, the Iberian Peninsula, by analyses of 201 populations from both regions genotyped with the same SNPs. These analyses detected four genetic groups, but all Moroccan accessions belonged to a common Iberian/Moroccan cluster that appeared highly differentiated from the remaining groups. Thus, we identified a genetic lineage with an isolated demographic history in the south-western Mediterranean region. The existence of this lineage was further supported by the study of several flowering genes and traits, which also found Moroccan accessions similar to the same Iberian group. Nevertheless, genetic diversity for neutral SNPs and flowering genes was higher in Moroccan than in Iberian populations of this lineage. Furthermore, we analyzed the genetic relationships between Morocco and other world regions by joint analyses of a worldwide collection of 337 accessions, which detected an additional weak relationship between North Africa and Asia.
Conclusions
The patterns of genetic diversity and structure of A. thaliana in Morocco show that North Africa is part of the species native range and support the occurrence of a glacial refugium in the Atlas Mountains. In addition, the identification of a genetic lineage specific of Morocco and the Iberian Peninsula indicates that the Strait of Gibraltar has been an A. thaliana migration route between Europe and Africa. Finally, the genetic relationship between Morocco and Asia suggests another migration route connecting north-western Africa and Asia.
Keywords: Arabidopsis thaliana, Population genetics, Natural variation, Genetic diversity, Genetic structure, Demographic history, North Africa, Mediterranean Basin, Glacial refugium/refugia
Background
Arabidopsis thaliana is a wild, annual, self-fertilizing plant with a broad geographic range as a native species in the Eurasian continent [1,2]. In the past decade, this species has become the main model plant, not only for molecular biology studies [3] but also for addressing the ecological and evolutionary bases of plant adaptation [4-6]. Deciphering the genetic structure of A. thaliana diversity across its geographic range is now a major aim because it explains its current ecological distribution, it reflects its demographic history, and it enables the precise design and analysis of experimental populations used to determine the molecular mechanisms of adaptive traits [6-8].
Currently, more than 6000 wild genotypes (accessions) of A. thaliana from different world regions have been collected. Studies of the genetic diversity and structure, at global (worldwide) and regional scales, have proposed several major events in A. thaliana demographic history in Europe [9-14]. In particular, high diversity has been described in the Mediterranean Peninsulas compared to Central and northern Europe, hence supporting the existence of multiple Pleistocene glacial refugia in southern Europe [15,16]. Further haplotype network analyses have led to the tentative suggestion of the Caucasus region as the ancestral A. thaliana centre of origin [15]. However, the largest diversity has been found in the Iberian Peninsula, whose strong geographic structure has prompted the hypothesis of multiple Iberian glacial refugia with differential contribution to the colonization of Europe [16]. Various global genetic analyses have identified two main postglacial colonization routes of Europe, from at least two refugia [9,12,16,17]. A glacial refugium in the Iberian Peninsula has been proposed to contribute to a west–east colonization of western and northern Europe, while an Asian refugium was likely the source for an east–west colonization of eastern and northern Europe. In agreement with this view, genetic structure analyses of northern European populations have detected several differentiated clusters, which support multiple sources of postglacial colonization [18-20]. In addition, a few studies have addressed A. thaliana history in East Asia and in regions outside the Eurasian continent. Nuclear and chloroplast analyses of A. thaliana populations from China have also shown substantial genetic variation and geographic structure, hence suggesting a rapid A. thaliana west–east expansion from Central Asia [21,22]. Moreover, genetic studies of North American and Japanese populations have detected no or weak geographic patterns, which indicate a very recent colonization from multiple sources [13,23,24].
Despite the unique resources available for A. thaliana, the scarcity of accessions from many regions located outside of Europe has hampered our knowledge of its demographic history in Africa and Asia. For instance, the centre of origin of A. thaliana is still unknown, since conclusive evidence for one of the two classical hypothetical locations, Europe-North Africa and Middle Asia, requires further samples and studies from those regions [2,15]. In particular, North Africa (from Morocco to Libya) is the extreme southern limit of A. thaliana distribution in the Mediterranean Basin, and it is assumed to be part of the native range [1,2]. However, the history of A. thaliana in this region remains virtually ignored since less than a handful of African accessions have been studied until now [10,14,15].
The relevance of northern Africa in structuring biodiversity, at global and regional scales, has been well documented for several plant species (reviewed in [25]). This is determined mainly by the unique biogeographic location of this region, which is part of one of the world’s largest biodiversity hotspot, the Mediterrranean Basin, and it contains migration routes from Europe, Africa and Asia [25,26]. A small number of phylogeographical and palaeoecological studies have identified several Pleistocene glacial refugia for perennial and annual plants in the African side of the Mediterranean Basin [25]. In addition, the central-marginal hypothesis for the geographic distribution of diversity across species’ ranges predicts lower genetic diversity and higher genetic differentiation in peripheral than central populations (reviewed in [27]). However, most studies supporting these expectations, including those of A. thaliana, have focused in the northern limit of species from the North hemisphere [27-29]. Therefore, additional studies estimating the genetic diversity and structure of plant species, especially herb plants, in southern limits like North Africa are needed to evaluate current hypotheses explaining plant evolutionary histories and their actual geographic distributions.
In this study we have addressed the genetic diversity and structure of A. thaliana in North Africa by developing a new collection of accessions derived from populations distributed throughout the species range in Morocco. In particular, we aim to determine, first, if North Africa has been recently colonized, or if it might contain genetically isolated and structured populations supporting the occurrence of A. thaliana as a native species. Second, we aim to detect genetic relationships between Morocco and other geographic regions, which might provide information about the demographic history in North Africa. To approach these questions, we have analysed this Moroccan collection together with a collection from the nearest European region, the Iberian Peninsula, and a worldwide collection. Population genetic analyses of genome-wide neutral markers and potentially adaptive flowering genes and traits, demonstrate a shared history in southern Europe and northern Africa. Together, these analyses identified the Strait of Gibraltar as a migration route between Europe and Africa, and suggest that the Atlas area was a Pleistocene glacial refugium for A. thaliana.
Results
Genetic diversity and structure in North Africa
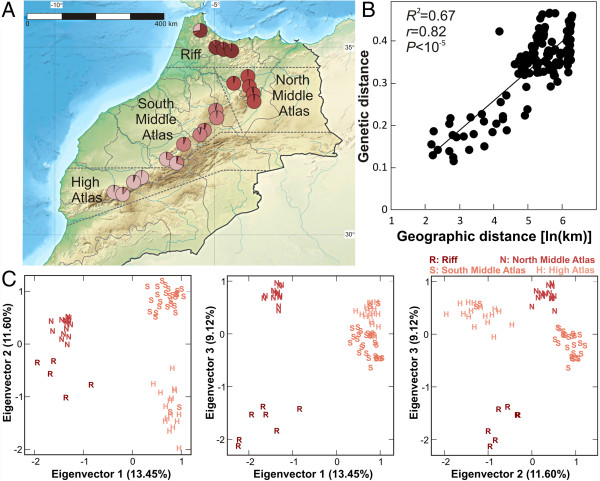
To determine the genetic diversity of A. thaliana in North Africa we sampled 151 individuals in 20 populations distributed across the major mountain ranges of Morocco (Figure 1A, Additional file 1: Figure S1, Additional file 2: Table S1). Genotyping of this collection with a genome-wide set of 249 presumably neutral single nucleotide polymorphisms (SNPs) detected a total of 65 different haplotypes. No haplotype was shared by several populations. Moreover, substantial variation was found among populations for the number of haplotypes per population, because two populations carried a single haplotype while two others contained seven or more genotypes (Table 1). Similar variation was detected for gene diversity (H S ) and allelic richness (R S ) per population, which showed up to a tenfold range of variation among populations. The amount of genetic variation per population displayed a weak geographic pattern since the percentage of polymorphic loci (PL) correlated negatively with latitude (r = −0.46; P = 0.046), and R S showed similar negative but marginal correlation (r = −0.4; P = 0.09). In agreement, four geographic subregions established according to latitude differed slightly for PL, R S and H S (0.08 > P > 0.04), populations from the Riff containing less genetic variation than populations from the High Atlas (Table 1).
Figure 1.
Genetic and geographic structure of A. thaliana in Morocco. A) Geographic location of Moroccan populations and their genetic relationships inferred with STRUCTURE. Each population is depicted as a pie chart quantifying membership proportions for four genetic clusters (K = 4). B) Correlation between geographic and genetic distances (average proportion of allelic differences) among populations. C) Scatter plots displaying paired combinations of the first three eigenvectors estimated by PCA of Moroccan genotypes. The four main groups detected by clustering analysis of these principal components are shown with the same colors as the STRUCTURE clusters of A.
Table 1.
Genetic diversity of A. thaliana populations from Morocco
| Population | Subregion 1 | N | N H | PL | H S | R S | R P |
|---|---|---|---|---|---|---|---|
| Zin |
1 |
9 |
2 |
14 |
0.031±0.007 |
1.03±0.01 |
0.026±0.013 |
| Bab |
1 |
4 |
2 |
3 |
0.015±0.008 |
1.02±0.01 |
0.008±0.008 |
| Bbe |
1 |
10 |
1 |
0 |
0±0 |
1±0 |
0.003±0.003 |
| Ket |
1 |
8 |
2 |
5 |
0.018±0.007 |
1.02±0.01 |
0.008±0.008 |
|
Mean |
1 |
7.8 |
1.8 |
5 |
0.016±0.006 |
1.02±0.01 |
0.011±0.008 |
| Taz |
2 |
6 |
4 |
12 |
0.052±0.013 |
1.06±0.01 |
0.002±0.002 |
| Tah |
2 |
10 |
2 |
16 |
0.078±0.016 |
1.08±0.02 |
0±0 |
| Bba |
2 |
3 |
3 |
14 |
0.062±0.014 |
1.07±0.02 |
0.005±0.005 |
| Meh |
2 |
10 |
3 |
13 |
0.027±0.006 |
1.03±0.01 |
0±0 |
| Tiz |
2 |
10 |
2 |
21 |
0.104±0.018 |
1.11±0.02 |
0±0 |
|
Mean |
2 |
7.8 |
2.8 |
15 |
0.065±0.011 |
1.07±0.02 |
0.001±0.001 |
| Ifr |
3 |
7 |
5 |
24 |
0.09±0.0150 |
1.1±0.02 |
0.007±0.006 |
| Azr |
3 |
10 |
6 |
30 |
0.109±0.016 |
1.11±0.02 |
0.007±0.004 |
| Agl |
3 |
8 |
7 |
23 |
0.084±0.014 |
1.09±0.02 |
0.002±0.002 |
| Khe |
3 |
10 |
2 |
18 |
0.057±0.011 |
1.06±0.01 |
0.001±0.001 |
| Elk |
3 |
10 |
6 |
13 |
0.039±0.009 |
1.04±0.01 |
0.009±0.008 |
| Oua |
3 |
1 |
1 |
- |
- |
- |
- |
| Til |
3 |
10 |
1 |
0 |
0±0 |
1±0 |
0±0 |
|
Mean |
3 |
9.2 |
4.5 |
18 |
0.063±0.013 |
1.07±0.02 |
0.004±0.004 |
| Elh |
4 |
10 |
10 |
42 |
0.15±0.0170 |
1.16±0.02 |
0.012±0.006 |
| Arb |
4 |
2 |
2 |
18 |
0.092±0.017 |
1.12±0.02 |
0±0 |
| Ait |
4 |
6 |
2 |
17 |
0.048±0.009 |
1.05±0.01 |
0.001±0.001 |
| Set |
4 |
7 |
2 |
20 |
0.049±0.009 |
1.05±0.01 |
0±0 |
| Mean | 4 | 6.25 | 4 | 24 | 0.085±0.013 | 1.09±0.01 | 0.003±0.002 |
The following parameters are shown: number of individuals (N), number of haplotypes (N H ), percentage of polymorphic loci (PL), gene diversity (H S ), allelic richness (R S ) and private allelic richness (R P ) per locus. Populations are arranged according to latitude, from north to south, and classified into four subregions as in Figure 1. H S , R S and R P are mean values ± SE.
1: 1: Riff; 2: North Middle Atlas; 3: South Middle Atlas; 4: High Atlas.
The genetic relationship among Moroccan populations was first analysed by constructing Neighbor-joining (NJ) trees of the 65 haplotypes (Additional file 1: Figure S2A), which showed that genotypes are genetically closer within than among populations. AMOVA analyses indicated that populations were highly differentiated, with 81.6% of the genetic variation appearing distributed among populations (F ST range: 0.26-1) and only 18.4% occurring within populations (P < 0.001). Mantel tests of pair-wise geographic distances and genetic distances, measured as F ST or as percentage of allele differences among populations, detected highly significant correlations (0.59 < r < 0.82; P < 0.001; Figure 1B) accounting for up to 67% of the variance. Therefore, A. thaliana genetic variation showed a continuous isolation by distance (IBD) geographic pattern across Morocco.
To further determine the genetic structure of A. thaliana in Morocco we analysed the different haplotypes using two clustering approaches, STRUCTURE and principal component (PCA) analyses. STRUCTURE analysis detected four major clusters that closely corresponded to the four geographic subregions (Figure 1, Additional file 1: Figure S2B). Clustering analyses using the first three principal components, which explained 13.5, 11.6 and 9.1% of the genetic variation, identified precisely the same four groups (Figure 1C, Additional file 2: Table S2). These genetic groups were also supported by NJ analyses since the four main NJ clades corresponded to such clusters (Additional file 1: Figure S2A). AMOVA analyses indicated that 43.1% of the genetic variation differentiates the four groups, whereas 40.2% differentiates populations within groups. Thus, A. thaliana genetic variation for neutral markers appears strongly geographically structured in Morocco.
Genetic diversity and structure in the south-western Mediterranean region
To establish the genetic relationships among A. thaliana populations in the south-western Mediterranean range, we analysed the 20 Moroccan populations together with a set of 181 previously sampled Iberian populations [30]. Genotyping of a single random individual from each population with the same genome-wide set of SNPs showed that all 201 samples were different genotypes. Gene diversity and allelic richness values indicated that A. thaliana diversity in Morocco was considerably lower than in the Iberian Peninsula (Table 2). Mantel tests of correlations between geographic and genetic distances detected significantly higher correlation coefficients and slopes of linear regressions in Morocco than in Iberia (P = 0.02; Additional file 2: Figure S3). However, the difference between these slopes was mainly determined by the larger genetic distances between geographically closer populations of Iberia. Therefore, the apparently stronger IBD pattern detected in Morocco might reflect higher Iberian diversity or a more complex Iberian geographic structure at lower spatial scales.
Table 2.
Genetic diversity of A. thaliana in different world regions
| World region | N | PL | H S | R S | R P |
|---|---|---|---|---|---|
| North America |
6 |
66 |
0.266±0.013 |
1.66±0.03 |
0±0 |
| Morocco |
20 |
54 |
0.192±0.013 |
1.50±0.03 |
0±0 |
| Iberia |
181 |
95 |
0.315±0.011 |
1.78±0.02 |
0.015±0.004 |
| British Isles |
11 |
81 |
0.299±0.012 |
1.77±0.02 |
0.005±0.003 |
| Central Europe |
51 |
92 |
0.324±0.011 |
1.79±0.02 |
0.003±0.001 |
| Fennoscandia |
13 |
79 |
0.308±0.012 |
1.76±0.03 |
0.004±0.003 |
| South Europe |
7 |
72 |
0.284±0.013 |
1.72±0.03 |
0.003±0.003 |
| East Europe |
6 |
63 |
0.247±0.013 |
1.63±0.03 |
0±0 |
| East Asia |
8 |
73 |
0.279±0.012 |
1.72±0.03 |
0.001±0.000 |
| Caucasus |
10 |
71 |
0.276±0.013 |
1.70±0.03 |
0.005±0.003 |
| Central Asia |
8 |
79 |
0.298±0.011 |
1.78±0.03 |
0.007±0.005 |
| Japan | 12 | 73 | 0.277±0.012 | 1.70±0.03 | 0.002±0.001 |
The following parameters are shown for world regions arranged from west to east: number of populations (N), percentage of polymorphic loci (PL), gene diversity (H S ), allelic richness (R S ) and private allelic richness (R P ) per locus. H S , R S and R P are mean values ± SE.
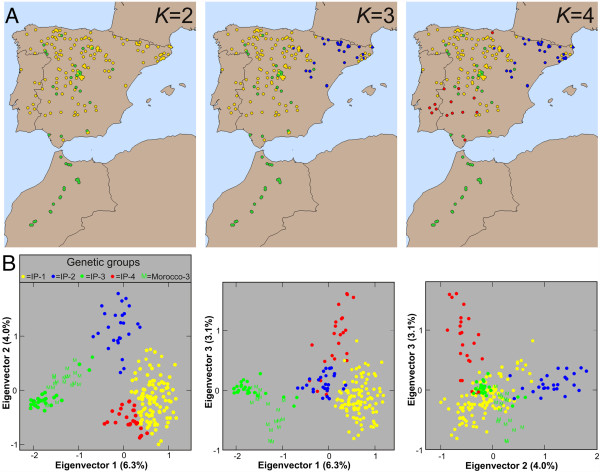
The genetic structure in this region was further analysed with STRUCTURE and PCA clustering methods. STRUCTURE analyses detected four different genetic groups showing distinct geographic distribution (Figure 2A, Additional file 1: Figure S4). Clusters were numbered as described in a previous study with a lower number of populations and SNPs [16], groups 1, 2 and 4 appearing distributed mainly in NW, NE and SW of the Iberian Peninsula. By contrast, group 3 displayed a broader geographic distribution because it included 24 Iberian accessions scattered throughout this region, as well as all Moroccan genotypes. The first three principal components detected by PCA accounted for 6.5, 4.1 and 3.1% of the genetic variation. Clustering analyses of these three components identified very similar groups to STRUCTURE (Figure 2B) since 92% of the accessions were assigned to the same group based on the major cluster membership of each accession (Additional file 2: Table S2). AMOVA analyses showed an average F ST differentiation among Iberian genetic groups of 0.28. However, group 3 displayed the largest differentiations (0.28 < F ST < 0.45; P < 0.001) in agreement with the early distinction of this cluster in the first (K = 2) analysis of STRUCTURE (Figure 2A). In addition, low differentiation (F ST = 0.24) and low mean allelic differences (12.3%) were estimated between Iberian and Moroccan groups of accessions belonging to cluster 3. Genetic diversity measurements also showed that group 3 contained the lowest amount of variation, although Morocco accessions contained higher diversity than Iberian accessions of this group (Table 3). Together, these analyses indicated that A. thaliana populations from Morocco are genetically related to populations assigned to group 3 from the Iberian Peninsula.
Figure 2.
Genetic and geographic structure of A. thaliana in the south-western Mediterranean region. A) Genetic group assignment of Moroccan and Iberian accessions based on majority membership proportions inferred with STRUCTURE, for K = 2, K = 3 and K = 4. B) Scatter plots displaying pair combinations of the first three eigenvectors estimated by PCA of Moroccan and Iberian genotypes. The four main groups detected by clustering analysis of these principal components are shown with the same colors as STRUCTURE clusters of A, except that genetic group 3 is divided into Iberia (IP-3) and Morocco (Morocco-3).
Table 3.
Genetic diversity in different A. thaliana clusters detected in Morocco and Iberia, and in worldwide analyses
| Analysis | Genetic cluster 1 | N | PL | H S | R S | R P |
|---|---|---|---|---|---|---|
| Morocco and Iberia |
Morocco |
20 |
57 |
0.202±0.013 |
1.52±0.03 |
0.008±0.003 |
| |
1 |
112 |
98 |
0.321±0.011 |
1.81±0.02 |
0.051±0.009 |
| |
2 |
31 |
85 |
0.273±0.012 |
1.72±0.02 |
0.041±0.009 |
| |
3 |
24 |
58 |
0.145±0.011 |
1.45±0.03 |
0.005±0.003 |
| |
4 |
14 |
64 |
0.195±0.012 |
1.57±0.03 |
0.015±0.005 |
| Worldwide |
1 |
159 |
95 |
0.311±0.011 |
1.78±0.02 |
0.016±0.004 |
| |
2 |
34 |
83 |
0.271±0.012 |
1.71±0.02 |
0.029±0.008 |
| |
3 |
28 |
63 |
0.147±0.010 |
1.46±0.03 |
0.003±0.002 |
| |
4 |
14 |
67 |
0.203±0.012 |
1.59±0.03 |
0.008±0.004 |
| |
5 |
68 |
93 |
0.329±0.011 |
1.81±0.02 |
0.033±0.007 |
| 6 | 34 | 73 | 0.235±0.012 | 1.61±0.03 | 0.003±0.002 |
The following parameters are shown for the genetic groups detected by STRUCTURE analyses of Iberian and Moroccan accessions and of all worldwide accessions: number of populations (N), percentage of polymorphic loci (PL), gene diversity (H S ), allelic richness (R S ) and private allelic richness (R P ) per locus. H S , R S and R P are mean values ± SE.
1: Genetic group 3 is divided into Moroccan and Iberian accessions in the top analysis.
Diversity and differentiation for flowering genes, traits and climatic variables in the south-western Mediterranean region
The genetic diversity of A. thaliana in the south-western Mediterranean range was also analysed for the nucleotide variation of four well-known flowering genes that are likely involved in adaptation (Methods). Overall, flowering genes showed larger diversity in Iberia than Morocco as estimated by the private allelic richness (Table 4). Genes showed similar nucleotide diversity patterns in Morocco and Iberia, with FRI and FLC displaying low silent diversities and CRY2 and PHYC high values (Table 4). However, analyses of the major known functional polymorphisms for these genes showed a lower frequency of predicted FRI loss-of-function mutations in Moroccan than Iberian accessions. In addition, as previously reported in Iberia, only one of the two major European haplogroups described in FLC was detected in Morocco (Table 4). Furthermore, the two major European haplogroups of CRY2 and PHYC were also found in Morocco at similar low frequencies to the Iberian Peninsula (Table 4). Interestingly, a new CRY2 haplogroup named as CRY2-C, differentiated by three aminoacid substitutions, was found at high frequency in Morocco (35%). This CRY2 haplogroup appeared at very low frequency in Iberia (1.1%) and explained the larger Moroccan CRY2 variation. Therefore, A. thaliana populations from Morocco showed a similar amount and pattern of diversity to the Iberian Peninsula for FLC and PHYC. However, Moroccan populations displayed higher and lower amounts of potentially functional diversity at CRY2 and FRI, respectively.
Table 4.
Genetic diversity of flowering genes in Morocco and Iberia
| Gene | Sequence length 1 | Genetic group 2 | Number of populations | Number of polymorphisms 3 | N H 3 | H S 3 | R S 3 | R P 3 | π silent | Haplogrups or truncations 4 | Haplogrup frequency (%) 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
FRI |
3470 |
M |
20 |
24 |
16 |
0.049±0.011 |
1.18±0.03 |
0.092±0.022 |
0.0023 |
Yes |
5 |
| |
|
IP |
178 |
87 |
97 |
0.058±0.009 |
1.24±0.03 |
0.181±0.023 |
0.0023 |
Yes |
12.2 |
| |
|
IP-1 |
111 |
69 |
54 |
0.059±0.009 |
1.24±0.03 |
0.079±0.011 |
0.0023 |
Yes |
17.9 |
| |
|
IP-2 |
29 |
26 |
14 |
0.029±0.007 |
1.13±0.02 |
0.028±0.010 |
0.0010 |
Yes |
6.5 |
| |
|
IP-3 |
24 |
35 |
22 |
0.055±0.011 |
1.21±0.03 |
0.098±0.018 |
0.0026 |
No |
0 |
| |
|
IP-4 |
14 |
19 |
7 |
0.046±0.011 |
1.15±0.03 |
0.032±0.014 |
0.0019 |
No |
0 |
|
FLC |
451 |
M |
20 |
4 |
5 |
0.126±0.058 |
1.31±0.13 |
0.314±0.135 |
0.0033 |
A |
100 |
| |
|
IP |
181 |
8 |
18 |
0.104±0.035 |
1.38±0.11 |
0.378±0.106 |
0.0039 |
A |
100 |
| |
|
IP-1 |
112 |
7 |
8 |
0.081±0.032 |
1.31±0.11 |
0.144±0.092 |
0.0032 |
A |
100 |
| |
|
IP-2 |
31 |
3 |
3 |
0.073±0.040 |
1.22±0.12 |
0.064±0.041 |
0.0022 |
A |
100 |
| |
|
IP-3 |
24 |
4 |
4 |
0.047±0.028 |
1.19±0.09 |
0.083±0.072 |
0.0025 |
A |
100 |
| |
|
IP-4 |
14 |
2 |
3 |
0.049±0.039 |
1.14±0.10 |
0.106±0.078 |
0.0027 |
A |
100 |
|
CRY2 |
1529 |
M |
20 |
8 |
6 |
0.104±0.036 |
1.29±0.09 |
0.210±0.078 |
0.0229 |
A, B |
B=5 |
| |
|
IP |
181 |
19 |
24 |
0.054±0.023 |
1.20±0.06 |
0.139±0.047 |
0.0145 |
A, B |
B=9.9 |
| |
|
IP-1 |
112 |
10 |
8 |
0.037±0.021 |
1.13±0.05 |
0.029±0.011 |
0.0075 |
A, B |
B=3.6 |
| |
|
IP-2 |
31 |
5 |
5 |
0.033±0.022 |
1.10±0.05 |
0.031±0.021 |
0.0052 |
A, B |
B=3.2 |
| |
|
IP-3 |
24 |
7 |
8 |
0.052±0.024 |
1.18±0.07 |
0.162±0.064 |
0.0023 |
A |
100 |
| |
|
IP-4 |
14 |
4 |
3 |
0.052±0.025 |
1.16±0.08 |
0.051±0.044 |
0.0229 |
A, B |
B=85.7 |
|
PHYC |
868 |
M |
19 |
6 |
7 |
0.065±0.033 |
1.24±0.11 |
0.121±0.059 |
0.0168 |
Ler, Col |
Col=5 |
| |
|
IP |
160 |
18 |
34 |
0.062±0.024 |
1.23±0.06 |
0.141±0.036 |
0.0106 |
Ler, Col |
Col=7.4 |
| |
|
IP-1 |
102 |
13 |
15 |
0.061±0.024 |
1.23±0.06 |
0.090±0.032 |
0.0092 |
Ler, Col |
Col=6.9 |
| |
|
IP-2 |
30 |
3 |
4 |
0.026±0.018 |
1.09±0.05 |
0.032±0.029 |
0.0039 |
Ler |
100 |
| |
|
IP-3 |
16 |
8 |
9 |
0.070±0.028 |
1.25±0.08 |
0.100±0.045 |
0.0158 |
Ler, Col |
Col=16.7 |
| IP-4 | 12 | 5 | 6 | 0.062±0.028 | 1.20±0.11 | 0.035±0.024 | 0.0148 | Ler, Col | Col=14.3 |
For each gene, the following parameters are shown for Morocco, the Iberian Peninsula and the four Iberian genetic groups: number of polymorphisms, number of haplotypes (N H ), gene diversity (H S ), allelic richness (R S ), private allelic richness (R P ), silent nucleotide diversity (π silent ), and the type and frequency of major haplogroups or functional alleles. H S , R S and R P are mean values ± SE.
1: Alignment length of all sequences.
2: Genetic groups correspond to the four main Iberian groups detected with STRUCTURE; IP: Iberian Peninsula; M: Morocco.
3: Only polymorphisms that are not in complete LD are included.
4: Accessions are classified as functional and loss-of-function truncations for FRI[30] or as the two major functional haplogroups described for FLC, CRY2 and PHYC[54-56]. Frequencies are given for truncations and for the minor frequency haplogroups.
The genetic relationships between Morocco and each of the four Iberian genetic groups detected with neutral markers were also analysed using flowering gene sequences. Gene diversity and allelic richness of Moroccan populations were most similar to those of Iberian genetic group 3, although Moroccan diversities were mostly higher (Table 4). In addition, in contrast to Morocco, Iberian group 3 was not segregating for FRI loss-of-function alleles and for the major CRY2 haplogroups (Table 4). Pair-wise analyses of F ST values between Morocco and the four Iberian genetic groups showed that group 3 displays the lowest genetic differentiation from Moroccan populations for most genes (0.04 < F ST < 0.27; Additional file 2: Table S3). Hence, A. thaliana populations from Morocco appeared also most similar to Iberian genetic group 3 for flowering genes.
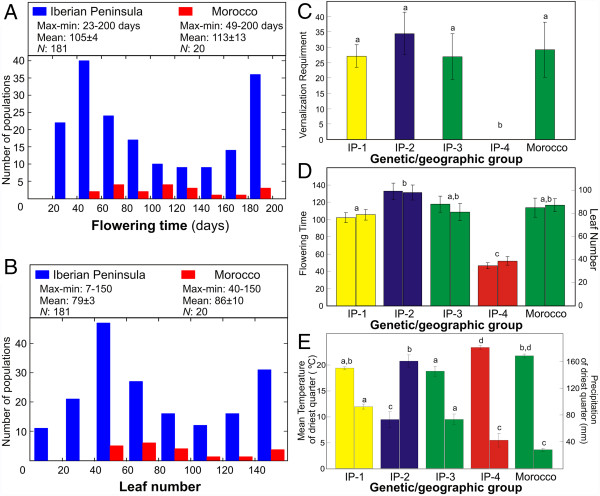
The genetic differentiation between Morocco and the Iberian Peninsula was further analysed by measuring three quantitative traits related with flowering induction: vernalization requirement, flowering time and leaf number (Figure 3). Four of the 20 Moroccan accessions (20%) showed an obligate vernalization requirement because they did not flower at all without a low temperature treatment but they flowered after two months at 4°C. This proportion was not significantly different from the 12% frequency observed in Iberia. However, the range of variation among Moroccan accessions for flowering time (49–200 days) and leaf number (40–150 leaves) was significantly smaller than the Iberian variation (23–200 days and 7–150 leaves) (Figure 3A and B). In particular, no Moroccan accession showed an extreme early flowering behaviour, in comparison with 17% of Iberian samples flowering in less than 45 days and with fewer than 32 leaves. Comparisons of flowering traits between Morocco and the four Iberian genetic groups showed that Moroccan accessions were most similar to Iberian group 3, although differences were only significantly different between Morocco and genetic group 4 (Figure 3C and D). In agreement, the genetic differentiation between Morocco and the rest of groups estimated by pair-wise Q ST values of flowering traits, varied between 0 for groups 1 or 3, and 0.66 for group 4 (P < 0.001).
Figure 3.
A. thaliana flowering and climatic variation in the south-western Mediterranean region. A) Flowering time and B) leaf number of Moroccan and Iberian accessions. C) Obligate vernalization requirement, D) flowering time (left column) and leaf number (right column), and E) mean temperature (left column) and precipitation (right column) of the driest quarter, in the four genetic groups detected with STRUCTURE in the Iberian Peninsula and in Morocco. In C-E, bars are the mean ± SE of the accessions assigned to each genetic group, except that group 3 is divided in Iberia (IP-3) and Morocco. Shared or different letters above bars indicate non-significant and significant differences between groups (P < 0.05) according to Tukey tests. Bar colors are similar to group colors shown in Figure 2.
Since A. thaliana flowering variation has been involved in climatic adaptation [30] we analyzed if the similarities observed between Morocco and Iberia for flowering traits and genes might be determined by the climatic environment of local populations. Comparisons of climatic variables showed that, on average, Moroccan populations are exposed to higher mean annual temperature (Morocco = 13.5 ± 18.7°C; Iberia = 11.7 ± 26.4°C) and lower annual precipitation than Iberian populations (Morocco = 626 ± 204 mm; Iberia = 652 ± 208 mm; Additional file 2: Table S1). Further comparisons of climatic variables between Morocco and the four Iberian genetic groups showed that the climatic distribution of Moroccan populations was most similar to that of Iberian genetic group 4 distributed in south-western Iberia (Figure 3E). Both, Moroccan populations and Iberian genetic group 4, appeared significantly associated with high temperatures and low summer precipitation. Therefore, the genetic similarity between Morocco and Iberia for flowering genes and traits do not seem to reflect shared adaptations to a similar climatic environment.
Genetic diversity and structure at a global scale
A. thaliana genetic relationships between the south-western Mediterranean region and the rest of world was also studied by analyzing the 201 Moroccan and Iberian accessions together with 136 genotypes representing populations from eleven additional world regions (Additional file 1: Figure S5). Genotyping of these 337 accessions with the 249 segregating SNPs showed that Morocco and the Iberian Peninsula contained the lowest and highest A. thaliana diversity, respectively (Table 2).
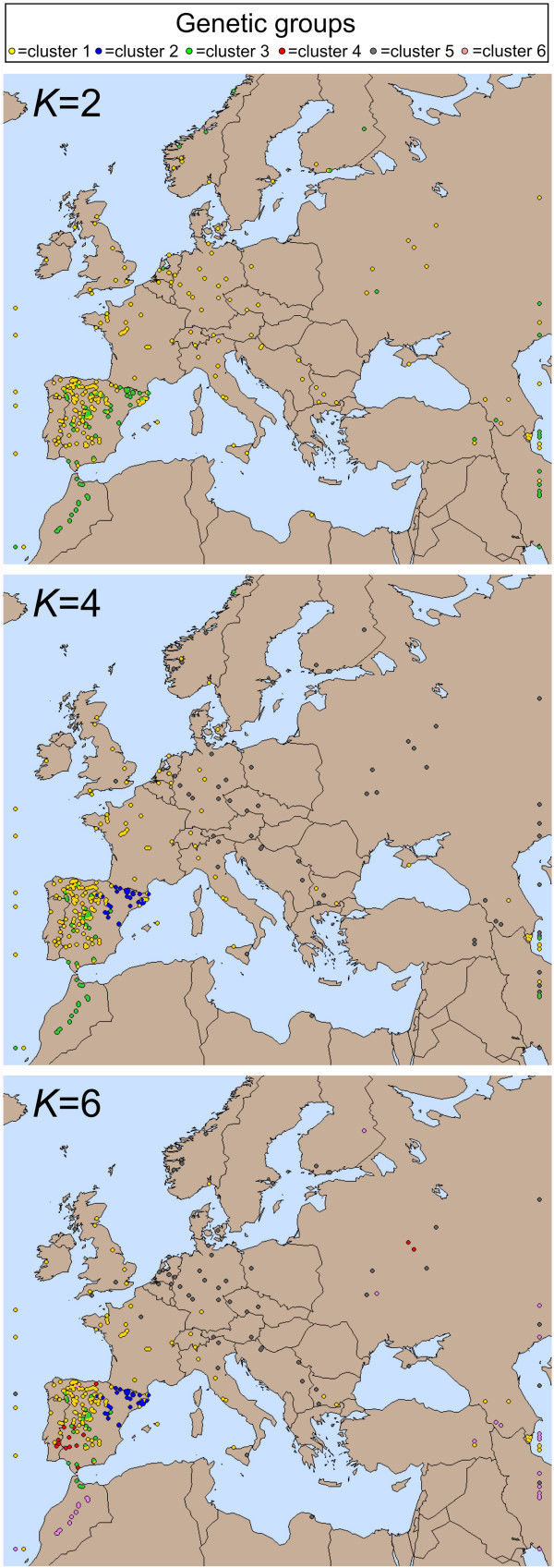
Analysis of the genetic structure of this A. thaliana worldwide collection using STRUCTURE detected six different clusters with distinct geographic patterns (Figure 4, Additional file 1: Figure S6). PCA analysis of the same genotypes found that the first five principal components accounted for 5.0 to 2.0% of the genetic variation. The subsequent clustering of these components detected similar genetic clusters to STRUCTURE (Additional file 1: Figure S7) since 80% of the accessions were assigned to equivalent genetic groups (Additional file 2: Table S2). Four of the genetic groups closely corresponded to the Iberian clusters identified in the previous analysis of the south-western Mediterranean region (Figure 2A, Figure 4). However, two additional clusters (5 and 6) were detected as distributed mainly in eastern Europe, Asia and Africa. Groups 1 and 5 showed the widest geographic distributions in Eurasia. Analysis of the average regional proportions of these groups (Additional file 1: Figure S6) showed that group 1 displays the highest frequency in Iberia and British Isles, and its Eurasian frequency decreases in a west–east direction. By contrast, group 5 displayed high frequency in Central, North and East Europe but it was nearly absent in Iberia and Morocco. Groups 6 and 4 showed an intermediate distribution, with group 6 appearing mainly in Asia and North Africa, while group 4 showed moderate frequency in Asia and East Europe. Groups 2 and 3 displayed restricted geographic distributions since group 3 was present almost exclusively in North Africa and Iberia, and group 2 mostly in the Iberian Peninsula.
Figure 4.
Genetic and geographic structure of A. thaliana at a global scale. Genetic group assignment of accessions is based on majority membership proportions inferred with STRUCTURE, for K = 2, K = 4 and K = 6. Accessions located outside the Eurasian map are shown at their corresponding latitude in the left (American accessions) and right (Asian accessions) edges.
F ST analyses among the six genetic groups rendered an average genetic differentiation of 0.22, but again group 3 displayed the largest differentiations from the rest of clusters (0.24 < F ST < 0.42; P < 0.001) and the lowest genetic diversity (Table 3). In agreement, group 3 was also the first cluster differentiated in the K = 2 analysis of STRUCTURE, and an identical cluster was detected in subsequent analyses from K = 2 to K = 4. This group contained the same 24 Iberian accessions and all 20 Moroccan accessions assigned to group 3 in the previous Mediterranean analysis. However, three and 16 Moroccan accessions were assigned to an Asian group in STRUCTURE analyses with K = 5 and K = 6, respectively. Thus, global analyses suggest that Moroccan populations of A. thaliana are closely related to those from Iberia but also weakly related to Asian populations.
Discussion
Low genetic diversity and strong geographic structure in Morocco reflect a homogeneous native history of A. thaliana in North Africa
In this study we have systematically sampled and analysed A. thaliana populations across Morocco, the southern Mediterranean limit of the species range. Populations were typically found at altitudes between 1000 and 2000 m.a.s.l. in the High Atlas, Middle Atlas and Riff mountains where natural vegetation still predominates (Additional file 1: Figure S1). This region contained less genetic variation than other world regions, for neutral markers, flowering genes and potentially adaptive quantitative traits. In particular, no extreme early flowering accession was found in Morocco, which was in agreement with the association of such flowering behavior with low altitude [30]. The amount of variation detected within Moroccan local populations was lower than that described for southern European Peninsulas proposed as Mediterranean glacial refugia [15,16]. Instead, the average intrapopulation variation of Morocco was similar to that estimated in Scandinavia, the northern limit of the native range reached after postglacial colonization [18,20], or in Japan, a non-native region recently colonized [23].
The strong A. thaliana geographic structure found in Morocco at different spatial scales supports an old history as part of the native range. First, populations appeared highly differentiated, the average F ST value of 0.81 estimated among Moroccan populations being similar to that reported in the northern limit of the species native range [18,20]. Second, in contrast to recently colonized regions such as Japan and North America [23,24] the strong pattern of isolation by distance across Morocco indicates limited geographic seed dispersal from local populations throughout Morocco. However, discrete genetic groups that are geographically structured were also detected consistently by different clustering methods. This indicates the presence of some spatial discontinuities in Morocco IBD pattern, which are likely caused by geographical (physical or environmental) barriers that further limit A. thaliana dispersal and contribute to subregional differentiation in Morocco. It must be emphasized that the detection of these clusters is not the result of uneven geographic sampling, as illustrated by the assignment of two populations from the South Middle Atlas subregion, to the genetic cluster of the High Atlas populations (Figure 1). In agreement with the central-marginal hypothesis for the geographic distribution of diversity across species’ ranges [27] Morocco region appears as a trailing edge of A. thaliana’s native range characterized by little recent admixture and genetically isolated and differentiated populations. This pattern of variation is likely to be determined not only by contemporary local adaptation of populations to patches of suitable habitat that are close to the species environmental tolerance limits, but also, as discussed below, by abiotic or biotic factors that affected the demographic history of A. thaliana in the past [28,29].
A Moroccan/Iberian specific genetic lineage reveals an A. thaliana history shared between south-western Europe and North Africa
The Iberian Peninsula has been proposed as a region containing several A. thaliana refugia during the last glaciation, which have played an important role in the postglacial colonization of Europe [9,15,16]. This has been supported by the large genetic diversity of this region and the strong geographic structure reported for multiple kinds of markers and for whole-genome sequences of Iberian accessions [14,16]. In agreement with previous results, our genome-wide SNP analysis finds Iberia as the world region containing the largest diversity, which appears structured into four genetic groups with distinct geographic distributions. Several analyses consistently show that Moroccan populations are genetically related mainly to one particular Iberian group. First, Morocco populations group with Iberian genetic group 3 by clustering analyses of neutral SNP markers. Second, Moroccan populations showed similar flowering behavior to group 3 from the Iberian Peninsula, which is characterized by the absence of early flowering accessions. Third, similar frequencies are found in Morocco and Iberia for the major functional haplotypes described in Europe for flowering genes. Accordingly, Morocco displayed the lowest differentiation from Iberian genetic group 3 for flowering genes and traits, as estimated by F ST and Q ST values. Thus, we identified an A. thaliana genetic lineage with a shared demographic history in the south-western Mediterranean region.
The Moroccan/Iberian specific distribution of this lineage provides new insights into A. thaliana history in North Africa and south-western Europe. On one hand, it indicates that the Strait of Gibraltar has been an A. thaliana migration route between the European and African continents. This is in agreement with the well-documented natural history of this region, which recognizes a biodiversity hotspot consisting of two areas with partially overlapping sets of species separated by the Strait of Gibraltar [31-34]. However, our results also suggest that rather limited A. thaliana genetic flow has occurred between Morocco and Iberia because this genetic lineage shows an isolated history from the remaining Iberian genetic groups, as indicated by its low genetic diversity and its strong differentiation. In addition, the western Mediterranean distribution of this lineage seems to be predominantly determined by A. thaliana demographic history rather than by adaptation to a similar climatic environment in this region. This conclusion is supported by: i) the wide geographic distribution of this group in Iberia and Morocco; ii) the substantial diversity of group 3 for some potentially adaptive haplogroups described for flowering genes such as CRY2 and PHYC; iii) the higher climatic similarity of Moroccan populations with the geographic range of southern Iberia occupied by genetic group 4 than with that occupied by genetic group 3.
On the other hand, the strongly isolated and structured distribution of this genetic lineage in the western Mediterranean region suggests that it was maintained as a refugium during the last glaciations. As reported for several plant and animal species [25], we hypothesize that the Atlas mountain range in north-western Africa could have been a Mediterranean glacial refugium for A. thaliana during the Pleistocene. Consequently, we speculate that this genetic lineage might have colonized northern Morocco and southern Spain during the postglacial period. Latitudinal declines in genetic diversity have been associated with recent northward colonization in temperate species [28,35-37]. In agreement with these observations, a weak latitudinal cline of the intrapopulation genetic variation is detected in Morocco, populations from the Riff area containing slightly lower variation than populations from the Atlas Mountains. Furthermore, both neutral markers and flowering genes (CRY2, FLC and PHYC) showed higher private allelic richness in Moroccan than Iberian populations assigned to this genetic lineage. Interestingly, it has been described for several forest plants that colonizations from glacial refugia do not necessarily involve a gradual diversity decrease away from the source populations [29]. In contrast, complex genetic patterns of admixed populations can be generated by contributions from different refugia. Accordingly, the hypothesis of an African Atlas refugium for A. thaliana is also compatible with the large genetic diversity and complex structure found in the Iberian Peninsula, since additional A. thaliana refugia in this region probably accounted for its overall larger intrapopulation and regional variation. In fact, previous analyses of Iberian intrapopulation variation using the same set of SNPs [38] shows that populations Gra and Mar belonging to genetic group 3 have similar low diversities to Moroccan populations reported here. Nevertheless, we cannot discard that a glacial refugium of this lineage was located in Central Iberia, and that this was the source for the subsequent colonization of north-western Africa.
Genetic relationships at global scale support a potential migration route between North Africa and Asia
The worldwide genetic and geographic structures detected in this study are in agreement with the previously proposed postglacial colonization routes of Europe during the Pleistocene [9,12,14,17]. In particular, the opposite longitudinal gradients displayed by the frequencies of the two most common genetic groups (Figure 4, clusters 1 and 5) support north-western Iberian and Asian refugia contributing to the colonization of Europe. On the contrary, the restricted geographic distribution of several genetic groups suggests that other Iberian refugia, as well as the newly proposed Moroccan and/or Iberian refugium, did not contribute substantially to such colonization [16]. In addition, the genetic relationship detected between Morocco and Asia, further suggests a potential A. thaliana migration route connecting north-western Africa and Asia. Accordingly, a colonization event of Morocco is hypothesized to have occurred from Asia through this northern African route, which could predate the latest glaciations or could have occurred later during the African humid period documented between 15,000 and 5,000 years BP [39].
Conclusions
The research presented here shows that A. thaliana displays low genetic diversity in Morocco but the genetic variation is strongly geographically structured, hence supporting a native history in North Africa. In addition, Morocco appears genetically related mainly to the Iberian Peninsula, which indicates a shared demographic history between south-western Europe and North Africa. Our work illustrates the relevance of systematic population genetic analyses of unknown geographic regions to infer the history of A. thaliana. Further comparative studies including underrepresented regions from the rest of North Africa, the Middle East and Central Asia are needed to test the propossed historical hypotheses. In particular, it is likely that the speculated A. thaliana refugia trace back to different Pleistocene dates as consequence of strong climatic fluctuations and occurrence of several species retraction/expansion episodes during this period [25]. Eventually, model-based analyses of future whole-genome sequences [14,40] will enable testing of alternative historical scenarios and temporal estimations of demographic events that occurred during A. thaliana’s evolutionary history.
Methods
Plant material
Twenty A. thaliana populations were sampled and geo-referenced during 2005–2009, in the main mountain ranges of Morocco (Figure 1, Additional file 1: Figure S1, Additional file 2: Table S1). Populations were spaced at an average distance of 247 ± 156 km, with a minimum and maximum of 6.5 km and 540 km, respectively. Climatic information from the locations of populations was obtained from http://www.worldclim.org, including monthly, maximum and minimum temperatures, total precipitations and 19 BIO variables [41]. Seeds of one to ten individuals were collected from each population, providing a total of 151 Moroccan samples. Materials are available through the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info).
A set of 181 Iberian A. thaliana accessions collected from different previously described populations [16,30] and a collection of 136 accessions representing different populations from the rest of the world geographic range were also analysed (Additional file 1:Figure S5, Additional file 2: Table S4).
Genome-wide SNP genotyping
Accessions were genotyped for a genome-wide set of 343 SNP loci previously selected as frequent polymorphisms in Central Europe (CE; [17]), the Iberian Peninsula (IP; [16,42]), or in worldwide collections (W; [43]) (Additional file 2: Table S5). SNPs were assessed by SNPlex and Veracode methods through the CEGEN genotyping service (http://www.cegen.org). A total of 249 SNPs showing less than 25% missing data (average of 3.9%) were used for population genetic analysis. For the analysis of Morocco genetic diversity, 130 SNPs (17 CE, 35 IP, and 78 W) segregating among the 151 samples were used after removing monomorphic (106 SNPs) or SNPs missing within entire Moroccan population samples (13 SNPs). For analyses of the 201 Moroccan and Iberian populations, 237 polymorphic SNP loci (44 CE, 80 IP, and 113 W) were used after removal of monomorphic loci. For analyses of the 337 Moroccan, Iberian, and worldwide populations, all 249 polymorphic SNPs (55 CE, 80 IP, and 114 W) were included. Genotypic data of all accessions are given in Additional file 2: Table S6.
Population genetic analyses
Analyses were done with all SNPs and with the three sets of markers separately. Similar results were observed for all sets of markers, indicating that there was no substantial effect on our analyses derived from potential SNP bias ascertainment. Therefore, only results obtained with the total SNP set are shown.
Genetic diversity was measured as number of haplotypes per population (N H ), percentage of polymorphic loci (PL) and gene diversity (H S ) using GenAlex v6.41 [44]. Mean allelic richness (R S ) per locus and mean private allelic richness (Rp) were estimated with rarefaction to a common sample population size using HP-RARE [45].
Genetic differentiation among populations was estimated by analysis of molecular variance (AMOVA) using Arlequin v3.5.1.2 with multilocus genotypes [46], which calculates F ST -like statistics and their significance from 20,000 permutations.
Isolation by distance (IBD) was tested using Mantel tests for correlation between paired population matrices of geographic distances and genetic distances using Arlequin v3.5.1.2 [47] with 10,000 permutations for significance tests.
Genetic relationships among genotypes were determined by neighbor-joining (NJ) analysis. Genetic distances were measured as pairwise allele differences among Moroccan haplotypes and 10,000 bootstrap permutations were applied to determine the significance of groups using MEGA v5.05 [48].
Discrete clustering of genetic structure was analyzed in three sets of accessions: the 65 different Moroccan haplotypes, the 201 Moroccan and Iberian accessions, and all 337 genotypes of the worldwide collection. Since we were mainly interested in detecting the major genetic groups that were consistently differentiated in each geographic region, we compared two distinct methods. First, genetic structure was inferred using the Bayesian model-based clustering algorithm implemented in STRUCTURE v2.3.3 [49]. Model settings included haploid multilocus genotypes, correlated allele frequencies between populations and a linkage model using genetic distances derived from TAIR10 SNP physical positions (Additional file 2: Table S5). Each run consisted of 50,000 burn-in MCMC iterations and 100,000 MCMC after-burning repetitions for parameter estimation. To determine the K number of ancestral populations and the ancestry membership proportions of each individual in each population, the algorithm was run 20 times for each defined number of groups (K value) from 1 to 10. The number of distinct genetic clusters was determined by testing the differences between the data likelihood for successive K values using Wilcoxon tests for two related samples. The final K number was estimated as the largest K value with significantly higher likelihood than that of K-1 runs (two-sided P < 0.005). This was supported by a high similarity among the ancestry membership matrices from different runs of the same K value (H’ > 0.84). The average symmetric similarity coefficient H’ among runs and the average matrix of ancestry membership proportions, derived from the 20 runs, were calculated with CLUMPP v1 [50].
Population structure was also determined by principal component analysis (PCA) as implemented in the smartpca module of EigenSoft v4.2 [51]. Allele frequency normalization and correction for linkage disequilibrium by regression against both neighbouring SNP markers was used. Significances of explanatory eigenvalues were tested using Tracy-Widom statistics as implemented in the twstats module of EigenSoft. To avoid including axes describing variance unrelated to population structure [51] only the 3 to 5 largest eigenvectors weighted by eigenvalues, each accounting for at least 2% of the variance, were used for subsequent clustering analyses. Distinct clusters were estimated using a hierarchical clustering of Euclidean distances with Ward’s minimized variance as implemented in the R package mclust v3.4.8 [52,53]. Support for the detected clusters was assessed with the relative height values indicative of the genetic distance between clusters. Genetic groups identified by STRUCTURE and PCA were compared by: i) calculating the percentage of accessions assigned to equivalent genetic groups, based on the largest membership proportion estimated for each accession by STRUCTURE and on the cluster assignment determined from PCA; ii) calculating the similarity coefficient H' between the STRUCTURE matrix of majority membership proportions and the matrix of cluster assignments derived after PCA.
The average membership proportions of each accession to the different genetic groups and the geographic distribution of the majority population assignments of accessions were plotted using R v2.13 and the R package maps [52].
Gene sequence analysis
Four flowering time genes known to contribute to the natural variation for flowering traits in A. thaliana were sequenced in one individual per Moroccan population: FRI and FLC involved in the vernalization pathway; CRY2 and PhyC affecting the photoperiod pathway. These genes have been previously studied in world-wide and Iberian collections, where several haplotypes have been associated with flowering and with climatic factors [30,54-56]. The gene regions that were sequenced correspond to the same fragments analysed in a previous Iberian study [30], including the complete FRI gene (3.5 kb), 0.7 kb of the first intron of FLC, 1.5 kb of CRY2 coding region and 0.9 kb of PHYC promoter. Sequencing was done as previously described [30] using an ABI PRISM 3700 DNA Analyser (Applied Biosystems, Foster City, CA, USA). DNA sequences were aligned using DNASTAR v8.0 (Lasergene, Madison, WI, USA). The 20 Moroccan sequences were analysed together with available sequences from 181 Iberian accessions [30] and nucleotide diversity was estimated using DnaSP v5 [57]. The subset of SNPs in these genes that did not show complete linkage disequilibrium were used to estimate other genetic diversity parameters as described above. Paired genetic distances between groups of accessions were measured as corrected mean number of allele differences and as F ST values using Arlequin v3.5.1.2. Genbank accession numbers of sequences generated in this study are KF275035-KF275114.
Flowering initiation analysis
Fifteen plants of one selected accession from each of the 20 Moroccan populations were grown in a glasshouse without vernalization using a randomized experimental design. Flowering initiation was quantified as flowering time (FT) and leaf number (LN) following ref. [30]. FT was measured as the number of days to opening of the first flower. LN was measured as the number of rosette and cauline leaves present at the flowering date. The experiment was continued for 200 days, when the proportion of non-flowering plants per accession was estimated. To avoid underestimation of flowering initiation due to removal of non-flowering individuals, values of 150 leaves and 200 days were given to such individuals, which are the maximum FT value observed in flowering plants and the corresponding LN estimated from the regression line of FT onto LN. Moroccan accessions were also grown and measured after a vernalization treatment of 8 weeks at 4°C to induce flowering [30]. Since all plants flowered in these conditions, the percentage of non-flowering plants without vernalization was taken as a measurement of the obligate vernalization requirement of each accession. Comparison of the flowering behavior between Moroccan and Iberian populations was carried out using available Iberian flowering data obtained in another experiment performed under similar conditions in the same glasshouse [30]. Genetic differentiation between groups of accessions for quantitative traits was measured as Q ST values estimated from analysis of variance. Between groups (V B ) and within groups (V W ) variances were estimated by the REML method of variance component analysis, and Q ST was calculated as V B /(V B + V W ).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ACB, JJM-Z, FXP and CA-B conceived the study. AH, FXP, JMM-Z and CA-B collected the samples. BM-V and CA-B generated the laboratory data. ACB, BM-V, FXP and CA-B analysed the data. ACB, FXP and CA-B wrote the paper with the input of all authors. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Moroccan populations surveyed in this study. Figure S2. Population structure of A. thaliana in Morocco. Figure S3. Isolation by distance structure of A. thaliana in the south-western Mediterranean region. Figure S4. Population structure of A. thaliana in the south-western Mediterranean region. Figure S5. Geographic distribution of 337 A. thaliana populations analysed in this study. Figure S6. Population structure of A. thaliana at a worldwide scale. Figure S7. Genetic and geographic structure of A. thaliana at global scale established by PCA.
Table S1. Geographical and ecological information of Moroccan populations surveyed in this study. Table S2. Comparison of STRUCTURE and PCA analyses. Table S3. Genetic differentiation between Morocco and Iberian genetic groups for flowering genes. Table S4. Worldwide A. thaliana accessions analysed in this work. Table S5. SNP markers genotyped in this study. Table S6. Genotypic data of the 468 accessions for the 249 SNPs analysed in this study.
Contributor Information
Adrian C Brennan, Email: acebrennan@ebd.csic.es.
Belén Méndez-Vigo, Email: bmendez@cnb.csic.es.
Abdelmajid Haddioui, Email: ahaddioui@yahoo.fr.
José M Martínez-Zapater, Email: zapater@icvv.es.
F Xavier Picó, Email: xpico@ebd.csic.es.
Carlos Alonso-Blanco, Email: calonso@cnb.csic.es.
Acknowledgements
The authors thank to Mercedes Ramiro and Jenifer Pozas for technical assistance and to EBD-CSIC for logistic support during field campaigns. This work was funded by grant BIO2010-15022 from the Ministerio de Ciencia and Innovación of Spain to CA-B. ACB was funded by Grant No. 264125 EcoGenes from FP7-REGPOT 2010–1.
References
- Al-Shehbaz IA, O’Kane SL. Taxonomy and phylogeny of Arabidopsis (Brassicaceae) Arabidopsis Book. 2002;1:E0001. doi: 10.1199/tab.0001. doi:10.1199/tab.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MH. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae) J Biogeogr. 2002;29:125–134. doi: 10.1046/j.1365-2699.2002.00647.x. [DOI] [Google Scholar]
- Koornneef M, Meinke D. The development of Arabidopsis as a model plant. Plant J. 2010;61:909–921. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjalmsson BJ. et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 2012;158:2–22. doi: 10.1104/pp.111.189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol. 2000;9:2109–2118. doi: 10.1046/j.1365-294X.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R. et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Stahl EA, Toomajian C, Nordborg M, Kreitman M, Bergelson J. Distribution of genetic variation within and among local populations of Arabidopsis thaliana over its species range. Mol Ecol. 2006;15:1405–1418. doi: 10.1111/j.1365-294X.2006.02884.x. [DOI] [PubMed] [Google Scholar]
- François O, Blum MG, Jakobsson M, Rosenberg NA. Demographic history of european populations of Arabidopsis thaliana. PLoS Genet. 2008;4:e1000075. doi: 10.1371/journal.pgen.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K. et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C. et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Beck JB, Schmuths H, Schaal BA. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects pleistocene glacial dynamics. Mol Ecol. 2008;17:902–915. doi: 10.1111/j.1365-294X.2007.03615.x. [DOI] [PubMed] [Google Scholar]
- Picó FX, Mendez-Vigo B, Martinez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian Peninsula. Genetics. 2008;180:1009–1021. doi: 10.1534/genetics.108.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Torjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor Appl Genet. 2006;112:1104–1114. doi: 10.1007/s00122-006-0212-7. [DOI] [PubMed] [Google Scholar]
- Stenoien HK, Fenster CB, Tonteri A, Savolainen O. Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Mol Ecol. 2005;14:137–148. doi: 10.1111/j.1365-294X.2004.02359.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Säll T, Lind-Halldén C, Halldén C. The evolutionary history of the common chloroplast genome of Arabidopsis thaliana and A. suecica. J Evol Biol. 2007;20:104–121. doi: 10.1111/j.1420-9101.2006.01217.x. [DOI] [PubMed] [Google Scholar]
- Lewandowska-Sabat AM, Fjellheim S, Rognli OA. Extremely low genetic variability and highly structured local populations of Arabidopsis thaliana at higher latitudes. Mol Ecol. 2010;19:4753–4764. doi: 10.1111/j.1365-294X.2010.04840.x. [DOI] [PubMed] [Google Scholar]
- He F, Kang D, Ren Y, Qu LJ, Zhen Y, Gu H. Genetic diversity of the natural populations of Arabidopsis thaliana in China. Heredity. 2007;99:423–431. doi: 10.1038/sj.hdy.6801020. [DOI] [PubMed] [Google Scholar]
- Yin P, Kang J, He F, Qu LJ, Gu H. The origin of populations of Arabidopsis thaliana in China, based on the chloroplast DNA sequences. BMC Plant Biol. 2010;10:22. doi: 10.1186/1471-2229-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todokoro S, Terauchi R, Kawano S. Microsatellite polymorphisms in natural populations of Arabidopsis thaliana in Japan. Jpn J Genet. 1995;70:543–554. doi: 10.1266/jjg.70.543. [DOI] [Google Scholar]
- Jorgensen S, Mauricio R. Neutral genetic variation among wild North American populations of the weedy plant Arabidopsis thaliana is not geographically structured. Mol Ecol. 2004;13:3403–3413. doi: 10.1111/j.1365-294X.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean basin. J Biogeogr. 2009;36:1333–1345. doi: 10.1111/j.1365-2699.2008.02051.x. [DOI] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Petit R, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M. et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Mendez-Vigo B, Pico FX, Ramiro M, Martinez-Zapater JM, Alonso-Blanco C. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 2011;157:1942–1955. doi: 10.1104/pp.111.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J. Plant diversity in the region of the strai of gilbraltar: a multilevel approach. Lagascalia. 1997;19:393–404. [Google Scholar]
- Médail F, Quézel P. Hot-spots analysis for conservation of plant biodiversity in the Mediterranean Basin. Ann Mo Bot Gard. 1997;84:112–127. doi: 10.2307/2399957. [DOI] [Google Scholar]
- Rodríguez-Sánchez F, Pérez-Barrales R, Ojeda F, Vargas P, Arroyo J. The strait of Gibraltar as a melting pot for plant biodiversity. Quat Sci Rev. 2007;27:2100–2117. [Google Scholar]
- Lavergne S, Hampe A, Arroyo J. In and out of Africa: how did the strait of Gibraltar affect plant species migration and local diversification? J Biogeogr. 2013;40:24–36. doi: 10.1111/j.1365-2699.2012.02769.x. [DOI] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the quaternary. Philos Trans R Soc Lond. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PR, McKay JK. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution. 2004;58:938–945. doi: 10.1111/j.0014-3820.2004.tb00428.x. [DOI] [PubMed] [Google Scholar]
- Guo Q. Incorporating latitudinal and central-marginal trends in assessing genetic variation across species ranges. Mol Ecol. 2012;21:5396–5403. doi: 10.1111/mec.12012. [DOI] [PubMed] [Google Scholar]
- Mendez-Vigo B, Gomaa NH, Alonso-Blanco C, Pico FX. Among- and within-population variation in flowering time of Iberian Arabidopsis thaliana estimated in field and glasshouse conditions. New Phytol. 2013;197:1332–1343. doi: 10.1111/nph.12082. [DOI] [PubMed] [Google Scholar]
- deMenocal P, Ortiz J, Guilderson T, Adkins J, Sarnthein M, Baker L, Yarusinsky M. Abrupt onset and termination of the African Humid Period: rapid climate responses to gradual insolation forcing. Quat Sci Rev. 2000;19:347–361. doi: 10.1016/S0277-3791(99)00081-5. [DOI] [Google Scholar]
- Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat Rev Genet. 2011;12:603–614. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- Gomaa NH, Montesinos-Navarro A, Alonso-Blanco C, Pico FX. Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol Ecol. 2011;20:3540–3554. doi: 10.1111/j.1365-294X.2011.05193.x. [DOI] [PubMed] [Google Scholar]
- Warthmann N, Fitz J, Weigel D. MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics. 2007;23:2784–2787. doi: 10.1093/bioinformatics/btm428. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowksi ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. doi: 10.1111/j.1471-8286.2004.00845.x. [DOI] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria; http://www.R-project.org/ [Google Scholar]
- Fraley C, Raftery AE. Technical Report no 504. Department of Statistics, University of Washington, USA; 2006. MCLUST Version 3 for R: normal mixture modeling and model-based clustering. [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics. 2004;167:1361–1369. doi: 10.1534/genetics.103.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Moroccan populations surveyed in this study. Figure S2. Population structure of A. thaliana in Morocco. Figure S3. Isolation by distance structure of A. thaliana in the south-western Mediterranean region. Figure S4. Population structure of A. thaliana in the south-western Mediterranean region. Figure S5. Geographic distribution of 337 A. thaliana populations analysed in this study. Figure S6. Population structure of A. thaliana at a worldwide scale. Figure S7. Genetic and geographic structure of A. thaliana at global scale established by PCA.
Table S1. Geographical and ecological information of Moroccan populations surveyed in this study. Table S2. Comparison of STRUCTURE and PCA analyses. Table S3. Genetic differentiation between Morocco and Iberian genetic groups for flowering genes. Table S4. Worldwide A. thaliana accessions analysed in this work. Table S5. SNP markers genotyped in this study. Table S6. Genotypic data of the 468 accessions for the 249 SNPs analysed in this study.