Abstract Abstract
The mitochondrial cytochrome c oxidase subunit I (COI) can serve as a fast and accurate marker for the identification of animal species, and has been applied in a number of studies on birds. We here sequenced the COI gene for 387 individuals of 147 species of birds from the Netherlands, with 83 species being represented by > 2 sequences. The Netherlands occupies a small geographic area and 95% of all samples were collected within a 50 km radius from one another. The intraspecific divergences averaged 0.29% among this assemblage, but most values were lower; the interspecific divergences averaged 9.54%. In all, 95% of species were represented by a unique barcode, with 6 species of gulls and skua (Larus and Stercorarius) having at least one shared barcode. This is best explained by these species representing recent radiations with ongoing hybridization. In contrast, one species, the Lesser Whitethroat Sylvia curruca showed deep divergences, averaging 5.76% and up to 8.68% between individuals. These possibly represent two distinct taxa, S. curruca and S. blythi, both clearly separated in a haplotype network analysis. Our study adds to a growing body of DNA barcodes that have become available for birds, and shows that a DNA barcoding approach enables to identify known Dutch bird species with a very high resolution. In addition some species were flagged up for further detailed taxonomic investigation, illustrating that even in ornithologically well-known areas such as the Netherlands, more is to be learned about the birds that are present.
Keywords: Aves, conservation, cytochrome c oxidase subunit I, COI, taxonomy
Introduction
DNA barcoding is used as an effective tool for both the identification of known species and the discovery of new ones (Hebert et al. 2003, 2010, Savolainen et al. 2005). The core idea of DNA barcoding is based on the fact that just a small portion of a single gene, comprising a 650 to 700 bp fragment from the first half of the mitochondrial cytochrome c oxidase subunit I gene (COI), shows a lower intraspecific than interspecific variation. An attribute which characterizes a threshold of variation for each taxonomic group, above which a group of individuals does not belong to the same species but instead forms an intraspecific taxon. In other words, the recognition of patterns in sequence diversity of a small fragment from the mtDNA genome has led to an alternative approach for species identification across phyla.
Initially, DNA barcodes were proposed for the Animal Kingdom in 2003, when Hebert and colleagues tested a single gene barcode to identify species and coined the term ‘DNA barcoding’ (Hebert et al. 2003). Since that time COI sequences have been used as identifiers in the majority of animal phyla including vertebrates (e.g. Hebert et al. 2004, Ward et al. 2005, Kerr et al. 2007, Smith et al. 2008, Nijman and Aliabadian 2010, Luo et al. 2011) and invertebrates (Hajibabaei et al. 2006, Bucklin et al. 2011, Hausmann et al. 2011). In recent years, the practical utility of DNA barcodes proved to be an appealing tool to help resolve taxonomic ambiguity (Hebert et al. 2004, 2010), to screen biodiversity (e.g. Plaisance et al. 2009, Naro-Maciel et al. 2009, Grant et al. 2011), and to support applications in conservation biology (Neigel et al. 2007, Rubinoff 2006, Dalton and Kotze 2011).
Birds are among the best-known classes of animals and thus provide a taxonomically good model for analyzing the applicability of DNA barcoding. In the last seven years some 30 scientific papers have been published on the DNA barcoding of bird species, which combined have been cited 500 times (V. Nijman, unpubl. data April 2013). Most of the studies have shown that from this small fragment of DNA, individuals have been identified down to species level for 94% of the species in Scandinavian birds (Johnsen et al. 2010), 96% in Nearctic birds (Kerr et al. 2009a), 98% in Holarctic birds (Aliabadian et al. 2009) and 99% in Argentinean and South Korean birds (Kerr et al. 2009a, Yoo et al. 2006). Species delineation relying on the use of theshold set to differentiate between intraspecific variation and interspecific divergence has been criticized as leading to too unacceptable high error rates especially in incompletely samples groups (Meyer and Paulay 2005). However, even the critics of DNA barcoding concede that DNA barcoding holds promise for identification in taxonomically well-understood and thoroughly sampled clades. Birds are taxonomically well-known, especially those of the Western Palearctic to which the Netherlands, our study area, forms part. As noted by Taylor and Harris (2012), compared to other taxa that have been subjected to DNA barcoding, DNA barcoding studies of birds tend to represent aggregations of very large number of bird species barcodes. These often include (near) cosmopolitan species with samples from distant geographic locations potentially increasing the amount of interspecific variation in COI.
Here we explore the efficiency of identifying species using DNA barcoding from a large set of sympatric bird species in the Netherlands. Compared to previous studies on birds, our study area covers a very small geographic area, allowing to directly test the functionality of DNA barcoding ‘in one’s backyard’.
Methods
Sampling
The Netherlands is a small, densely populated country in northwestern Europe, with a land surface area of some 34 000 km2, and ornithologically it is arguable one of the best-covered countries (Sovon 2002). The tissue samples used for sequencing were collected from breeding areas in the Netherlands, excluding oversees dependencies. Given the small size of the country some 95% of the samples were collected within a 50 km-radius of each other. Samples were part of the tissue collection of the Zoological Museum of Amsterdam (ZMA), which were recently relocated and deposited in the Naturalis Biodiversity Center, Leiden. Most were collected in the period 2000–2012 by a network of volunteers, ringers, airport staff, and bird asylums; no birds were specifically collected or killed to be included in the collection of the ZMA. Species and subspecies identification was based on morphology and when necessary, external measurements. These identifications were done by authors HvB and CSR, with the help of Tineke G. Prins. Individual birds were frozen upon arrival to be thawed and skinned at a later date, and indeed many birds arrived frozen. Samples were mostly taken from the bird’s pectorial muscles, because of its size and easy access, and stored in 96% ethanol. Species nomenclature follows the taxonomy of Dickinson (2003). The complete list of sampled specimens including information about vouchers and trace files is available from the project ‘Aves of the Netherlands’ at the BOLD website (http://www.barcodinglife.com/).
PCR and sequencing
The tissue samples were subsampled and subjected to DNA extraction using DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s protocol. PCR and sequencing reactions were performed, mainly following the same protocols described in Förschler et al. (2010), but with some minor modifications. Polymerase chain reaction (PCR) amplifications were initially performed using standard primers BirdF1 (TTCTCCAACCACAAAGACATTGGCAC) and BirdR1 (ACGTGGGAGATAATTCCAAATCCTG). When amplification was unsuccessful, alternate reverse primer BirdR2 (ACTACATGTGAGATGATTCCGAATCCAG) was used in combination with BirdF1 or alternate primer pair CO1-ExtF (ACGCTTTAACACTCAGCCATCTTACC) and CO1-ExtR (AACCAGCATATGAGGGTTCGATTCCT) was used (Hebert et al. 2004, Johnsen et al. 2010). All PCRs were run under the following thermal cycle program: 3 min at 94 °C followed by 40 cycles of 15 s at 94 °C, 30 s at 50 °C and 40 s at 72 °C, and a final elongation of 5 min at 72 °C. For each reaction the PCR mixture consisted of 2.5 µl Qiagen Coral Load 10 × PCR buffer, 1.0 µl of each 10 mM primer, 0.5 µl 2.5 mM dNTPs, 0.25 µl 5 U/µl QiagenTaq DNA polymerase, 18.75 µl milliQ and 1.0 µl template DNA for a total volume of 25 µl. Bi-directional sequencing was performed for all specimens at Macrogen. We checked the possible amplification of pseudogenes (Numts) by translating the protein coding genes into amino acids sequences, but we did not observe any unexpected stop codons, frameshifts or unusual amino acidic substitutions. Furthermore we amplified a longer sequence of the COI gene with primers (CO1-ExtF and CO1-ExtR) for selected samples, and also here we did not see any indication of pseudogene co-amplification. Lijtmaer et al. (2012) found that, in birds, full-length COI pseudogenes are uncommon noting that they might be more frequently encountered when working with avian blood samples as opposed to muscle tissue samples (as used in here).
Data analysis
Sequences shorter than 500 bp and containing more than 10 ambiguous nucleotides were excluded from the analyses. All sequences have been deposited in GenBank (Accession numbers KF946551 to KF946937). A full list of the museum vouchers, for all specimens applied in this study, is provided in Appendix – Table 1.
For all sequence comparisons, the Kimura 2-parameter (K2P) model was used, because it is shown to be the best metric to compare closely related taxa (Nei and Kumar 2000, but for a contrasting view see Srivathsan and Meier 2012). Average intraspecific distances were calculated for those species that were represented by at least two specimens using MEGA5 software (Tamura et al. 2011).
For a group of birds that expressed a larger than expected intraspecific variation, the Sylvia warblers, we created a phylogenetic tree and created a haplotype network. We chose GTR+G+I as the best-fitting model of nucleotide substitution based on its Akaike’s information criterion as implemented in JModelTest v0.1.1 (Posada 2008). A maximum likelihood (ML) tree was constructed in PAUP* v4.0b10 (Swofford 2002) using a heuristic search with the tree-bisection-reconnection branch-swapping algorithm and random addition of taxa. Relative branch support was evaluated with 500 bootstrap replicates (Felsenstein 1985). A minimum spanning haplotype network was constructed using a statistical parsimony network construction approach as implemented in TCS software package (Clement et al. 2000). This programme calculates the number of mutational steps by which pairwise haplotypes differ and computes the probability of parsimony (Templeton et al. 1992) for pairwise differences until the probability exceeds 0.95. The number of mutational differences associated with the probability just before the 0.95 cut-off point is then the maximum number of mutational connections between pairs of sequences justified by the parsimony criterion; these justified connections are applied in the haplotype network (Clement et al. 2000).
Results
A total of 387 sequences for 141 species (representing at least 158 taxa) were retrieved, including 52% of the breeding bird species in the Netherlands (Supplementary table 1). The average number of sequences per species was 3.36 (range 1-13), with 83 species (59%) represented by more than two sequences. The mean K2P-divergence within species bears no significant relationship with sample sizes, i.e. number of sequences per species (R2 = 0.001, p = 0.465). The mean intraspecific K2P-distance was 0.29% (range 0-8.68%) some 30 times lower than the mean intrageneric K2P-distances (9.54%, range 0-27.71%) (Table 1, Figure 1).
Table 1.
Comparisons of K2P-pairwise distances within various taxonomic levels for 83 species of birds from the Netherlands for which two or more sequences were available. Distances are expressed in percentages.
| Individuals | Taxa | Comparisons | Distances | |||
|---|---|---|---|---|---|---|
| Minimum | Mean ± S.E.M. | Maximum | ||||
| Within Species | 340 | 83 | 805 | 0 | 0.294±0.001 | 8.683 |
| Within Genera | 203 | 23 | 794 | 0 | 9.544±0.004 | 15.849 |
| Within Families | 282 | 20 | 2519 | 5.809 | 14.467±0.001 | 20.473 |
Figure 1.
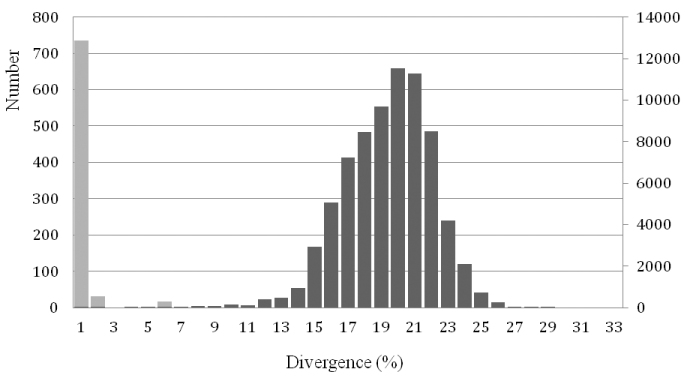
Comparisons of K2P-pairwise distances based on the COI gene of 141 species of birds from the Netherlands, showing a clear barcoding gap. Interspecific distances are indicated with light grey bars and intraspecific distances with dark grey bars. Left Y-axis: numbers of intraspecific comparisons; Right Y-axis: numbers of interspecific comparisons.
In general, 95% of species (134 species) showed a unique DNA barcode (these included the 58 species for which we only sequenced single individuals), while six congeneric species shared the same barcode and the mean intraspecific distance of them fell well below the threshold of species based on distance-based criterion (Hebert et al. (2003) 10 × rule). These congeneric species mostly included circumpolar species with close morphological similarities (Table 2).
Table 2.
Bird species (Charadriiformes) from the Netherlands with one or more shared DNA barcodes (K2P-distances of 0%). For a detailed breakdown of the individual samples involved see Appendix – Table 2.
| Family | Species | Nearest species | Mean K2P-distance (%) |
|---|---|---|---|
| Laridae | Herring Gull Larus argentatus | Yellow-legged Gull Larus michahellis | 0.14 |
| Lesser Black-backed Gull Larus fuscus | Caspian Gull Larus cachinnans | 0 | |
| Iceland Gull Larus glaucoides | Caspian Gull Larus cachinnans | 0 | |
| Glaucous Gull Larus hyperboreus | Yellow-legged Gull Larus michahellis | 0.58 | |
| Yellow-legged Gull Larus michahellis | Caspian Gull Larus cachinnans | 0 | |
| Stercorariidae | Great Skua Stercorarius skua | Pomarine Skua Stercorarius pomarinus | 0.30 |
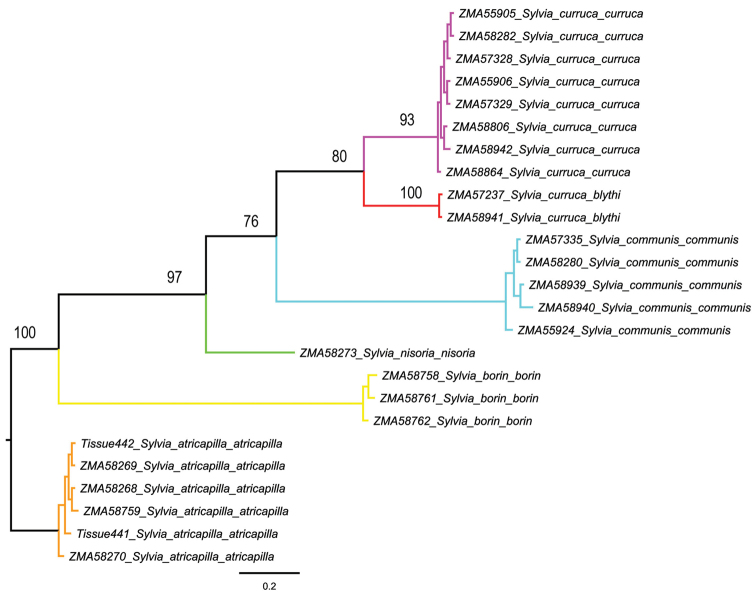
Although most species possessed low intraspecific distances, one species showed high intraspecific K2P-distances clearly above the threshold of 2 to 3 per cent sequence divergence in our data set. This is the Lesser Whitethroat Sylvia curruca, with a mean interspecific divergence of 5.76% and a maximum interspecific distance of 8.68%. Two subspecies occur in the Netherlands, i.e. the Western Lesser Whitethroat Sylvia curruca curruca and, as a migrant, the Northeastern Lesser Whitethroat Sylvia curruca blythi. Both are morphologically somewhat distinct, with compared to the nominate Sylvia curruca blythi having a paler top of the head, separated from face by a white supercilium, and geographically the nominate occupies the western part of the species range and Sylvia curruca blythi the eastern part. A maximum likelihood tree for these two taxa based on K2P-model is presented in Figure 2. Two different haplotype networks, one each for Sylvia curruca curruca and Sylvia curruca blythi were recovered by TCS (Figure 3), and given the large genetic distances between their haplotypes, the two taxa are not included in the same haplotype network.
Figure 2.
Phylogenetic relationships of two putative subspecies of Lesser Whitethroat, i.e. the Western Lesser Whitethroat Sylvia curruca curruca and the Northeastern Lesser Whitethroat Sylvia curruca blythi from the Netherlands, based on analysis of 694 bp of the mitochondrial cytochrome c oxidase subunit I gene (COI). Bootstrap values are given for the maximum likelihood (ML) analysis.
Figure 3.
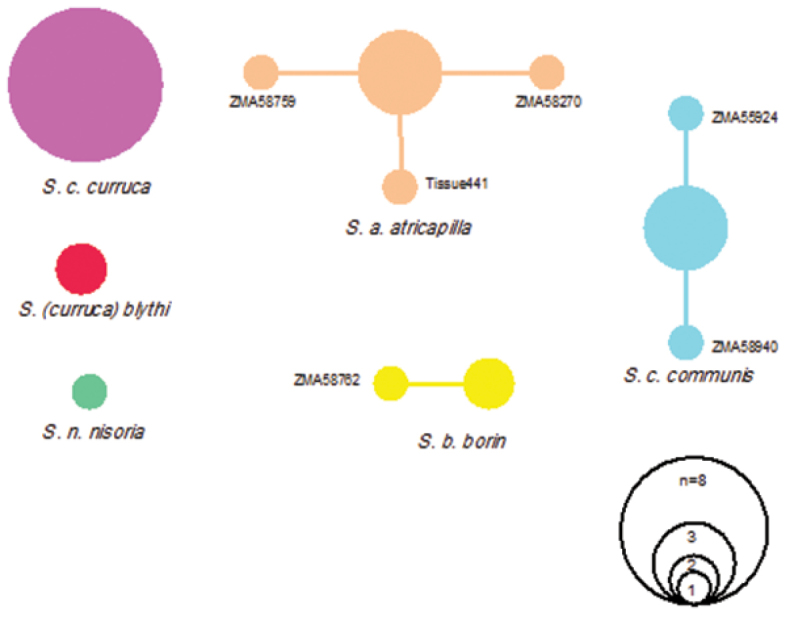
Haplotype networks constructed with statistical parsimony based on 694 bp of the mitochondrial cytochrome c oxidase subunit I gene (COI) of the Sylvia group (25 individuals). Each circle represents one haplotype; size of circles is proportional to haplotype frequency.
Discussion
We here present the results of a modest effort to barcode the avifauna of the Netherlands. In terms of DNA barcoding of birds, the Netherlands form the southernmost part of one of the most densely sampled regions globally (Lijtmaer et al. 2012: figure 1). In addition, many of the species that overwinter in the country originate equally well-sampled regions to the north. As such our study adds to a growing number of studies allowing us to build up comprehensive public libraries of bird barcodes. Combined these allow us to explore new lines of scientific inquiry and practical applications (Hebert et al. 2010, Lijtmaer et al. 2012, but see Ebach and Carvalho 2010). The collection of our samples was done as part of the museum’s standard collection management of newly obtained material, and as such sample collection was inexpensive and required little effort in terms of manpower. All birds were collected and processed in the Netherlands and did not require specific permits other than the ones already required to curate the collections.
Recently, Taylor and Harris (2012) expressed the opinion that proponents of DNA barcoding consistently fail to recognize its limitations (including, but not restricted to, the functioning of COI as a universal barcoding gene, whether its use is to be restricted to species identification only or whether it has a role in species discovery and delimitation and the failure to have sufficient systems in place to deal with the large amounts of data generated), do not evolve their methodologies, and do not embrace the possibilities that next-generation sequencing offers. We agree that DNA barcoding will not offer a panacea for all the issues Taylor and Harris (2012) raised, or indeed some of its earlier critics (Will et al. 2005, Moritz and Cicero 2004) but we point out that for this was probably never the intention of DNA barcoding when envisaged some ten years ago. Irrespective of the aims and goals of DNA barcoding as a ‘global enterprise’ (Ebach and Carvalho 2010), we found it a useful tool in our studies on birds (cf. Baker et al. 2009). The bird collection of the Zoological Museum Amsterdam, and our sample reported in this study, was well-curated by knowledgeable staff, with a very high degree of taxonomic certainty attached to each individual specimen. We see immense value to having a DNA barcoding dataset linked to this reference collection. As such this work has added to the growing library of DNA barcodes of bird species of the world and subsequent improvement in our knowledge of biodiversity.
The mean intraspecific divergences found in the birds of the Netherlands (0.29%, based on 147 species) is congruent with that of for instance Argentina (0.24%, 500 species), North America (0.23%, 643 species) and the Holarctic (0.24%, 566 species) (Kerr et al. 2009a, Aliabadian et al. 2009). More importantly, like other studies on birds, the efficiency of DNA barcode sequences to identify species is high, showing a clear barcoding gap (Figure 1), and overall it seems that for birds typically 95% or more of the species can be identified (Hebert et al. 2003, Johnsen et al. 2010, Kerr et al. 2009a, b, Yoo et al. 2006, Aliabadian et al. 2009).
Most DNA barcoding studies of birds flag a small number of deep divergences (e.g. Johnsen et al. 2010, Kerr et al. 2009b, Aliabadian et al. 2009, Nijman and Aliabadian 2013), in our study involving the two subspecies of Sylvia curruca, where the two lineages diverge almost 6%. Similar results were found by Olsson et al. (2013) when analyzing the cytochrome b gene for these two taxa, with distances in the order of 11-14%. Based on COI sequences, the two taxa appear to be sister taxa, albeit with a relatively low support (Figure 3), but no other members of the Sylvia curruca were included in the analysis. In contrast, having included a range of other members of this complex, Olsson et al. (2013) found curruca and blythi not to be sister taxa. Olsson et al. (2013: 81) concluded that while “due to their morphological similarity it is unclear where their ranges meet, [o]ur data suggest that blythi is a valid taxon, not closely related to curruca. It has its closest relatives to the south-east [Asia], and may have colonised the eastern taiga from this direction, ultimately coming into contact with curruca”. When it comes to drawing conclusion from their work with respect to taxomomy, Olsson et al. (2013) were, in our view correctly, cautious. They noted that the Sylvia curruca complex comprised up to 13 taxa with little consensus as to circumscription and taxonomic rank. Of these, morphologically some taxa are very similar, including Sylvia curruca curruca and Sylvia curruca blythi, and the apparent conflict between morphology and phylogeny (based in their case on cyt b and in our study on COI) can be explained in different ways. One would be to accept the single mitochondrial gene trees at face value in which case the morphological similarities in pelage coloration may be a result of parallel evolution possibly in response to adaptations to similar temperate forest habitats – both taxa are then best treated as different species. Alternatively, the mitochondrial gene trees do not reflect the species tree and, based on morphological similarities, Sylvia curruca curruca and Sylvia curruca blythi are best treated as sister taxa (either as one or two species). Their divergent position on the mitochondrial gene tree, and the large genetic distances between these taxa, are due to ancient mitochondrial introgression. In either case, working with single mitochondrial markers cannot not resolve this issue and a more integrative approach ideally involving the analysis of nuclear genes is paramount.
Those cases where we found species sharing the same DNA barcodes were small in number but not insignificant. Seven of the eight cases involved closely related gulls with partially overlapping ranges, or allopatric distributions, that are part of a recent Holarctic radiation (Liebers-Helbig et al. 2010). Alternatively, the sharing of DNA barcodes may be due to hybridization or, perhaps less likely, misidentification. Likewise, skuas are part of a recent radiation with, just like gulls, frequent hybridization between species (Ritz 2009). DNA barcoding using a relative slowly evolving maternally inherited gene, with, compared to other mitochondrial genes, small amounts of rate heterogeneity (Pacheco et al. 2011), will, on its own, not be able to differentiate between these taxa.
We conclude that DNA barcoding approach makes it possible to identify known Dutch bird species with a very high resolution. Although some species were flagged for further detailed taxonomic investigation, our study reaffirms once more that a short segment of COI gene can be used to handle large number of taxa and aid in detecting overlooked taxa and hybridizing species with low deep barcode divergences.
Acknowledgements
We thank Tineke G Prins, involved in the sampling and administering of the bird specimens over the years in the Zoological Museum Amsterdam, for her commitment and hard work, and Miguel Vences, formerly of the Zoological Museum Amsterdam and currently at the Technical University Braunschweig, as the initiator of this project. Hans Breeuwer, Betsy Voetdijk, Peter Kuperus, and Lin Dong are thanked for their support and advice in the molecular laboratory of the Evolutionary Biology Department, University of Amsterdam. Finally, we thank the editors of this special issue for their patience, guidance and support, and two sets of reviewers for constructive comments: combined their efforts greatly improved the quality and clarity of the work. Our molecular work is funded in part by the Fonds Economische Structuurversterking. We dedicate this paper to the memory of Jan Wattel, former curator of birds at the Zoological Museum Amsterdam, who passed away in March 2013.
Appendix
Supplementary table 1.
List of all Dutch birds that have been sequenced in this study, with voucher numbers and collection localities. Note that specimens from which only tissue samples have been taken have not been given a collection number, sine loco refers to specimens collected in the Netherlands but without a precise named collection locality. Localities in the province of Friesland are listed with their Dutch name first, followed by their Frisian name. Coordinates are given in decimal degrees.
| Species or subspecies | ZMA number | Preparation | Locality | Coordinates | Access numbers |
|---|---|---|---|---|---|
| Accipiter gentilis gentilis | ZMA58297 | skin | Zaandam | 52.25N, 4.49E | KF946551 |
| Accipiter gentilis gentilis | ZMA58724 | skin | De Rips | 51.32N, 5.48E | KF946552 |
| Accipiter nisus nisus | ZMA58243 | skin | Malden | 51.47N, 5.52E | KF946553 |
| Accipiter nisus nisus | ZMA58245 | skin | Helden | 51.21N, 5.55E | KF946554 |
| Accipiter nisus nisus | ZMA58246 | skin | Reuver | 51.17N, 6.04E | KF946555 |
| Accipiter nisus nisus | ZMA58247 | skin | Culemborg | 51.55N, 5.15E | KF946556 |
| Accipiter nisus nisus | ZMA58248 | skin | Amsterdam | 52.21N, 4.53E | KF946557 |
| Accipiter nisus nisus | ZMA58741 | skin | Amsterdam | 52.21N, 4.53E | KF946558 |
| Accipiter nisus nisus | ZMA58742 | skin | Montfort | 51.07N, 5.56E | KF946559 |
| Accipiter nisus nisus | ZMA58743 | skin | Belfeld | 51.18N, 6.08E | KF946560 |
| Accipiter nisus nisus | ZMA58744 | skin | Laren | 52.11N, 6.22E | KF946561 |
| Accipiter nisus nisus | ZMA58745 | skin | Almere | 52.22N, 5.13E | KF946562 |
| Accipiter nisus nisus | ZMA58746 | skin | Venlo | 51.21N, 6.11E | KF946563 |
| Acrocephalus palustris | ZMA56679 | skin | Harderbroek reserve | 52.22N, 5.35E | KF946564 |
| Acrocephalus palustris | ZMA58811 | skin | Castricum | 52.32N, 4.36E | KF946565 |
| Acrocephalus schoenobaenus | ZMA58278 | skin | Almere | 52.22N, 5.13E | KF946566 |
| Acrocephalus schoenobaenus | ZMA58809 | skin | Almere | 52.22N, 5.13E | KF946567 |
| Acrocephalus schoenobaenus | ZMA58810 | skin | Castricum | 52.32N, 4.36E | KF946568 |
| Acrocephalus schoenobaenus | ZMA58862 | skin | Wassenaar | 53.08N, 5.53E | KF946569 |
| Acrocephalus scirpaceus scirpaceus | ZMA58277 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946570 |
| Acrocephalus scirpaceus scirpaceus | ZMA58725 | skin | Schermerhorn | 52.36N, 4.54E | KF946571 |
| Acrocephalus scirpaceus scirpaceus | ZMA58727 | skin | Lelystad | 52.29N, 5.24E | KF946572 |
| Acrocephalus scirpaceus scirpaceus | ZMA58728 | skin | Lelystad | 52.29N, 5.24E | KF946573 |
| Acrocephalus scirpaceus scirpaceus | ZMA58729 | skin | Castricum | 52.32N, 4.36E | KF946574 |
| Acrocephalus scirpaceus scirpaceus | ZMA58863 | skin | Lauwersmeer | 53.22N, 6.14E | KF946575 |
| Acrocephalus scirpaceus scirpaceus | ZMA58937 | skin | Lelystad | 52.29N, 5.24E | KF946576 |
| Acrocephalus scirpaceus scirpaceus | ZMA58938 | skin | Purmerend | 52.28N, 4.58E | KF946577 |
| Aegithalos caudatus europaeus | ZMA57353 | skin | Westenschouwen | 51.41N, 3.42E | KF946578 |
| Aegithalos caudatus europaeus | ZMA57354 | skin | Westenschouwen | 51.41N, 3.42E | KF946579 |
| Aegithalos caudatus europaeus | ZMA57356 | skin | Hilversum | 52.13N, 5.09E | KF946580 |
| Aegithalos caudatus europaeus | ZMA58804 | skin | Castricum | 52.32N, 4.36E | KF946581 |
| Alcedo atthis ispida | ZMA56216 | skin | Haelen | 51.13N, 5.56E | KF946582 |
| Alcedo atthis ispida | ZMA57341 | skin | Purmerland | 52.28N, 4.55E | KF946583 |
| Alcedo atthis ispida | ZMA57342 | skin | Alkmaar | 52.38N, 4.44E | KF946584 |
| Alcedo atthis ispida | ZMA57343 | skin | Utrecht | 52.03N, 5.08E | KF946585 |
| Alcedo atthis ispida | ZMA58869 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946586 |
| Alle alle alle | ZMA58842 | skin | Amsterdam | 52.21N, 4.53E | KF946587 |
| Alle alle alle | ZMA58917 | skin | Amsterdam | 52.21N, 4.53E | KF946588 |
| Alle alle alle | ZMA58918 | skin | Den Helder | 52.55N, 4.46E | KF946589 |
| Anas acuta | ZMA58228 | skin | Vlieland Island | 53.15N, 4.59E | KF946590 |
| Anas strepera strepera | ZMA58913 | skin | Driebond Polder | 53.11N, 6.37E | KF946591 |
| Anthus spinoletta spinoletta | ZMA58279 | skin | Lelystad | 52.29N, 5.24E | KF946592 |
| Anthus spinoletta spinoletta | ZMA64552 | skin | Castricum | 52.32N, 4.36E | KF946593 |
| Anthus trivialis trivialis | Tissue553 | DNA sample | Castricum | 52.32N, 4.36E | KF946594 |
| Apus apus apus | ZMA58717 | skin | Tegelen | 51.19N, 6.09E | KF946595 |
| Ardea cinerea cinerea | Tissue434 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946596 |
| Ardea cinerea cinerea | Tissue435 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946597 |
| Asio flammeus flammeus | ZMA58253 | skin | Texel Island | 53.04N, 4.43E | KF946598 |
| Asio otus otus | Tissue455 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946599 |
| Asio otus otus | ZMA58233 | skin | Purmerend | 52.28N, 4.58E | KF946600 |
| Asio otus otus | ZMA58234 | skin | Zutphen | 52.07N, 6.12E | KF946601 |
| Athene noctua vidalii | ZMA58493 | skin | Heerhugowaard | 52.4N, 4.51E | KF946602 |
| Athene noctua vidalii | ZMA58294 | skin | Blerick | 51.21N, 6.08E | KF946603 |
| Bombycilla garrulus garrulus | ZMA56300 | skin | Amsterdam | 52.21N, 4.53E | KF946604 |
| Bombycilla garrulus garrulus | ZMA56301 | wings | Texel Island | 53.04N, 4.43E | KF946605 |
| Bombycilla garrulus garrulus | ZMA58301 | wings | Hellendoorn | 52.23N, 6.26E | KF946606 |
| Bombycilla japonica | ZMA58302 | skin | Amsterdam | 52.21N, 4.53E | KF946607 |
| Buteo buteo buteo | Tissue461 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946608 |
| Buteo buteo buteo | ZMA58238 | skin | Wieringermeer | 52.54N, 5.01E | KF946609 |
| Buteo buteo buteo | ZMA58239 | skin | De Rips | 51.32N, 5.48E | KF946610 |
| Buteo buteo buteo | ZMA58781 | wing | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946611 |
| Buteo buteo buteo | ZMA58828 | skin | Wartena | 52.12N, 4.3E | KF946612 |
| Buteo buteo buteo | ZMA58920 | wings | Rolde | 52.58N, 6.38E | KF946613 |
| Calidris alpina alpina | ZMA58700 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946614 |
| Calonectris diomedea borealis | ZMA57255 | skin | Lith | 51.47N, 5.26E | KF946615 |
| Carduelis cannabina cannabina | ZMA58911 | skin | Noordijk | 52.08N, 6.34E | KF946616 |
| Carduelis carduelis | ZMA58866 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946617 |
| Carduelis chloris chloris | ZMA57337 | skin | Cadier en Keer | 50.49N, 5.46E | KF946618 |
| Carduelis chloris chloris | ZMA58947 | skin | Goor | 52.14N, 6.34E | KF946619 |
| Carduelis flammea cabaret | ZMA57248 | skin | Kennemerduinen | 52.42N, 4.58E | KF946620 |
| Carduelis flammea cabaret | ZMA58283 | skin | Westenschouwen | 51.41N, 3.42E | KF946621 |
| Carduelis flammea flammea | ZMA57251 | skin | Kennemerduinen | 52.42N, 4.58E | KF946622 |
| Carduelis flammea flammea | ZMA64564 | skin | Castricum | 52.32N, 4.36E | KF946623 |
| Carduelis flavirostris | ZMA57253 | skin | Castricum | 52.32N, 4.36E | KF946624 |
| Carduelis flavirostris | ZMA57254 | skin | Castricum | 52.32N, 4.36E | KF946625 |
| Carduelis spinus | ZMA55904 | skin | Nijverdal | 52.22N, 6.28E | KF946626 |
| Carduelis spinus | ZMA57256 | skin | Westenschouwen | 51.41N, 3.42E | KF946627 |
| Carduelis spinus | ZMA58286 | skin | Hellendoorn | 52.23N, 6.26E | KF946628 |
| Certhia brachydactyla megarhyncha | ZMA57322 | skin | Hellendoorn | 52.23N, 6.26E | KF946629 |
| Certhia brachydactyla megarhyncha | ZMA57323 | skin | Lekkerkerk | 51.53N, 4.41E | KF946630 |
| Certhia brachydactyla megarhyncha | ZMA57325 | skin | Wageningen | 51.58N, 5.38E | KF946631 |
| Certhia brachydactyla megarhyncha | ZMA57326 | skin | Zeist | 52.05N, 5.16E | KF946632 |
| Certhia brachydactyla megarhyncha | ZMA57327 | skin | Heiloo | 52.36N, 4.44E | KF946633 |
| Certhia brachydactyla megarhyncha | ZMA58805 | skin | Castricum | 52.32N, 4.36E | KF946634 |
| Certhia brachydactyla megarhyncha | ZMA58949 | skin | Lekkerkerk | 51.53N, 4.41E | KF946635 |
| Certhia brachydactyla megarhyncha | ZMA64563 | skin | Castricum | 52.32N, 4.36E | KF946636 |
| Charadrius hiaticula | Tissue452 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946637 |
| Circus aeruginosus aeruginosus | ZMA58780 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946638 |
| Circus aeruginosus aeruginosus | ZMA58826 | skin | Eibergen | 52.06N, 6.37E | KF946639 |
| Circus aeruginosus aeruginosus | ZMA58874 | wings | Zuid-Flevoland | 52.26N, 5.16E | KF946640 |
| Coccothraustes coccothraustes | ZMA56212 | skin | Laag Keppel | 51.59N, 6.13E | KF946641 |
| Corvus corax corax | ZMA57144 | skin | Appelscha/Appelskea | 52.55N, 5.2E | KF946642 |
| Coturnix coturnix coturnix | ZMA58775 | skin | Deventer | 52.15N, 6.11E | KF946643 |
| Coturnix coturnix coturnix | ZMA58776 | skin | Het Bildt | 53.17N, 5.4E | KF946644 |
| Cuculus canorus canorus | ZMA56681 | skin | Bergen | 52.4N, 4.41E | KF946645 |
| Cuculus canorus canorus | ZMA64549 | skin | Alkmaar | 52.38N, 4.44E | KF946646 |
| Delichon urbicum | ZMA56215 | skin | Sea | KF946647 | |
| Delichon urbicum urbicum | ZMA55919 | skin | Nieuwegein | 52.01N, 5.05E | KF946648 |
| Delichon urbicum urbicum | ZMA58300 | wings | Lage Zwaluwe | 51.42N, 4.42E | KF946649 |
| Delichon urbicum urbicum | ZMA58870 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946650 |
| Dendrocopos major pinetorum | ZMA58803 | skin | Oudkerk/Aldtsjerk | 53.15N, 5.53E | KF946651 |
| Dryocopus martius martius | ZMA58766 | skin | Tegelen | 51.19N, 6.09E | KF946652 |
| Emberiza citrinella citrinella | ZMA57257 | skin | Westenschouwen | 51.41N, 3.42E | KF946653 |
| Emberiza melanocephala | ZMA56996 | skin | Bovenkerk | 52.17N, 4.49E | KF946654 |
| Emberiza pusilla | ZMA58859 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946655 |
| Emberiza pusilla | ZMA58860 | skin | Vlieland Island | 53.15N, 4.59E | KF946656 |
| Emberiza schoeniclus schoeniclus | ZMA58857 | skin | Noordpolderzijl | 53.25N, 6.34E | KF946657 |
| Emberiza schoeniclus schoeniclus | ZMA58858 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946658 |
| Erithacus rubecula rubecula | Tissue436 | DNA sample | Castricum | 52.32N, 4.36E | KF946659 |
| Erithacus rubecula rubecula | Tissue437 | DNA sample | Castricum | 52.32N, 4.36E | KF946660 |
| Erithacus rubecula rubecula | ZMA58274 | skin | Bloemendaal | 52.24N, 4.33E | KF946661 |
| Erithacus rubecula rubecula | ZMA58740 | skin | Doldersum | 52.52N, 6.17E | KF946662 |
| Falco columbarius aesalon | ZMA58840 | skin | Texel Island | 53.04N, 4.43E | KF946663 |
| Falco columbarius aesalon | ZMA60127 | skin | Spaarndam | 52.24N, 4.41E | KF946664 |
| Falco peregrinus peregrinus | ZMA58872 | skin | Haarlem | 52.23N, 4.37E | KF946665 |
| Falco subbuteo subbuteo | ZMA56231 | skin | Zundert | 51.28N, 4.38E | KF946666 |
| Falco subbuteo subbuteo | ZMA56232 | skin | Heerhugowaard | 52.4N, 4.51E | KF946667 |
| Falco subbuteo subbuteo | ZMA58241 | skin | Hoogland | 52.1N, 5.21E | KF946668 |
| Falco subbuteo subbuteo | ZMA58242 | skin | Texel Island | 53.04N, 4.43E | KF946669 |
| Falco subbuteo subbuteo | ZMA58841 | skin | Amsterdam | 52.21N, 4.53E | KF946670 |
| Falco tinnunculus tinnunculus | Tissue456 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946671 |
| Falco tinnunculus tinnunculus | ZMA58296 | skin | Zaandam | 52.25N, 4.49E | KF946672 |
| Falco tinnunculus tinnunculus | ZMA58752 | skin | Maasbree | 51.21N, 6.03E | KF946673 |
| Falco tinnunculus tinnunculus | ZMA58754 | skin | Boekend | 51.22N, 6.06E | KF946674 |
| Falco tinnunculus tinnunculus | ZMA58774 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946675 |
| Falco tinnunculus tinnunculus | ZMA58837 | skin | Westzaan | 52.26N, 4.46E | KF946676 |
| Falco tinnunculus tinnunculus | ZMA58838 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946677 |
| Falco tinnunculus tinnunculus | ZMA58839 | wings | Reutum | 52.23N, 6.5E | KF946678 |
| Falco vespertinus | ZMA58773 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946679 |
| Ficedula hypoleuca muscipeta | ZMA55913 | skin | Otterlo | 52.04N, 5.5E | KF946680 |
| Ficedula hypoleuca muscipeta | ZMA57239 | skin | Markelo | 52.14N, 6.3E | KF946681 |
| Ficedula hypoleuca muscipeta | ZMA57320 | skin | Garderen | 52.12N, 5.43E | KF946682 |
| Ficedula hypoleuca | ZMA58865 | skin | Eemshaven | 53.26N, 6.52E | KF946683 |
| Fratercula arctica grabae | ZMA56226 | skin | Texel Island | 53.04N, 4.43E | KF946684 |
| Fratercula arctica grabae | ZMA58226 | skin | Texel Island | 53.04N, 4.43E | KF946685 |
| Fratercula arctica grabae | ZMA58227 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946686 |
| Fringilla coelebs coelebs | ZMA58948 | skin | Goor | 52.14N, 6.34E | KF946687 |
| Fringilla montifringilla | Tissue449 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946688 |
| Fulmarus glacialis auduboni | ZMA56235 | wings | Hondsbossche Zeewering | 52.44N, 4.38E | KF946689 |
| Fulmarus glacialis glacialis | ZMA60119 | skin | Neeltje Jans | 51.37N, 3.41E | KF946690 |
| Fulmarus glacialis glacialis | ZMA60120 | skin | Texel Island | 53.04N, 4.43E | KF946691 |
| Fulmarus glacialis glacialis | ZMA60121 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946692 |
| Fulmarus glacialis glacialis | ZMA60123 | skin | Ameland Island | 53.27N, 5.39E | KF946693 |
| Fulmarus glacialis glacialis | ZMA60124 | skin | Ameland Island | 53.27N, 5.39E | KF946694 |
| Fulmarus glacialis glacialis | ZMA60125 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946695 |
| Fulmarus glacialis glacialis | ZMA60126 | skin | Petten | 52.46N, 4.38E | KF946696 |
| Fulmarus glacialis | ZMA58737 | skin | Vlieland Island | 53.15N, 4.59E | KF946697 |
| Gallinula chloropus chloropus | Tissue105 | DNA sample | Wijde Wormer | 52.28N, 4.53E | KF946698 |
| Gallinula chloropus chloropus | Tissue110 | DNA sample | Wijde Wormer | 52.28N, 4.53E | KF946699 |
| Garrulus glandarius glandarius | ZMA58306 | wings | Amsterdam | 52.21N, 4.53E | KF946700 |
| Gavia immer | Tissue214 | DNA sample | Bergen aan Zee | 52.39N, 4.37E | KF946701 |
| Haematopus ostralegus ostralegus | Tissue458 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946702 |
| Haematopus ostralegus ostralegus | Tissue459 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946703 |
| Hirundo rustica rustica | Tissue450 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946704 |
| Hirundo rustica rustica | Tissue451 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946705 |
| Hirundo rustica rustica | ZMA56214 | skin | Amstelveen | 52.18N, 4.53E | KF946706 |
| Hirundo rustica rustica | ZMA58289 | skin | Appelscha/Appelskea | 52.55N, 5.2E | KF946707 |
| Hirundo rustica rustica | ZMA58290 | skin | Appelscha/Appelskea | 52.55N, 5.2E | KF946708 |
| Hirundo rustica rustica | ZMA58696 | skin | Rijswijk | 51.57N, 5.21E | KF946709 |
| Hirundo rustica rustica | ZMA58802 | skin | Noordbergum/Noardburgum | 53.13N, 6E | KF946710 |
| Jynx torquilla torquilla | ZMA56213 | skin | Aarle-Rixtel | 51.3N, 5.39E | KF946711 |
| Jynx torquilla torquilla | ZMA57330 | skin | Limmen | 52.34N, 4.41E | KF946712 |
| Jynx torquilla torquilla | ZMA58303 | wings | Belfeld | 51.18N, 6.08E | KF946713 |
| Jynx torquilla torquilla | ZMA58873 | skin | Wilnis | 52.11N, 4.54E | KF946714 |
| Larus argentatus argenteus | ZMA58921 | wings | Eemshaven | 53.26N, 6.52E | KF946715 |
| Larus argentatus | Tissue433 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946716 |
| Larus cachinnans | ZMA64547 | skin | Vlieland Island | 53.15N, 4.59E | KF946717 |
| Larus fuscus graelsii | Tissue432 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946718 |
| Larus fuscus intermedius | Tissue327 | DNA-sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946719 |
| Larus fuscus intermedius | ZMA55932 | skin | Neeltje Jans | 51.37N, 3.41E | KF946720 |
| Larus fuscus intermedius | ZMA56230 | skin | Europoort | 51.56N, 4.05E | KF946721 |
| Larus fuscus intermedius | ZMA58834 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946722 |
| Larus glaucoides glaucoides | ZMA58836 | wings | Texel Island | 53.04N, 4.43E | KF946723 |
| Larus hyperboreus | ZMA56221 | skin | Texel Island | 53.04N, 4.43E | KF946724 |
| Larus melanocephalus | ZMA57226 | skin | Wijdenes | 52.37N, 5.1E | KF946725 |
| Larus michahellis michahellis | ZMA58835 | skin | Afsluitdijk | 52.57N, 5.04E | KF946726 |
| Limosa lapponica lapponica | ZMA58202 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946727 |
| Limosa lapponica lapponica | ZMA58203 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946728 |
| Limosa lapponica taymyrensis | ZMA58204 | skin | Paesens | 53.24N, 6.06E | KF946729 |
| Limosa lapponica taymyrensis | ZMA58205 | skin | Paesens | 53.24N, 6.06E | KF946730 |
| Limosa lapponica taymyrensis | ZMA58206 | skin | Paesens | 53.24N, 6.06E | KF946731 |
| Limosa lapponica taymyrensis | ZMA58207 | skin | Paesens | 53.24N, 6.06E | KF946732 |
| Limosa lapponica taymyrensis | ZMA58208 | skin | Paesens | 53.24N, 6.06E | KF946733 |
| Limosa lapponica taymyrensis | ZMA58782 | wings | Castricum | 52.32N, 4.36E | KF946734 |
| Limosa lapponica taymyrensis | ZMA58783 | wings | Castricum | 52.32N, 4.36E | KF946735 |
| Limosa limosa limosa | Tissue457 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946736 |
| Limosa limosa limosa | ZMA57227 | skin | Holysloot | 52.24N, 5.01E | KF946737 |
| Limosa limosa limosa | ZMA58229 | skin | Waterland | 52.07N, 4.19E | KF946738 |
| Limosa limosa limosa | ZMA58230 | skin | Edam | 52.32N, 5.01E | KF946739 |
| Limosa limosa limosa | ZMA58231 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946740 |
| Limosa limosa limosa | ZMA58232 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946741 |
| Locustella luscinioides luscinioides | ZMA64557 | skin | Castricum | 52.32N, 4.36E | KF946742 |
| Locustella naevia naevia | ZMA56675 | skin | Almere | 52.22N, 5.13E | KF946743 |
| Locustella naevia naevia | ZMA56678 | skin | Almere | 52.22N, 5.13E | KF946744 |
| Locustella naevia naevia | ZMA57235 | skin | Westenschouwen | 51.41N, 3.42E | KF946745 |
| Locustella naevia naevia | ZMA58812 | skin | Castricum | 52.32N, 4.36E | KF946746 |
| Locustella naevia naevia | ZMA58936 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946747 |
| Locustella naevia naevia | ZMA60132 | skin | Kennemerduinen | 52.42N, 4.58E | KF946748 |
| Locustella naevia naevia | ZMA60133 | skin | Kennemerduinen | 52.42N, 4.58E | KF946749 |
| Locustella naevia naevia | ZMA64556 | skin | Castricum | 52.32N, 4.36E | KF946750 |
| Loxia curvirostra curvirostra | ZMA57246 | skin | Eesveen | 52.5N, 6.06E | KF946751 |
| Loxia curvirostra curvirostra | ZMA57247 | skin | Leersum | 52.01N, 5.25E | KF946752 |
| Luscinia megarhynchos megarhynchos | ZMA58798 | skin | Amsterdam | 52.21N, 4.53E | KF946753 |
| Lymnocryptes minimus | ZMA55930 | skin | Heerhugowaard | 52.4N, 4.51E | KF946754 |
| Lymnocryptes minimus | ZMA58293 | skin | Uitgeest | 52.31N, 4.42E | KF946755 |
| Milvus milvus milvus | ZMA58307 | wings | Grolloo | 52.55N, 6.39E | KF946756 |
| Milvus milvus milvus | ZMA58824 | wings | Susteren | 51.03N, 5.52E | KF946757 |
| Milvus milvus milvus | ZMA58825 | skin | Heurne | 51.54N, 6.34E | KF946758 |
| Motacilla alba yarrellii | ZMA58946 | skin | Haastrecht | 51.59N, 4.46E | KF946759 |
| Motacilla cinerea cinerea | ZMA57241 | skin | Westenschouwen | 51.41N, 3.42E | KF946760 |
| Motacilla cinerea cinerea | ZMA58266 | skin | Westenschouwen | 51.41N, 3.42E | KF946761 |
| Motacilla cinerea cinerea | ZMA58267 | skin | Westenschouwen | 51.41N, 3.42E | KF946762 |
| Motacilla cinerea cinerea | ZMA58945 | skin | Westenschouwen | 51.41N, 3.42E | KF946763 |
| Muscicapa striata striata | ZMA57336 | skin | Ilpendam | 52.27N, 4.56E | KF946764 |
| Numenius arquata arquata | Tissue431 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946765 |
| Numenius arquata arquata | ZMA58765 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946766 |
| Numenius arquata arquata | ZMA58829 | skin | Heemskerk | 52.3N, 4.36E | KF946767 |
| Oenanthe oenanthe leucorhoa | ZMA58868 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946768 |
| Oenanthe oenanthe oenanthe | ZMA58275 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946769 |
| Oenanthe oenanthe oenanthe | ZMA58800 | skin | Noordbergum/Noardburgum | 53.13N, 6E | KF946770 |
| Oriolus oriolus oriolus | ZMA58288 | skin | Heteren | 51.57N, 5.45E | KF946771 |
| Oriolus oriolus oriolus | ZMA58305 | wings | Zundert | 51.28N, 4.38E | KF946772 |
| Pandion haliaetus haliaetus | ZMA58823 | wing | Vlieland Island | 53.15N, 4.59E | KF946773 |
| Panurus biarmicus biarmicus | ZMA57318 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946774 |
| Panurus biarmicus biarmicus | ZMA58262 | skin | Lelystad | 52.29N, 5.24E | KF946775 |
| Panurus biarmicus biarmicus | ZMA58263 | skin | Lelystad | 52.29N, 5.24E | KF946776 |
| Panurus biarmicus biarmicus | ZMA58264 | skin | Lelystad | 52.29N, 5.24E | KF946777 |
| Panurus biarmicus biarmicus | ZMA58265 | skin | Lelystad | 52.29N, 5.24E | KF946778 |
| Panurus biarmicus biarmicus | ZMA58854 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946779 |
| Panurus biarmicus biarmicus | ZMA58855 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946780 |
| Panurus biarmicus biarmicus | ZMA58856 | skin | Oostvaardersdijk | 52.29N, 5.23E | KF946781 |
| Parus ater ater | Tissue555 | DNA sample | Castricum | 52.32N, 4.36E | KF946782 |
| Parus ater ater | ZMA56219 | skin | Huizen | 52.17N, 5.14E | KF946783 |
| Parus ater ater | ZMA57242 | skin | Arnhem | 51.58N, 5.53E | KF946784 |
| Parus ater ater | ZMA57243 | skin | Amsterdam | 52.21N, 4.53E | KF946785 |
| Parus ater ater | ZMA58867 | skin | Amsterdam | 52.21N, 4.53E | KF946786 |
| Parus ater ater | ZMA64562 | skin | Castricum | 52.32N, 4.36E | KF946787 |
| Parus caeruleus caeruleus | Tissue438 | DNA sample | Castricum | 52.32N, 4.36E | KF946788 |
| Parus caeruleus caeruleus | Tissue439 | DNA sample | Castricum | 52.32N, 4.36E | KF946789 |
| Parus caeruleus caeruleus | Tissue440 | DNA sample | Castricum | 52.32N, 4.36E | KF946790 |
| Parus caeruleus caeruleus | ZMA58944 | wing | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946791 |
| Parus cristatus mitratus | ZMA56677 | skin | Nijverdal | 52.22N, 6.28E | KF946792 |
| Parus cristatus mitratus | ZMA57245 | skin | Hoog Buurlo | 52.1N, 5.5E | KF946793 |
| Parus major major | ZMA58796 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946794 |
| Parus major major | ZMA58797 | skin | Castricum | 52.32N, 4.36E | KF946795 |
| Parus palustris palustris | ZMA57244 | skin | Castricum | 52.32N, 4.36E | KF946796 |
| Parus palustris palustris | ZMA64561 | skin | Goor | 52.14N, 6.34E | KF946797 |
| Passer domesticus domesticus | ZMA58799 | skin | Cadier en Keer | 50.49N, 5.46E | KF946798 |
| Passer domesticus domesticus | ZMA60138 | skin | Lekkerkerk | 51.53N, 4.41E | KF946799 |
| Passer montanus montanus | ZMA58851 | skin | Zuidhorn | 53.14N, 6.23E | KF946800 |
| Passer montanus montanus | ZMA58852 | skin | Zuidhorn | 53.14N, 6.23E | KF946801 |
| Passer montanus montanus | ZMA58853 | skin | Zuidhorn | 53.14N, 6.23E | KF946802 |
| Passer montanus montanus | ZMA58950 | skin | Zuidhorn | 53.14N, 6.23E | KF946803 |
| Perdix perdix perdix | ZMA58738 | skin | Texel Island | 53.04N, 4.43E | KF946804 |
| Perdix perdix perdix | ZMA58739 | skin | Petten | 52.46N, 4.38E | KF946805 |
| Pernis apivorus | ZMA58827 | wings | Vledder | 52.53N, 6.13E | KF946806 |
| Phalacrocorax aristotelis aristotelis | ZMA58224 | skin | Wijk aan Zee | 52.28N, 4.34E | KF946807 |
| Philomachus pugnax | ZMA56680 | skin | Graftermeer polder | 52.33N, 4.48E | KF946808 |
| Philomachus pugnax | ZMA58250 | skin | Lelystad | 52.29N, 5.24E | KF946809 |
| Phoenicopterus chilensis | ZMA56683 | skin | Ransdorp | 52.23N, 4.59E | KF946810 |
| Phoenicurus phoenicurus phoenicurus | ZMA55914 | skin | Westenschouwen | 51.41N, 3.42E | KF946811 |
| Phylloscopus collybita collybita | ZMA55917 | skin | Nijverdal | 52.22N, 6.28E | KF946812 |
| Phylloscopus collybita collybita | ZMA55918 | wings | Leveroy | 51.14N, 5.5E | KF946813 |
| Phylloscopus collybita collybita | ZMA56217 | skin | Hoogland | 52.1N, 5.21E | KF946814 |
| Phylloscopus trochilus | ZMA58284 | skin | Lelystad | 52.29N, 5.24E | KF946815 |
| Phylloscopus trochilus | ZMA58710 | skin | Almere | 52.22N, 5.13E | KF946816 |
| Phylloscopus trochilus | ZMA58713 | skin | Egmond aan Zee | 52.37N, 4.38E | KF946817 |
| Phylloscopus trochilus | ZMA58714 | skin | Lekkerkerk | 51.53N, 4.41E | KF946818 |
| Phylloscopus trochilus | ZMA58715 | skin | Texel Island | 53.04N, 4.43E | KF946819 |
| Phylloscopus trochilus | ZMA58716 | skin | Castricum | 52.32N, 4.36E | KF946820 |
| Phylloscopus trochilus | ZMA58861 | skin | Castricum | 52.32N, 4.36E | KF946821 |
| Phylloscopus trochilus | ZMA58933 | wings | Goor | 52.14N, 6.34E | KF946822 |
| Phylloscopus trochilus | ZMA58934 | skin | Eemshaven | 53.26N, 6.52E | KF946823 |
| Picus viridis viridis | ZMA58718 | skin | Breda | 51.33N, 4.46E | KF946824 |
| Picus viridis viridis | ZMA58719 | skin | Haaksbergen | 52.08N, 6.4E | KF946825 |
| Picus viridis viridis | ZMA58720 | skin | Alkmaar | 52.38N, 4.44E | KF946826 |
| Picus viridis viridis | ZMA58721 | skin | Roggel | 51.17N, 5.54E | KF946827 |
| Picus viridis viridis | ZMA58722 | skin | Bergen | 52.4N, 4.41E | KF946828 |
| Plectrophenax nivalis insulae | ZMA56672 | skin | Castricum | 52.32N, 4.36E | KF946829 |
| Pluvialis apricaria | ZMA58213 | skin | Winsum | 53.09N, 5.38E | KF946830 |
| Pluvialis apricaria | ZMA58214 | skin | Winsum | 53.09N, 5.38E | KF946831 |
| Pluvialis apricaria | ZMA58215 | skin | Dronrijp/Dronryp | 53.11N, 5.4E | KF946832 |
| Pluvialis squatarola squatarola | ZMA56224 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946833 |
| Pluvialis squatarola squatarola | ZMA56225 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946834 |
| Puffinus gravis | ZMA64542 | skin | Sexbierum/Seisbierrum | 53.14N, 5.28E | KF946835 |
| Pyrrhula pyrrhula europoea | ZMA56673 | skin | Castricum | 52.32N, 4.36E | KF946836 |
| Pyrrhula pyrrhula europoea | ZMA58793 | skin | Castricum | 52.32N, 4.36E | KF946837 |
| Pyrrhula pyrrhula europoea | ZMA58794 | skin | Castricum | 52.32N, 4.36E | KF946838 |
| Pyrrhula pyrrhula europoea | ZMA58795 | skin | Castricum | 52.32N, 4.36E | KF946839 |
| Pyrrhula pyrrhula europoea | ZMA60137 | wings | Kennemerduinen | 52.42N, 4.58E | KF946840 |
| Rallus aquaticus aquaticus | ZMA58763 | skin | Lauwersmeer | 53.22N, 6.14E | KF946841 |
| Recurvirostra avosetta | ZMA58216 | skin | Petten | 52.46N, 4.38E | KF946842 |
| Regulus ignicapilla ignicapilla | Tissue448 | DNA sample | Castricum | 52.32N, 4.36E | KF946843 |
| Regulus ignicapilla ignicapilla | ZMA57360 | skin | Zundert | 51.28N, 4.38E | KF946844 |
| Regulus ignicapilla ignicapilla | ZMA58807 | skin | Castricum | 52.32N, 4.36E | KF946845 |
| Regulus ignicapilla ignicapilla | ZMA58808 | skin | Castricum | 52.32N, 4.36E | KF946846 |
| Regulus regulus regulus | ZMA64560 | skin | Castricum | 52.32N, 4.36E | KF946847 |
| Riparia riparia riparia | ZMA58871 | skin | Zeewolde | 52.21N, 5.34E | KF946848 |
| Saxicola rubetra | ZMA60131 | skin | Kennemerduinen | 52.42N, 4.58E | KF946849 |
| Saxicola rubetra | ZMA64555 | skin | Castricum | 52.32N, 4.36E | KF946850 |
| Somateria mollissima mollissima | ZMA58912 | skin | Lauwersoog | 53.24N, 6.12E | KF946851 |
| Stercorarius longicaudus | ZMA58779 | wings | Afsluitdijk | 52.57N, 5.04E | KF946852 |
| Stercorarius longicaudus | ZMA64546 | skin | Petten | 52.46N, 4.38E | KF946853 |
| Stercorarius parasiticus | ZMA56229 | skin | Vlieland Island | 53.15N, 4.59E | KF946854 |
| Stercorarius parasiticus | ZMA56684 | wings | Terschelling Island | 53.26N, 5.29E | KF946855 |
| Stercorarius parasiticus | ZMA58778 | skin | Den Oever | 52.56N, 5.02E | KF946856 |
| Stercorarius parasiticus | ZMA58830 | skin | Den Helder | 52.55N, 4.46E | KF946857 |
| Stercorarius pomarinus | Tissue211 | DNA sample | Texel Island | 53.04N, 4.43E | KF946858 |
| Stercorarius pomarinus | ZMA55929 | skin | Hondsbossche Zeewering | 52.44N, 4.38E | KF946859 |
| Stercorarius skua skua | ZMA64545 | skin | Egmond aan Zee | 52.37N, 4.38E | KF946860 |
| Sterna albifrons albifrons | ZMA58832 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946861 |
| Sterna hirundo hirundo | ZMA58915 | skin | Eemshaven | 53.26N, 6.52E | KF946862 |
| Sterna paradisaea | ZMA58831 | skin | Amsterdam | 52.21N, 4.53E | KF946863 |
| Streptopelia decaocto decaocto | ZMA58923 | wing | Hoogkerk | 53.12N, 6.3E | KF946864 |
| Streptopelia turtur turtur | ZMA58757 | skin | Texel Island | 53.04N, 4.43E | KF946865 |
| Sylvia atricapilla atricapilla | Tissue441 | DNA sample | Castricum | 52.32N, 4.36E | KF946866 |
| Sylvia atricapilla atricapilla | Tissue442 | DNA sample | Castricum | 52.32N, 4.36E | KF946867 |
| Sylvia atricapilla atricapilla | ZMA58268 | skin | Bloemendaal | 52.24N, 4.33E | KF946868 |
| Sylvia atricapilla atricapilla | ZMA58269 | skin | Bloemendaal | 52.24N, 4.33E | KF946869 |
| Sylvia atricapilla atricapilla | ZMA58270 | skin | Bloemendaal | 52.24N, 4.33E | KF946870 |
| Sylvia atricapilla atricapilla | ZMA58759 | skin | Cadier en Keer | 50.49N, 5.46E | KF946871 |
| Sylvia borin borin | Tissue443 | DNA sample | Castricum | 52.32N, 4.36E | KF946872 |
| Sylvia borin borin | ZMA58758 | skin | Groningen | 53.14N, 6.35E | KF946873 |
| Sylvia borin borin | ZMA58761 | skin | Almere | 52.22N, 5.13E | KF946874 |
| Sylvia borin borin | ZMA58762 | skin | Purmerend | 52.28N, 4.58E | KF946875 |
| Sylvia communis communis | ZMA55924 | wing | Asten | 51.21N, 5.48E | KF946876 |
| Sylvia communis communis | ZMA57335 | skin | Almere | 52.22N, 5.13E | KF946877 |
| Sylvia communis communis | ZMA58280 | skin | Breda | 51.33N, 4.46E | KF946878 |
| Sylvia communis communis | ZMA58939 | skin | Castricum | 52.32N, 4.36E | KF946879 |
| Sylvia communis communis | ZMA58940 | skin | Bloemendaal | 52.24N, 4.33E | KF946880 |
| Sylvia curruca blythi | ZMA58941 | skin | Houten | 52.01N, 5.1E | KF946881 |
| Sylvia curruca blythi | ZMA57237 | skin | Rotterdam | 51.57N, 4.32E | KF946882 |
| Sylvia curruca curruca | ZMA55905 | skin | Westenschouwen | 51.41N, 3.42E | KF946883 |
| Sylvia curruca curruca | ZMA55906 | skin | Amsterdam | 52.21N, 4.53E | KF946884 |
| Sylvia curruca curruca | ZMA57328 | skin | Almere | 52.22N, 5.13E | KF946885 |
| Sylvia curruca curruca | ZMA57329 | skin | Texel Island | 53.04N, 4.43E | KF946886 |
| Sylvia curruca curruca | ZMA58282 | skin | Zeewolde | 52.21N, 5.34E | KF946887 |
| Sylvia curruca curruca | ZMA58806 | skin | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946888 |
| Sylvia curruca curruca | ZMA58864 | skin | Eemshaven | 53.26N, 6.52E | KF946889 |
| Sylvia curruca curruca | ZMA58942 | skin | Bloemendaal | 52.24N, 4.33E | KF946890 |
| Sylvia nisoria nisoria | ZMA58273 | skin | Westenschouwen | 51.41N, 3.42E | KF946891 |
| Tringa ochropus | ZMA64544 | skin | Castricum | 52.32N, 4.36E | KF946892 |
| Tringa totanus totanus | ZMA58212 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946893 |
| Troglodytes troglodytes troglodytes | Tissue447 | DNA sample | Castricum | 52.32N, 4.36E | KF946894 |
| Troglodytes troglodytes troglodytes | ZMA58281 | skin | Bloemendaal | 52.24N, 4.33E | KF946895 |
| Turdus iliacus iliacus | ZMA58287 | skin | Bloemendaal | 52.24N, 4.33E | KF946896 |
| Turdus merula merula | ZMA56669 | skin | Haarlem | 52.23N, 4.37E | KF946897 |
| Turdus merula merula | ZMA56670 | skin | Bergen | 52.4N, 4.41E | KF946898 |
| Turdus merula merula | ZMA57345 | skin | Zwolle | 52.3N, 6.06E | KF946899 |
| Turdus merula merula | ZMA58731 | skin | Alkmaar | 52.38N, 4.44E | KF946900 |
| Turdus merula merula | ZMA58732 | skin | Maasbree | 51.21N, 6.03E | KF946901 |
| Turdus merula merula | ZMA58733 | skin | Maasbree | 51.21N, 6.03E | KF946902 |
| Turdus merula merula | ZMA58734 | skin | Steijl | 51.2N, 6.07E | KF946903 |
| Turdus merula merula | ZMA58736 | skin | Schiermonnikoog Island | 53.29N, 6.11E | KF946904 |
| Turdus philomelos philomelos | Tissue453 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946905 |
| Turdus philomelos philomelos | Tissue454 | DNA sample | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946906 |
| Turdus torquatus torquatus | ZMA56222 | skin | Texel Island | 53.04N, 4.43E | KF946907 |
| Turdus torquatus torquatus | ZMA56671 | skin | Castricum | 52.32N, 4.36E | KF946908 |
| Turdus torquatus torquatus | ZMA58693 | skin | Apeldoorn | 52.1N, 5.58E | KF946909 |
| Turdus torquatus torquatus | ZMA58694 | skin | Vlieland Island | 53.15N, 4.59E | KF946910 |
| Turdus torquatus torquatus | ZMA58695 | skin | Zuilichem | 51.48N, 5.07E | KF946911 |
| Turdus torquatus torquatus | ZMA64554 | skin | Texel Island | 53.04N, 4.43E | KF946912 |
| Turdus viscivorus viscivorus | ZMA60130 | skin | Kennemerduinen | 52.42N, 4.58E | KF946913 |
| Tyto alba alba | ZMA56233 | skin | Burgerbrug | 52.45N, 4.42E | KF946914 |
| Tyto alba guttata | ZMA56682 | skin | Wierden | 52.22N, 6.34E | KF946915 |
| Tyto alba guttata | ZMA58235 | skin | Texel Island | 53.04N, 4.43E | KF946916 |
| Tyto alba guttata | ZMA58236 | skin | Ouderkerk aan de Amstel | 52.17N, 4.56E | KF946917 |
| Tyto alba guttata | ZMA58843 | skin | Westzaan | 52.26N, 4.46E | KF946918 |
| Tyto alba guttata | ZMA58844 | skin | Zaanstreek | 52.28N, 4.44E | KF946919 |
| Tyto alba guttata | ZMA58845 | skin | Roodkerk/Readtsjerk | 53.15N, 5.55E | KF946920 |
| Tyto alba guttata | ZMA58846 | skin | Garijp/Garyp | 53.1N, 5.57E | KF946921 |
| Tyto alba guttata | ZMA58847 | skin | Middenmeer | 52.48N, 4.59E | KF946922 |
| Tyto alba guttata | ZMA58848 | wings | Leeuwarden/Ljouwert | 53.13N, 5.45E | KF946923 |
| Tyto alba guttata | ZMA58919 | skin | Texel Island | 53.04N, 4.43E | KF946924 |
| Tyto alba guttata | ZMA64550 | skin | Purmerend | 52.28N, 4.58E | KF946925 |
| Tyto alba guttata | ZMA64551 | skin | Goor | 52.14N, 6.34E | KF946926 |
| Uria aalge albionis | ZMA56227 | skin | Amsterdam | 52.21N, 4.53E | KF946927 |
| Uria aalge albionis | ZMA58218 | skin | Vlieland Island | 53.15N, 4.59E | KF946928 |
| Uria aalge albionis | ZMA58916 | skin | Petten | 52.46N, 4.38E | KF946929 |
| Vanellus vanellus | ZMA58784 | wing | Valkenburg | 52.09N, 4.25E | KF946930 |
| Vanellus vanellus | ZMA58785 | wing | Valkenburg | 52.09N, 4.25E | KF946931 |
| Vanellus vanellus | ZMA58786 | wing | Valkenburg | 52.09N, 4.25E | KF946932 |
| Vanellus vanellus | ZMA58787 | wing | Valkenburg | 52.09N, 4.25E | KF946933 |
| Vanellus vanellus | ZMA58788 | wing | Valkenburg | 52.09N, 4.25E | KF946934 |
| Vanellus vanellus | ZMA58789 | wing | Valkenburg | 52.09N, 4.25E | KF946935 |
| Vanellus vanellus | ZMA58790 | wing | Valkenburg | 52.09N, 4.25E | KF946936 |
| Vanellus vanellus | ZMA58791 | wing | Valkenburg | 52.09N, 4.25E | KF946937 |
Supplementary table 2.
Bird species (gulls Larus and skuas Stercorarius) from the Netherlands with low (< 1.1%) K2P-mean intraspecific distances.
| Collection number and species | Collection number and species | Distance (%) | ||
|---|---|---|---|---|
| #ZMA58835 | Larus michahellis | #Tissue327 | Larus fuscus | 0 |
| #ZMA58835 | Larus michahellis | #Tissue432 | Larus fuscus | 0 |
| #ZMA58835 | Larus michahellis | #ZMA55932 | Larus fuscus | 0 |
| #ZMA58835 | Larus michahellis | #ZMA56230 | Larus fuscus | 0 |
| #ZMA64547 | Larus cachinnans | #Tissue327 | Larus fuscus | 0 |
| #ZMA64547 | Larus cachinnans | #Tissue432 | Larus fuscus | 0 |
| #ZMA64547 | Larus cachinnans | #ZMA55932 | Larus fuscus | 0 |
| #ZMA64547 | Larus cachinnans | #ZMA56230 | Larus fuscus | 0 |
| #ZMA64547 | Larus cachinnans | #ZMA58835 | Larus michahellis | 0 |
| #ZMA58921 | Larus argentatus | #ZMA55932 | Larus fuscus | 0.14 |
| #ZMA58921 | Larus argentatus | #ZMA58835 | Larus michahellis | 0.14 |
| #ZMA58921 | Larus argentatus | #Tissue432 | Larus fuscus | 0.15 |
| #ZMA58921 | Larus argentatus | #ZMA56230 | Larus fuscus | 0.15 |
| #ZMA64547 | Larus cachinnans | #ZMA58834 | Larus fuscus | 0.15 |
| #ZMA64547 | Larus cachinnans | #ZMA58921 | Larus argentatus | 0.15 |
| #ZMA58921 | Larus argentatus | #Tissue327 | Larus fuscus | 0.16 |
| #ZMA55932 | Larus fuscus | #Tissue433 | Larus argentatus | 0.29 |
| #ZMA58835 | Larus michahellis | #Tissue433 | Larus argentatus | 0.29 |
| #Tissue433 | Larus argentatus | #Tissue432 | Larus fuscus | 0.30 |
| #ZMA56230 | Larus fusca | #Tissue433 | Larus argentatus | 0.30 |
| #ZMA64545 | Stercorarius skua | #ZMA55929 | Stercorarius pomarinus | 0.30 |
| #ZMA58836 | Larus glaucoides | #Tissue432 | Larus fuscus | 0.31 |
| #ZMA58836 | Larus glaucoides | #ZMA55932 | Larus fuscus | 0.31 |
| #ZMA58836 | Larus glaucoides | #ZMA56230 | Larus fuscus | 0.31 |
| #ZMA58836 | Larus glaucoides | #ZMA58835 | Larus michahellis | 0.31 |
| #ZMA64547 | Larus cachinnans | #Tissue433 | Larus argentatus | 0.31 |
| #ZMA64547 | Larus cachinnans | #ZMA58836 | Larus glaucoides | 0.31 |
| #Tissue433 | Larus argentatus | #Tissue327 | Larus fuscus | 0.32 |
| #ZMA58836 | Larus glaucoides | #Tissue327 | Larus fuscus | 0.32 |
| #ZMA64545 | Stercorarius skua | #Tissue211 | Stercorarius pomarinus | 0.43 |
| #ZMA58835 | Larus michahellis | #ZMA58834 | Larus fuscus | 0.45 |
| #ZMA58836 | Larus glaucoides | #ZMA58834 | Larus fuscus | 0.46 |
| #ZMA58921 | Larus argentatus | #ZMA58836 | Larus glaucoides | 0.46 |
| #ZMA56221 | Larus hyperboreus | #ZMA55932 | Larus fuscus | 0.58 |
| #ZMA58835 | Larus michahellis | #ZMA56221 | Larus hyperboreus | 0.58 |
| #ZMA56221 | Larus hyperboreus | #Tissue432 | Larus fuscus | 0.60 |
| #ZMA56230 | Larus fuscus | #ZMA56221 | Larus hyperboreus | 0.60 |
| #ZMA58921 | Larus argentatus | #ZMA58834 | Larus fuscus | 0.60 |
| #ZMA64547 | Larus cachinnans | #ZMA56221 | Larus hyperboreus | 0.61 |
| #ZMA58836 | Larus glaucoides | #Tissue433 | Larus argentatus | 0.62 |
| #ZMA56221 | Larus hyperboreus | #Tissue327 | Larus fuscus | 0.64 |
| #ZMA58921 | Larus argentatus | #ZMA56221 | Larus hyperboreus | 0.73 |
| #ZMA58834 | Larus fuscus | #Tissue433 | Larus argentatus | 0.75 |
| #ZMA56221 | Larus hyperboreus | #Tissue433 | Larus argentatus | 0.87 |
| #ZMA58836 | Larus glaucoides | #ZMA56221 | Larus hyperboreus | 0.93 |
| #ZMA58834 | Larus fuscus | #ZMA56221 | Larus hyperboreus | 1.06 |
References
- Aliabadian M, Kaboli M, Nijman V, Vences M. (2009) Molecular identification of birds: performance of distance-based DNA barcoding in three genes to delimit parapatric species. PLoS ONE 4: e4119. doi: 10.1371/journal.pone.0004119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AJ, Tavares ES, Elbourne RF. (2009) Countering criticisms of single mitochondrial DNA gene barcoding in birds. Molecular Ecology Resources 2009, 9: 257–267. doi: 10.1111/j.1755-0998.2009.02650.x [DOI] [PubMed] [Google Scholar]
- Bucklin A, Steinke D, Blanco-Bercial L. (2011) DNA barcoding of marine metazoa. Annual Review of Marine Science 3: 471-508. doi: 10.1146/annurev-marine-120308-080950 [DOI] [PubMed] [Google Scholar]
- Clement X, Posada D, Crandall K. (2000) TCS: A computer program to estimate gene genealogies. Molecular Ecology 9: 1657-1659. doi: 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Dalton DL, Kotze A. (2011) DNA barcoding as a tool for species identification in three forensic wildlife cases in South Africa. Forensic Science International 207: e51–e54. doi: 10.1016/j.forsciint.2010.12.017 [DOI] [PubMed] [Google Scholar]
- Dickinson EC. (2003) The Howard & Moore Complete Checklist of the Birds of the World, 3rd Edition Christopher Helm, London. [Google Scholar]
- Ebach MC, Carvalho MRD. (2010) Anti-intellectualism in the DNA barcoding enterprise. Zoologia (Curitiba) 27: 165-178. doi: 10.1590/S1984-46702010000200003 [DOI] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791. doi: 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Förschler MI, Khoury F, Bairlein F, Aliabadian M. (2010) Phylogenetic analyses of the Mourning Wheatear complex. Molecular Phylogenetics and Evolution 56: 758-767. doi: 10.1016/j.ympev.2010.03.022 [DOI] [PubMed] [Google Scholar]
- Grant RA, Griffiths HJ, Steinke D, Wadley V, Linse K. (2011) Antarctic DNA barcoding, a drop in the ocean? Polar Biology 34: 775–780. doi: 10.1007/s00300-010-0932-7 [DOI] [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the USA 103: 968-971. doi: 10.1073/pnas.0510466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann A, Haszprunar G, Hebert PD. (2011) DNA barcoding the geometrid fauna of Bavaria (Lepidoptera): successes, surprises, and questions. PLoS ONE 6: e17134. doi: 10.1371/journal.pone.0017134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, de Waard JR. (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B (Supplement) 270: S96–S99. doi: 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. (2004) Identification of birds through DNA barcodes. PLoS Biology 2: 1657-1663. doi: 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, de Waard JR, Landry JF. (2010) DNA barcodes for 1/1000 of the animal kingdom. Biology Letters 6: 359-362. doi: 10.1098/rsbl.2009.0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen A, Rindal E, Ericson PGP, Zuccon D, Kerr KCR, Stoeckle MY, Lifjeld D. (2010) DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. Journal of Ornithology 151: 565-578. doi: 10.1007/s10336-009-0490-3 [DOI] [Google Scholar]
- Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN. (2007) Comprehensive DNA barcode coverage of North American birds. Molecular Ecology Notes 7: 535-543. doi: 10.1111/j.1471-8286.2007.01670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr KCR, Lijtmaer DA, Barreira AS, Hebert PDN, Tubaro PL. (2009a) Probing evolutionary patterns in Neotropical birds through DNA barcodes. PLoS ONE 4: e4379. doi: 10.1371/journal.pone.0004379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr KC, Birks SM, Kalyakin MV, Red’kin YA, Koblik EA, Hebert PD. (2009b) Filling the gap-COI barcode resolution in eastern Palearctic birds. Frontiers in Zoology 6(1): 29-42. doi: 10.1186/1742-9994-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebers-Helbig D, Sternkopf V, Helbig AJ, de Knijff P. (2010) The Herring Gull complex (Larus argentatus-fuscus-cachinnans) as a model group for recent Holarctic vertebrate radiations. In: Glaubrecht M. (Ed) Evolution in Action, Springer, Berlin, 351–371. doi: 10.1007/978-3-642-12425-9_17 [DOI] [Google Scholar]
- Lijtmaer DA, Kerr KC, Stoeckle MY, Tubaro PL. (2012) DNA barcoding birds: from field collection to data analysis. In: Kress WJ, Erickson DL. (Eds) DNA Barcodes: Methods and Protocols, Springer, New York, 127–152. doi: 10.1007/978-1-61779-591-6_7 [DOI] [PubMed] [Google Scholar]
- Luo A, Zhang A, Ho SY, Xu W, Zhang Y, Shi W, Cameron SL, Zhu C. (2011) Potential efficacy of mitochondrial genes for animal DNA barcoding: a case study using eutherian mammals. BMC Genomics 12(1): 84. doi: 10.1186/1471-2164-12-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CP, Paulay G. (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biology 3: e422. doi: 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Cicero C. (2004) DNA barcoding: promise and pitfalls. PLoS Biology 2: 1529-1531. doi: 10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro-Maciel E, Reid B, Fitzsimmons NN, Le M, DeSalle R, Amato G. (2009) DNA barcodes for globally threatened marine turtles: a registry approach to documenting biodiversity. Molecular Ecology Resources 10: 252-263. doi: 10.1111/j.1755-0998.2009.02747.x [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press, Oxford. [Google Scholar]
- Neigel J, Domingo A, Stake J. (2007) DNA barcoding as a tool for coral reef conservation. Coral Reefs 26: 487-499. doi: 10.1007/s00338-007-0248-4 [DOI] [Google Scholar]
- Nijman V, Aliabadian M. (2010) Performance of distance-based DNA barcoding in the molecular identification of Primates. Comptes rendus Biologies 333: 11-16. doi: 10.1016/j.crvi.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Nijman V, Aliabadian M. (2013) DNA barcoding as a tool for elucidating species delineation in wide-ranging species as illustrated by owls (Tytonidae and Strigidae). Zoological Science 30(11): 1005-1009. doi: 10.2108/zsj.30.1005 [DOI] [PubMed] [Google Scholar]
- Olsson U, Leader PJ, Carey GJ, Khan AA, Svensson L, Alström P. (2013) New insights into the intricate taxonomy and phylogeny of the Sylvia curruca complex. Molecular Phylogenetics and Evolution 67: 72-85. doi: 10.1016/j.ympev.2012.12.023 [DOI] [PubMed] [Google Scholar]
- Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA. (2011) Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Molecular Biology and Evolution 28: 1927-1942. doi: 10.1093/molbev/msr014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance L, Knowlton N, Paulay G, Meyer C. (2009) Reef-associated crustacean fauna: biodiversity estimates using semi-quantitative sampling and DNA barcoding. Coral Reefs 28: 977-986. doi: 10.1007/s00338-009-0543-3 [DOI] [Google Scholar]
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253-1256. doi: 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Ritz M. (2009) Speciation and hybridization in skuas (Catharacta spp.). PhD dissertation, Friedrich Schiller University, Jena.
- Rubinoff D. (2006) Utility of mitochondrial DNA barcodes in species conservation. Conservation Biology 20: 1026-1033. doi: 10.1111/j.1523-1739.2006.00372.x [DOI] [PubMed] [Google Scholar]
- Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. (2005) Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philosophical Transactions of the Royal Society B 360: 1805-1811. doi: 10.1098/rstb.2005.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders GW. (2005) Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society B 360: 1879-1888. doi: 10.1098/rstb.2005.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Samson RA, de Waard JR, Houbraken J, Levesque CA, Moncalvo JM, Louis-Seize G, Hebert PDN. (2007) Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences of the USA 104: 3901–3906. doi: 10.1073/pnas.0611691104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Poyarkov NA, Hebert PDN. (2008) CO1 DNA barcoding amphibians: take the chance, meet the challenge. Molecular Ecology Resources 8: 235-246. doi: 10.1111/j.1471-8286.2007.01964.x [DOI] [PubMed] [Google Scholar]
- SOVON (2002) Atlas van de Nederlandse broedvogels 1998–2000. SOVON, Nijmegen. [Google Scholar]
- Srivathsan A, Meier R. (2012) On the inappropriate use of Kimura‐2‐parameter (K2P) divergences in the DNA-barcoding literature. Cladistics 28: 190-194. doi: 10.1111/j.1096-0031.2011.00370.x [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*. Phylogenetic Analysis Using Parsimony (and other methods), Version 4b10. Sinauer Associates, Sunderland, Massachusets. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731-2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HR, Harris WE. (2012) An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Molecular Ecology Resources 12: 377-388. doi: 10.1111/j.1755-0998.2012.03119.x [DOI] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. (2005) DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B 360: 1847-1857. doi: 10.1098/rstb.2005.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will KW, Mishler BD, Wheeler QD. (2005) The perils of DNA barcoding and the need for integrative taxonomy. Systematic Biology 5: 844-51. doi: 10.1080/10635150500354878 [DOI] [PubMed] [Google Scholar]
- Yoo HS, Eah JY, Kim JS, Kim YJ, Min MS, Paek WK, Lee H, Kim CB. (2006) DNA barcoding Korean birds. Molecules and Cells 22: 323-327. [PubMed] [Google Scholar]