Abstract
Exploring the re-emergence of embryonic signaling pathways may reveal important information for cancer biology. Nodal is a transforming growth factor-β (TGF-β)-related morphogen that plays a critical role during embryonic development. Nodal signaling is regulated by the Cripto-1 co-receptor and another TGF-β member, Lefty. Although these molecules are poorly detected in differentiated tissues, they have been found in different human cancers. Poor prognosis of glioblastomas justifies the search for novel signaling pathways that can be exploited as potential therapeutic targets. Because our intracranial glioblastoma rat xenograft model has revealed importance of gene ontology categories related to development and differentiation, we hypothesized that increased activity of Nodal signaling could be found in glioblastomas. We examined the gene expressions of Nodal, Cripto-1, and Lefty in microarrays of invasive and angiogenic xenograft samples developed from four patients with glioblastoma. Protein expression was evaluated by immunohistochemistry in 199 primary glioblastomas, and expression levels were analyzed for detection of correlations with available clinical information. Gene expression of Nodal, Lefty, and Cripto-1 was detected in the glioblastoma xenografts. Most patient samples showed significant levels of Cripto-1 detected by immunohistochemistry, whereas only weak to moderate levels were detected for Nodal and Lefty. Most importantly, the higher Cripto-1 scores were associated with shorter survival in a subset of younger patients. These findings suggest for the first time that Cripto-1, an important molecule in developmental biology, may represent a novel prognostic marker and therapeutic target in categories of younger patients with glioblastoma.
Introduction
Glioblastomas are highly invasive primary brain tumors with a largely unknown etiology that are difficult to surgically eradicate [1,2]. The therapeutic effects of radiation and the cytotoxic drug temozolomide, the routinely used treatment today, are limited, as glioblastomas inevitably recur and most patients die within 2 years of diagnosis [3–9]. To improve prognosis, increased biologic knowledge, more differentiated diagnostics, and novel therapeutic strategies are needed.
Deregulated embryonic developmental features are suggested to be involved in cancer initiation and progression. The expression of morphogens, signaling molecules that govern the formation and differentiation of tissues and organs, during development is precisely regulated and controlled by specific mediators and cues from the environment that includes temporal and spatial expression of effectors and inhibitor molecules. In cancer, the balance of regulators may be disrupted and lead to aberrant expression of pluripotency-associated genes and proteins [10]. Aggressive tumor cells may therefore show characteristics similar to embryonic progenitors [11]. Furthermore, cell fate regulation in embryonic development and oncogenic activity in several cancers seems to share common signaling pathways [12–14].
Several tumor types are now suggested to be initiated and maintained by stem-like cells having capacity for self-renewal, propagation, and potential for multilineage differentiation [15,16]. Cancer stem-like cells seem to be resistant to conventional therapy and are believed to contribute to recurrence after therapy [17,18]. In addition, a high proportion of stem-like cells and also “stemness” signatures in different tumors have been associated with poorer clinical outcome [19–23].
Exploring in cancer the re-emergence of signaling pathways that are active in normal stem cell self-renewal should have the potential to increase tumor biology knowledge and thus open areas to deduce new therapeutic targets. The Nodal pathway is one of the signaling pathways shown to be required for the maintenance of undifferentiated embryonic stem cells [24–27]. Nodal is an embryonic morphogen that belongs to the transforming growth factor-β (TGF-β) superfamily. It is involved in the formation of different germ layers (embryonic cell layers) and influences the establishment of the left-right axis of different organ systems in the body [28–30]. Nodal signaling can be regulated by cofactors such as the epidermal growth factor-like member Cripto-1 and another TGF-β member Lefty. More specifically, Nodal ligand can bind to the Cripto co-receptor and a complex of type I and type II activin receptors (ALK4/7 and ActRIIB) and trigger phosphorylation events that can activate Smad2/3 and facilitate binding to Smad4 [31]. Human Cripto-1, also defined as teratocarcinoma-derived growth factor 1, is a cell membrane protein that can be secreted [32,33]. Lefty functions as an antagonist of the Nodal signaling pathway [34]. Nodal, Lefty, and Cripto-1 are critical for early embryonic development but poorly detected in normal adult tissues. However, an increased expression of both Nodal and Cripto-1 has been detected in different human tumors [11,32,33,35–38].
Our group has developed a human glioblastoma xenograft model that characterizes essential aspects of the disease.Multicellular spheroids from patient biopsies are generated and implanted intracranially in nude rats. The model reflects highly invasive and prominent angiogenic glioblastoma characteristics, where non-angiogenic tumors can switch to angiogenic tumors after serial in vivo intracranial passaging [39,40]. This model has also revealed the importance of gene ontology (GO) categories connected to development and differentiation as well as chemoresistant characteristics associated with an infiltrative stem-like phenotype [41].
Knowing the poor prognosis and limited effect of the current treatment in glioblastomas, we aim to contribute to development of increased pathobiologic understanding and to unravel potential targets for glioblastoma therapy. As there is increased evidence for similarities between features of embryonic development and factors involved in tumor initiation and progression, we hypothesized that increased activity in the Nodal signaling pathway could be found in glioblastomas. To evaluate this, we analyzed GO categories related to development and examined the gene expression of the morphogen Nodal and its related genes Cripto-1 and Lefty in the xenograft model. We also evaluated potential clinical relevance by analyzing the association between the protein expression of these molecules and patient survival. Our findings suggest for the first time that Cripto-1, an important molecule in developmental biology, may represent a novel prognostic marker and therapeutic target in categories of younger patients with glioblastoma.
Materials and Methods
Tissue Culture
Tumor fragments were obtained at surgery from patients with glioblastoma. The collection of tumor tissue was approved by the Regional Ethics Committee at Haukeland University Hospital (Bergen, Norway). The patients gave their informed consent to specimen collection. Biopsy spheroids were prepared as described previously [42]. Briefly, biopsy tissue were minced by surgical blades and transferred to agar-coated flasks containing standard tissue culture serum-supplemented medium. Such spheroid cultures are suggested to induce limited selection of cells compared to neurosphere cultures in serum-free medium [43]. After 1 to 2 weeks in culture, spheroids with diameters between 200 and 300 µm were selected for intracerebral implantation [40].
In Vivo Experiments
Biopsy spheroids were stereotactically implanted into the right brain hemisphere of nude rats as described earlier [40,44,45]. Tumor growth was monitored using an MRI Magnetom Vision Plus 1.5T T scanner (Siemens, Erlangen, Germany) and a small loop finger coil as previously described [46]. The animals were killed when symptoms developed, and the brains were removed. The tumors were excised, and new spheroids were generated and transplanted into new animals [39,40]. Brains were also fixed in 4% formaldehyde, or tissues were snap frozen in liquid N2 for further studies. All procedures were approved by the National Animal Research Authority (Oslo, Norway).
Gene Expression Analysis
Total RNA was extracted using the RNeasy Midi Kit (Qiagen GmbH, Hilden, Germany), and cDNAs and labeled cRNAs were generated as previously described [47,48] using the Applied Biosystems Chemiluminescent RT-IVT Labeling Kit (Applied Biosystems, Foster City, CA). The Applied Biosystems Human Genome Survey Microarray, Chemiluminescence Detection Kit, and Applied Biosystems 1700 Chemiluminescent Microarray Analyzer were used according to the manufacturer's instructions for ABI1700 DNA oligonucleotide microarrays (37k). Hybridizations were performed for corresponding low- and high-generation tumors from four patients. The resulting files from the image processing software were imported into the analysis software J-Express [49] (http://jexpress.bioinfo.no). Array data are available in the public repository ArrayExpress (http://www.ebi.ac.uk/arrayexpress; Accession No. E-MTAB-1185; Table W1). Controls, flagged spots, and weak spots (S/N < 3) were removed. All arrays were quantile normalized. Similar samples have previously been analyzed by two additional microarray platforms (Agilent Technologies 16k cDNA and 44k oligonucleotide arrays) and by real-time quantitative polymerase chain reaction [39]. Overrepresentation statistics of GO categories were calculated on the ranked gene list produced by a paired significance analysis of microarrays of data from low- and high-generation tumor samples. The top 1000 differentially expressed genes of the 18,269 present in total in the normalized data set was used as a representative top list to investigate for overrepresentation of GO categories, using a Fisher's exact test as implemented in J-Express.
Patient Material and Histopathology
Tumor biopsies originated from patients with glioma undergoing surgery at Haukeland University Hospital in the period from 1998 to 2008. Formalin-fixed and paraffin-embedded tissue specimens from 243 glioma biopsies were available. Hematoxylin and eosin-stained sections for all patients were examined, without knowledge of the clinical course, to verify the glioblastoma diagnosis (World Health Organization grade IV) according to the World Health Organization classification [50]. The patients gave their written informed consents to specimen collection, and the ethical board in Norway approved the study.
Tissue Microarray and Immunohistochemistry
Cylinders, 1 mm in diameter, were punched from representative areas of the tumors and mounted in recipient paraffin blocks using a standard precision instrument (Manual Tissue Arrayer MTA-1; Beecher Instruments, Inc, Sun Prairia, WI). A total of 729 cylinders were mounted, three representative samples of most tumors in addition to control samples from human tonsil, liver, gray matter, white matter, glioblastoma, and medulloblastoma. Five-micrometer-thick sections were prepared from the tissue microarray blocks and stained with hematoxylin and eosin to confirm that the specimens were suitable for immunohistochemical evaluation.
Five-micrometer-thick, formalin-fixed, paraffin-embedded tissue sections were also prepared for immunohistochemistry, carried out on a Microm HMS 710i Automated Immunostainer (Thermo Fisher Scientific/Richard-Allan Scientific, Fremont, CA). Briefly, following antigen retrieval (citrate buffer, pH 6.0) and blocking steps (H2O2, avidin, biotin, and Background Sniper for 10 minutes each), sections were incubated in mouse anti-human Nodal antibody (Abcam, Cambridge, MA; ab55676) in 1:50 dilution for 60 minutes, followed by biotinylated goat anti-mouse IgG secondary antibody (Biocare Medical, LLC, Pike Lane Concord, CA; GM601H) and then streptavidinperoxidase (Thermo Fisher Scientific Lab Vision, TS-125-HR) for 20 minutes. For the Cripto-1 detection, sections were incubated in rabbit anti-human Cripto-1 (teratocarcinoma-derived growth factor 1) antibody (Rockland, Gilbertsville, PA; 600-401-997) in 1:400 dilutions for 55 minutes followed by biotinylated goat anti-rabbit IgG secondary antibody (Biocare Medical, LLC; GR602 H). For the Lefty detection, sections were incubated in goat anti-Lefty (M-20) polyclonal antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, CA; sc-7408) in 1:50 dilutions for 60 minutes followed by biotinylated mouse anti-goat IgG secondary antibody (Biocare Medical, LLC; MG610 H). Color was developed with 3,3′-diaminobenzidine substrate (Thermo Scientific Lab Vision, TA-125-HDX), and sections were counterstained with hematoxylin (Biocare Medical, LLC; NM-HEM). As negative controls, adjacent serial sections were incubated with ChromPure mouse, rabbit, or goat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA; ChromPure Mouse IgG 015-000-003, ChromPure Rabbit IgG 011-000-003, and ChromPure Goat IgG 005-000-003) at the same concentration as the primary antibodies.
After staining, the specimens were evaluated semiquantitatively and blinded for the follow-up/clinical data by a method developed and validated in melanomas [51]. As the molecules analyzed are both cell surface localized and secreted into the surrounding matrix, the overall staining of the entire tissue was evaluated and given the following expression scores: negative (0), weak (1), moderate (2), and strong (3). A minimum of two spots from each patient was represented, and the highest staining score from each patient was chosen. The protein expression scores were also categorized in the following two groups: low expression (scores 0–1) and high expression (scores 2–3).
Statistics
A filter to select only the primary glioblastomas (tumors developed from lower grade and recurrent tumors were not included) was used, and frequency analyses of age, age classes, and protein expression were performed. The overall survival time (observation days) was defined as the time from operation to death (postoperative survival time) or to a censored time point for the eight patients still alive (censored survival time). Univariate survival data and curves were estimated by the Kaplan-Meier method [52] using the log-rank test [53] for disclosing differences between categories of each variable (gender, age classes, and different protein score groups). Variables were further analyzed by use of the Cox proportional hazards model [54]. To assess associations of variables to survival time, a backward stepwise selection process was used to identify a final multiple Cox proportional hazards regression model. The significance level was set at P ≤ .05. All statistical calculations were conducted using the Statistical Package for the Social Sciences (SPSS) software (PASW Statistics 18.0).
Results
Gene Expression in the Xenograft Model
We have earlier reported that GO categories connected to developmental aspects and negative regulators of differentiations are revealed in our glioblastoma xenograft model especially in the highly invasive phenotype [41]. Further evaluation of the analysis of statistical overrepresented GO categories from the top 1000 differential expressed genes in the paired significance analysis of microarrays from low (highly invasive)- and high (highly angiogenic)-generation tumor samples notified that 5 of the 51 GO categories with a P value < .05 were morphogenesis categories (data not shown).
As a follow-up, we examined the gene expression of the morphogen Nodal and the related genes Cripto-1 and Lefty in the highly invasive and in the highly angiogenic tumors developed after serial intracranial passaging. Nodal, Cripto-1, and Lefty were found expressed in both the invasive and angiogenic phenotypes. When comparing gene expression intensity in the two phenotypes, the Nodal gene expression intensity was found decreased in three of four patients in the highly angiogenic phenotype compared to the invasive phenotype (Figure 1A). The expression of the co-receptor Cripto-1 gene was found slightly increased in the angiogenic phenotype in a similar number of patients (Figure 1B). Furthermore, the decrease in Nodal expression in the highly angiogenic tumors corresponded with an increase in the expression of its inhibitor Lefty in samples from two of the patients (Figure 1C). Our findings suggest potential roles for these genes in glioblastomas. However, glioblastomas are known to be highly heterogeneous, and microenvironmental interplay and epigenetic changes may influence the expression of specific genes and proteins. To study the potential functional and clinical relevance of these genes, we examined the protein expression of Nodal, Cripto-1, and Lefty in samples from 243 patients with glioblastoma.
Figure 1.
Gene expression of Nodal (A), Cripto-1 (B), and Lefty (C) in samples from highly invasive versus angiogenic phenotypes in glioblastoma xenografts developed from four (1–4) glioblastoma patient biopsies. The diagrams illustrate log2-transformed gene expression intensity. The data were extracted from globally normalized (quantile normalization) microarray data. ArrayExpress Data Archive and corresponding patient sample codes are given in Table W1. The expression levels of Nodal, Lefty, and Cripto-1 are in the range of 0.3 to 0.7 compared to the levels of housekeeping genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin, beta (ACTB).
Patient Samples and Protein Expression
We wanted to assess the protein expression of our genes of interest in a cohort as homogeneous as possible.We therefore chose to exclude the patients with recurrent tumors and tumors developed from lower grade. Of the 243 glioma patient biopsies examined, the 199 patients with primary glioblastoma were selected. Only these patients, 115 males and 84 females, were included in the following analyses. The age range was 30 to 84 years, and the median age at operation was 65 years. The numbers of patients in different age classes are given in Table 1. Median postoperative survival time was 259 days (37 weeks/8.6 months). There was no significant difference between males (277 days) and females (229 days) in median survival time (log-rank test P value = .439).
Table 1.
Age and Sex Distribution of 199 Patients with Primary Glioblastoma Operated at Haukeland University Hospital in the Period from 1998 to 2008.
| Frequency | Percentage | |
| Sex | ||
| Male | 115 | 57.8 |
| Female | 84 | 42.2 |
| Age at operation in years* | ||
| 30 to 39 | 6 | 3.0 |
| 40 to 49 | 19 | 9.5 |
| 50 to 59 | 40 | 20.1 |
| 60 to 69 | 76 | 38.2 |
| 70 to 79 | 47 | 23.6 |
| 80 to 89 | 11 | 5.5 |
Median, 65 years; range, 30 to 84 years.
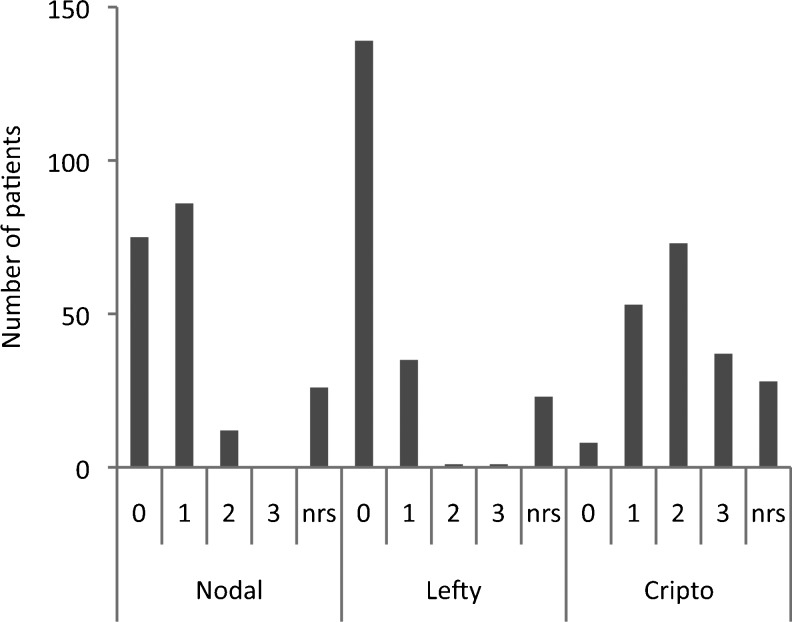
Protein expression of Nodal, Lefty, and Cripto-1 was assessed by immunohistochemistry on formalin-fixed and paraffin-embedded tissue microarray specimens (Figure 2). A variable expression of the different proteins examined was found in the primary glioblastoma specimens (Figure 3). The molecules analyzed in this study are both cytoplasmic and cell surface localized and also secreted into the surrounding matrix, revealing a rather diffuse staining pattern. The staining pattern was homogeneous within the tumor tissue in most samples, but the staining intensity varied between the different patients. Overall, a negative-to-weak expression of Nodal and Lefty and a stronger expression of Cripto-1 were found in these tumors (Figure 4). The study demonstrated moderate-to-strong expression of Cripto-1 protein in more than 50% of the primary glioblastomas examined, and only 4% were negative for this protein. In contrast, we could only verify a moderate expression of Nodal in 6% of the patients. Lefty was found moderately expressed in one patient and strongly expressed in another.
Figure 2.
Examples of immunostaining showing moderate (2) Nodal intensity staining (A) and strong (3) Lefty (C) and Cripto-1 (D) staining and a negative control IgG (B) in human glioblastoma biopsy sections. The arrowhead points to a mitotic cell in the specimen stained for Cripto-1.
Figure 3.
Different degrees of protein expression of Nodal, Lefty, and Cripto-1 in human glioblastomas. Nodal: scores 0 (A), 1 (B), and 2 (C). Negative control IgG (D). Lefty: scores 0 (E), 1 (F), 2 (G), and 3 (H). Cripto-1: scores 0 (I), 1 (J), 2 (K), and 3 (L).
Figure 4.
Numbers of patients with glioblastoma with different scores of Nodal, Lefty, and Cripto-1 protein expression. nrs denotes non-representative specimen/staining for evaluation.
We also asked whether high or low expression of any of the three markers co-associates with high or low expression of any of the other two. Both patients with high Lefty expression had a low Nodal expression and a high Cripto-1 expression. (This might suggest a possible inhibitory effect of Lefty on Nodal expression in these two patients.) When selecting for patients with high (scores 2–3) Nodal expression, there were 12 patients with expression score 2 (no patient had a Nodal expression score 3). Eight of these patients had a high (scores 2–3) Cripto-1 expression (four score 2 and four score 3), and all had a low (scores 0–1) Lefty expression.
In conclusion, protein expression studies of the important Nodal signaling molecules Nodal, Lefty, and Cripto-1 revealed convincing importance of Cripto-1, possibly independent of Nodal and Lefty in our glioblastoma patient cohort.
Protein Expression, Age, and Survival
To investigate if specific protein expression categories were associated with survival time, univariate survival analysis (Kaplan-Meier method) was performed. The protein expression scores were categorized in two groups, low (scores 0–1) and high (scores 2–3) expression. We were unable to reveal any significant association between protein expression and postoperative survival time in non-stratified analyses of observation days by low/high protein expression categories of Nodal, Lefty, or Cripto-1 (Table 2).
Table 2.
Frequency and Postoperative Survival of Patients with Primary Glioblastoma with Low and High Protein Expression Categories.
| Protein Expression | Number of Patients | Percentages of Patients | Median Survival Estimate (Days) | SE | 95% CI | Log-Rank Test P Value |
| Nodal | .595 | |||||
| Low (scores 0–1) | 161 | 80.9 | 241 | 26.1 | (190.0, 292.0) | |
| High (scores 2–3) | 12 | 6.0 | 285 | 13.0 | (259.5, 310.5) | |
| Missing | 26 | 13.1 | ||||
| Lefty | .162 | |||||
| Low (scores 0–1) | 174 | 87.4 | 259 | 28.2 | (203.7, 314.3) | |
| High (scores 2–3) | 2 | 1.0 | 86 | |||
| Missing | 23 | 11.6 | ||||
| Cripto-1 | .161 | |||||
| Low (scores 0–1) | 61 | 30.7 | 259 | 54.1 | (153.0, 365.1) | |
| High (scores 2–3) | 110 | 55.3 | 242 | 35.9 | (171.6, 312.4) | |
| Missing | 28 | 14.1 |
Univariate survival analyses (Kaplan-Meier method) of 199 patients operated at Haukeland University Hospital in the period from 1998 to 2008.
SE indicates standard error; CI, confidence interval.
There was a significant influence of age in non-stratified survival analysis (log-rank test P value < .001). When stratified for age (<60 and ≥60), patients <60 years had a significantly longer (107 days) median postoperative survival when they expressed low levels of Cripto-1 compared to high levels (log-rank test P value = .044). In contrast, for patients ≥60 years, no significant differences related to differential Cripto-1 expression was shown (P = .597). For patients below 50 years, the difference in median survival was 242 days (8 months; P = .013) in favor of Cripto-1 low-expressing tumors.
Both continuous age and age decades had a significant effect on postoperative survival time of the patients with glioblastoma, and the Cox regression analyses revealed interaction effect of age and Cripto-1 expression on survival (P = .006) (Table 3). The table expresses that 10 years increased age reduces the hazard ratio for high Cripto-1 by a factor of 0.63 as also shown in the corresponding Cox regression plots in Figure 5. These results showed a worse effect on survival with high Cripto-1 in younger patients compared to older patients.
Table 3.
Results from Cox Regression of 171 Patients with Primary Glioblastoma Operated at Haukeland University Hospital in the Period from 1998 to 2008.
| HR | 95% CI | LR Test P Value | |
| Age/10 years (continuous) | 2.01 | (1.53, 2.64) | <.001 |
| Cripto-1 | .006 | ||
| Low (scores 0–1) | 1.00 | Reference | |
| High (scores 2–3) | 18.85 | (2.20, 161.26) | |
| Cripto-1 high x age/10 | 0.63 | (0.45, 0.88) | .006 |
HR indicates hazard ratio; CI, confidence interval; LR, likelihood ratio.
Figure 5.
Illustration of decreasing effect on survival of high Cripto-1 protein expression score with increasing age. Survival functions and hazard ratios were estimated from 171 patients with primary glioblastoma, who were operated at Haukeland University Hospital in the period from 1998 to 2008, by a Cox regression model with interaction term between age and Cripto-1 score (likelihood test P = .006). The Cox regression plots illustrate the survival for individuals with the age of 40, 50, 60, and 70 years, respectively, as estimated by the Cox regression model.
No additional significant associations could be added by looking at different combinations of protein expression of the respective molecules Nodal, Cripto-1, or Lefty.
Discussion
In this study, we report, for the first time, considerable levels of Cripto-1 in more than 50% of the patients in a glioblastoma cohort including 199 primary tumors. In contrast, Nodal and its inhibitor Lefty exhibited very limited protein expression in these samples. The study revealed that younger patients with glioblastoma with high Cripto-1 protein expression were associated with a significantly shorter postoperative median survival compared to patients in the same age range with low Cripto-1 expressing tumors. The expression of Cripto-1, known to be important in maintaining stem cell pluripotency, and its association with age and survival are new and surprising findings in glioblastoma that can contribute to the identification of young patients with potentially worse prognosis than other young patients with glioblastoma, who, in general, have a more favorable prognosis after diagnosis compared to older patients.
Cripto-1 in Human Tumors
Cripto-1, known as a co-receptor for the TGF-β-related morphogen Nodal, is shown to play important roles in embryonic development and tumor progression [55]. Expression of Cripto-1 mRNA and/or protein has been found in human tumors including colorectal, breast, gastric, pancreatic, ovarian, and lung carcinomas [32,33] as well as in uveal [56] and cutaneous melanomas [57]. Interestingly, high levels of Cripto-1 have been associated with aggressive clinicopathologic features and poor prognosis in colorectal, gastric, and breast cancers [58–60]. The proportion of patients with Cripto-1-positive glioblastoma found in this study is comparable to those reported for other cancers [61].
Potential Nodal-Independent Mechanisms of Cripto-1 in Glioblastomas
To our knowledge, protein expression and side-by-side comparison of the expression of both Nodal and Cripto-1 in a large homogeneous diagnosed tumor patient cohort have not earlier been evaluated. Although other studies have indicated a role for Nodal in brain tumor invasion [36] and angiogenesis [62], our study showed a considerable protein expression of Cripto-1 and very limited expression of Nodal and Lefty in the tumors, suggesting a potential functional role for Cripto-1 independent of Nodal and Lefty in primary glioblastomas.
It is known that Cripto-1 can act as a cofactor for Nodal and interact with Nodal signaling at different levels [32], but it has also been shown to act as a growth factor independent of Nodal [63–65]. For instance, Cripto-1 can activate mitogen-activated protein kinase and Akt pathways independently of Nodal by directly binding to a membrane-associated heparan sulfate proteoglycan, glypican-1 [64], or to the heat shock glucose-regulated protein (GRP78) [66–68]. The binding can phosphorylate the tyrosine kinase c-Src, which in turn can trigger Nodal-independent pathways that influence epithelial-to-mesenchymal transition, migration, and invasion as well as proliferation and survival [32,64,69].
Although c-Src is suggested to be required for Cripto-1 activation of mitogen-activated protein kinase and Akt, immunohistochemical staining of glioblastoma biopsy specimens from our patients could not verify whether Cripto-1 signaling is occurring through these pathways (data not shown). However, this does not rule out the involvement of Cripto-1 in alternative signaling pathways [68].
Cripto-1 and Angiogenesis
Cripto-1-induced enhancement of migration, invasion, proliferation, and survival functions has been shown in epithelial cells, carcinoma, sarcoma, melanoma, and endothelial cells, indicating a role for Cripto-1 also as an angiogenic molecule in the formation of vascular structures in tumors [33,57,70,71]. Cripto-1 expression can be regulated by hypoxia-inducible factor-1a, and it has therefore been suggested that hypoxic conditions in tumors may trigger Cripto-1 expression that can contribute to increased tumor vascularization [72]. Glioblastomas are highly invasive and angiogenic tumors. In our xenograft model, we are able to study the distinct characteristic phenotypes. Interestingly, we observed slight increased Cripto-1 gene expression intensity in the animal angiogenic phenotype, indicating a possible contribution of Cripto-1 in glioblastoma angiogenesis. It would be interesting to determine the stage at which Cripto-1 would contribute to tumor angiogenesis in glioblastoma. In our patient cohort, the association between evolution of tumor angiogenesis and Cripto-1 levels could not be determined since the tumor samples were already highly vascularized.
Molecular Classification of Glioblastomas and Targets for Treatment
Heterogeneous glioblastoma populations (including both primary and secondary glioblastomas as well as some grade III tumors) have recently been found to cluster into four molecular subtypes, namely, proneural, neural, classic and mesenchymal, based on gene expression profiles [73,74]. Although these molecular subclasses predict prognosis and show different treatment responses, no specific targeted treatments have yet been suggested. In a more treatment-focused perspective, a stem cell-related “self-renewal” signature associated with resistance to concomitant chemotherapy in glioblastomas has been reported and suggested as a guide to future therapy and marker development for individualized treatment [17]. In that respect, the increased GRP78 mRNA expression found to be involved in chemoresistance and inversely correlated with survival in glioblastomas [75] is of interest, especially since Cripto-1/GRP78 interaction has been suggested to regulate this novel pathway in stem cell and cancer stem cell activities [76,77]. Approaches to target Cripto-1 expression or function should therefore become attractive for glioblastomas.
Inhibition of Cripto-1
On the basis of the known properties of Cripto-1 as having a critical role during early embryogenesis in stem cell renewal and pluripotency and to be expressed both in stem-like and angiogenic cancer phenotypes, targeting Cripto-1 could have the potential to eliminate both differentiated tumor cells and an undifferentiated subpopulation of cancer stem-like cells suggested to be important in tumor initiation and self-renewal [37]. Cripto antibodies repress tumor cell growth [78–80], and a humanized antibody is now being evaluated in a phase I clinical trial in cancer patients (NCT 00674947). To prepare for intracranial in vivo studies, and because we have detected Cripto-1-positive neurons (data not shown), potential unfavorable effects in normal tissue of inhibiting Cripto-1 protein expression have to be considered. In fact, Cripto has been indicated to have an important role in preventing neuronal differentiation in embryonic stem cells [81] and has been suggested as a target for improving embryonic stem cell-based therapy in Parkinson's disease [82,83]. These findings support that inhibition of Cripto-1 also in patients with brain tumors can be appropriate even with the existence of Cripto-1-positive neurons, because increased neuronal differentiation should be a desired effect.
Summary and Future Prospects
In our study, statistical considerations revealed a significant age-related association between expression of the pluripotency factor Cripto-1 and survival. These results suggest that Cripto-1 protein expression in younger patients with primary glioblastoma may represent a novel negative prognostic marker and a potential therapeutic target in these patients. Our work underline the necessity of examining a large number of patients to find and evaluate potential targets with clinical impact in glioblastomas, tumors that reveal some common morphologic features, but also show genetic and molecular heterogeneity to a large extent. Given the role of Cripto-1 in embryonic stem cells, glioblastoma cells in younger patients may retain a higher level of plasticity compared to older patients, which could translate into increased responsiveness to Cripto-1-dependent effects, such as proliferation, angiogenesis, and epithelial-to-mesenchymal transition. Here, we report a potential clinical important finding that warrants further studies, where survival in a larger group of younger aged patients with glioblastoma should be evaluated on the basis of Cripto-1 gene and protein expression, O6-methylguanine-DNA methyltransferase methylation status, performance score, and treatment to verify Cripto-1 as a therapeutic target that could influence patient outcome. In addition, possible biologic samples to scrutinize specific biologic mechanisms behind clinical differences should be obtained to design the most effective inhibitors. The use of appropriate tumor material and models will be crucial for delineating clinical important mechanisms, where development of tailored therapy for specific subgroups of patients requires models that reflect both the genomics and proteomics of the tumors and also mimic the microenvironment present.
Supplementary Material
Acknowledgments
We thank Laila Vårdal for production of the tissue microarrays (TMAs), Per Øyvind Enger and Jian Wang for doing animal experiments, Kai Ove Skaftnesmo and Per Øystein Sakariassen for collecting xenograft RNA, Anne M. Øyan and Karl-Henning Kalland for microarray data acquisition and analysis, Hrvoje Miletic for providing additional immunostained TMAs, and Rolf Bjerkvig for contributions to conception of the study, financial support, and comments.
Footnotes
The work was supported by the Norwegian Cancer Society, the Norwegian Research Council, Innovest AS, Strategic Research Programme, Helse-Vest, Haukeland University Hospital (Bergen, Norway), and the National Institutes of Health grants CA121205 and CA59702 to M.J.C.H.
This article refers to supplementary material, which is designated by Table W1 and is available online at www.transonc.com.
References
- 1.Ohgaki H, Kleihues P. Cost of migration: invasion of malignant gliomas and implications for treatment. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Reardon DA, Wen PY. Therapeutic advances in the treatment of glioblastoma: rationale and potential role of targeted agents. Oncologist. 2006;11:152–164. doi: 10.1634/theoncologist.11-2-152. [DOI] [PubMed] [Google Scholar]
- 7.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 9.Tran B, Rosenthal MA. Survival comparison between glioblastoma multiforme and other incurable cancers. J Clin Neurosci. 2010;17:417–421. doi: 10.1016/j.jocn.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Postovit LM, Costa FF, Bischof JM, Seftor EA, Wen B, Seftor RE, Feinberg AP, Soares MB, Hendrix MJ. The commonality of plasticity underlying multipotent tumor cells and embryonic stem cells. J Cell Biochem. 2007;101:908–917. doi: 10.1002/jcb.21227. [DOI] [PubMed] [Google Scholar]
- 11.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med. 2006;12:925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 12.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 13.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 14.Tysnes BB, Bjerkvig R. Cancer initiation and progression: involvement of stem cells and the microenvironment. Biochim Biophys Acta. 2007;1775:283–297. doi: 10.1016/j.bbcan.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 16.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 17.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Ramena G, Elble RC. The role of cancer stem cells in relapse of solid tumors. Front Biosci (Elite Ed) 2012;4:1528–1541. doi: 10.2741/e478. [DOI] [PubMed] [Google Scholar]
- 19.Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 20.Zeppernick F, Ahmadi R, Campos B, Dictus C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B, Herold-Mende CC. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 21.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26:2846–2853. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- 24.Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 25.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 26.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 27.Saha S, Ji L, de Pablo JJ, Palecek SP. TGFβ/Activin/Nodal pathway in inhibition of human embryonic stem cell differentiation by mechanical strain. Biophys J. 2008;94:4123–4133. doi: 10.1529/biophysj.107.119891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schier AF, Shen MM. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 29.Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 30.Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 31.Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianco C, Strizzi L, Normanno N, Khan N, Salomon DS. Cripto-1: an oncofetal gene with many faces. Curr Top Dev Biol. 2005;67:85–133. doi: 10.1016/S0070-2153(05)67003-2. [DOI] [PubMed] [Google Scholar]
- 33.Strizzi L, Bianco C, Normanno N, Salomon D. Cripto-1: a multifunctional modulator during embryogenesis and oncogenesis. Oncogene. 2005;24:5731–5741. doi: 10.1038/sj.onc.1208918. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Shen MM. Two modes by which Lefty proteins inhibit nodal signaling. Curr Biol. 2004;14:618–624. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- 35.Strizzi L, Postovit LM, Margaryan NV, Lipavsky A, Gadiot J, Blank C, Seftor RE, Seftor EA, Hendrix MJ. Nodal as a biomarker for melanoma progression and a new therapeutic target for clinical intervention. Expert Rev Dermatol. 2009;4:67–78. doi: 10.1586/17469872.4.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CC, Jan HJ, Lai JH, Ma HI, Hueng DY, Lee YC, Cheng YY, Liu LW, Wei HW, Lee HM. Nodal promotes growth and invasion in human gliomas. Oncogene. 2010;29:3110–3123. doi: 10.1038/onc.2010.55. [DOI] [PubMed] [Google Scholar]
- 37.Bianco C, Salomon DS. Targeting the embryonic gene Cripto-1 in cancer and beyond. Expert Opin Ther Pat. 2010;20:1739–1749. doi: 10.1517/13543776.2010.530659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strizzi L, Hardy KM, Margaryan NV, Hillman DW, Seftor EA, Chen B, Geiger XJ, Thompson EA, Lingle WL, Andorfer CA, et al. Potential for the embryonic morphogen Nodal as a prognostic and predictive biomarker in breast cancer. Breast Cancer Res. 2012;14:R75. doi: 10.1186/bcr3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakariassen PØ, Prestegarden L, Wang J, Skaftnesmo KO, Mahesparan R, Molthoff C, Sminia P, Sundlisaeter E, Misra A, Tysnes BB, et al. Angiogenesis-independent tumor growth mediated by stem-like cancer cells. Proc Natl Acad Sci USA. 2006;103:16466–16471. doi: 10.1073/pnas.0607668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Miletic H, Sakariassen PO, Huszthy PC, Jacobsen H, Brekkå N, Li X, Zhao P, Mørk S, Chekenya M, et al. A reproducible brain tumour model established from human glioblastoma biopsies. BMC Cancer. 2009;9:465. doi: 10.1186/1471-2407-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johannessen TC, Wang J, Skaftnesmo KO, Sakariassen PØ, Enger PØ, Petersen K, Øyan AM, Kalland KH, Bjerkvig R, Tysnes BB. Highly infiltrative brain tumours show reduced chemosensitivity associated with a stem cell-like phenotype. Neuropathol Appl Neurobiol. 2009;35:380–393. doi: 10.1111/j.1365-2990.2008.01008.x. [DOI] [PubMed] [Google Scholar]
- 42.Bjerkvig R, Tønnesen A, Laerum OD, Backlund EO. Multicellular tumor spheroids from human gliomas maintained in organ culture. J Neurosurg. 1990;72:463–475. doi: 10.3171/jns.1990.72.3.0463. [DOI] [PubMed] [Google Scholar]
- 43.Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PØ, Miletic H, Thorsen F, Bjerkvig R. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012;14:979–993. doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engebraaten O, Hjortland GO, Hirschberg H, Fodstad O. Growth of precultured human glioma specimens in nude rat brain. J Neurosurg. 1999;90:125–132. doi: 10.3171/jns.1999.90.1.0125. [DOI] [PubMed] [Google Scholar]
- 45.Mahesparan R, Read TA, Lund-Johansen M, Skaftnesmo KO, Bjerkvig R, Engebraaten O. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta Neuropathol. 2003;105:49–57. doi: 10.1007/s00401-002-0610-0. [DOI] [PubMed] [Google Scholar]
- 46.Thorsen F, Ersland L, Nordli H, Enger PO, Huszthy PC, Lundervold A, Standnes T, Bjerkvig R, Lund-Johansen M. Imaging of experimental rat gliomas using a clinical MR scanner. J Neurooncol. 2003;63:225–231. doi: 10.1023/a:1024241905888. [DOI] [PubMed] [Google Scholar]
- 47.Halvorsen OJ, Oyan AM, Bø TH, Olsen S, Rostad K, Haukaas SA, Bakke AM, Marzolf B, Dimitrov K, Stordrange L, et al. Gene expression profiles in prostate cancer: association with patient subgroups and tumour differentiation. Int J Oncol. 2005;26:329–336. [PubMed] [Google Scholar]
- 48.Petersen K, Oyan AM, Rostad K, Olsen S, Bø TH, Salvesen HB, Gjertsen BT, Bruserud O, Halvorsen OJ, Akslen LA, et al. Comparison of nucleic acid targets prepared from total RNA or poly(A) RNA for DNA oligonucleotide microarray hybridization. Anal Biochem. 2007;366:46–58. doi: 10.1016/j.ab.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Stavrum AK, Petersen K, Jonassen I, Dysvik B. Analysis of gene-expression data using J-Express. Curr Protoc Bioinformatics. 2008;Chapter 7 doi: 10.1002/0471250953.bi0703s21. Unit 7.3. [DOI] [PubMed] [Google Scholar]
- 50.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu L, Harms PW, Pouryazdanparast P, Kim DS, Ma L, Fullen DR. Expression of the embryonic morphogen Nodal in cutaneous melanocytic lesions. Mod Pathol. 2010;23:1209–1214. doi: 10.1038/modpathol.2010.101. [DOI] [PubMed] [Google Scholar]
- 52.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 53.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 54.Cox D. Regression models and life tables. J R Stat Soc Series B. 1972;34:187–220. [Google Scholar]
- 55.Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, Salomon DS. Role of Cripto-1 in stem cell maintenance and malignant progression. Am J Pathol. 2010;177:532–540. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallikarjuna K, Vaijayanthi P, Krishnakumar S. Cripto-1 expression in uveal melanoma: an immunohistochemical study. Exp Eye Res. 2007;84:1060–1066. doi: 10.1016/j.exer.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 57.De Luca A, Lamura L, Strizzi L, Roma C, D'Antonio A, Margaryan N, Pirozzi G, Hsu MY, Botti G, Mari E, et al. Expression and functional role of CRIPTO-1 in cutaneous melanoma. Br J Cancer. 2011;105:1030–1038. doi: 10.1038/bjc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyoshi N, Ishii H, Mimori K, Sekimoto M, Doki Y, Mori M. TDGF1 is a novel predictive marker for metachronous metastasis of colorectal cancer. Int J Oncol. 2010;36:563–568. doi: 10.3892/ijo_00000530. [DOI] [PubMed] [Google Scholar]
- 59.Zhong XY, Zhang LH, Jia SQ, Shi T, Niu ZJ, Du H, Zhang GG, Hu Y, Lu AP, Li JY, et al. Positive association of up-regulated Cripto-1 and downregulated E-cadherin with tumour progression and poor prognosis in gastric cancer. Histopathology. 2008;52:560–568. doi: 10.1111/j.1365-2559.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- 60.Gong YP, Yarrow PM, Carmalt HL, Kwun SY, Kennedy CW, Lin BP, Xing PX, Gillett DJ. Overexpression of Cripto and its prognostic significance in breast cancer: a study with long-term survival. Eur J Surg Oncol. 2007;33:438–443. doi: 10.1016/j.ejso.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Yoon HJ, Hong JS, Shin WJ, Lee YJ, Hong KO, Lee JI, Hong SP, Hong SD. The role of Cripto-1 in the tumorigenesis and progression of oral squamous cell carcinoma. Oral Oncol. 2011;47:1023–1031. doi: 10.1016/j.oraloncology.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 62.Hueng DY, Lin GJ, Huang SH, Liu LW, Ju DT, Chen YW, Sytwu HK, Chang C, Huang SM, Yeh YS, et al. Inhibition of Nodal suppresses angiogenesis and growth of human gliomas. J Neurooncol. 2011;104:21–31. doi: 10.1007/s11060-010-0467-3. [DOI] [PubMed] [Google Scholar]
- 63.Bianco C, Adkins HB, Wechselberger C, Seno M, Normanno N, De Luca A, Sun Y, Khan N, Kenney N, Ebert A, et al. Cripto-1 activates Nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial cells. Mol Cell Biol. 2002;22:2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bianco C, Strizzi L, Rehman A, Normanno N, Wechselberger C, Sun Y, Khan N, Hirota M, Adkins H, Williams K, et al. A Nodal- and ALK4-independent signaling pathway activated by Cripto-1 through Glypican-1 and c-Src. Cancer Res. 2003;63:1192–1197. [PubMed] [Google Scholar]
- 65.Bianco C, Mysliwiec M, Watanabe K, Mancino M, Nagaoka T, Gonzales M, Salomon DS. Activation of a Nodal-independent signaling pathway by Cripto-1 mutants with impaired activation of a Nodal-dependent signaling pathway. FEBS Lett. 2008;582:3997–4002. doi: 10.1016/j.febslet.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelber JA, Panopoulos AD, Shani G, Booker EC, Belmonte JC, Vale WW, Gray PC. Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene. 2009;28:2324–2336. doi: 10.1038/onc.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gray PC, Vale W. Cripto/GRP78 modulation of the TGF-β pathway in development and oncogenesis. FEBS Lett. 2012;586:1836–1845. doi: 10.1016/j.febslet.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagaoka T, Karasawa H, Castro NP, Rangel MC, Salomon DS, Bianco C. An evolving web of signaling networks regulated by Cripto-1. Growth Factors. 2012;30:13–21. doi: 10.3109/08977194.2011.641962. [DOI] [PubMed] [Google Scholar]
- 69.Strizzi L, Bianco C, Normanno N, Seno M, Wechselberger C, Wallace-Jones B, Khan NI, Hirota M, Sun Y, Sanicola M, et al. Epithelial mesenchymal transition is a characteristic of hyperplasias and tumors in mammary gland from MMTV-Cripto-1 transgenic mice. J Cell Physiol. 2004;201:266–276. doi: 10.1002/jcp.20062. [DOI] [PubMed] [Google Scholar]
- 70.Bianco C, Strizzi L, Ebert A, Chang C, Rehman A, Normanno N, Guedez L, Salloum R, Ginsburg E, Sun Y, et al. Role of human Cripto-1 in tumor angiogenesis. J Natl Cancer Inst. 2005;97:132–141. doi: 10.1093/jnci/dji011. [DOI] [PubMed] [Google Scholar]
- 71.Strizzi L, Bianco C, Hirota M, Watanabe K, Mancino M, Hamada S, Raafat A, Lawson S, Ebert AD, D'Antonio A, et al. Development of leiomyosarcoma of the uterus in MMTV-CR-1 transgenic mice. J Pathol. 2007;211:36–44. doi: 10.1002/path.2083. [DOI] [PubMed] [Google Scholar]
- 72.Bianco C, Cotten C, Lonardo E, Strizzi L, Baraty C, Mancino M, Gonzales M, Watanabe K, Nagaoka T, Berry C, et al. Cripto-1 is required for hypoxia to induce cardiac differentiation of mouse embryonic stem cells. Am J Pathol. 2009;175:2146–2158. doi: 10.2353/ajpath.2009.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 74.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee HK, Xiang C, Cazacu S, Finniss S, Kazimirsky G, Lemke N, Lehman NL, Rempel SA, Mikkelsen T, Brodie C. GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro Oncol. 2008;10:236–243. doi: 10.1215/15228517-2008-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J, Karlsson S. Hematopoietic stem cells are regulated by Cripto, as an intermediary of HIF-1α in the hypoxic bone marrow niche. Ann N Y Acad Sci. 2012;1266:55–62. doi: 10.1111/j.1749-6632.2012.06564.x. [DOI] [PubMed] [Google Scholar]
- 77.Wu MJ, Jan CI, Tsay YG, Yu YH, Huang CY, Lin SC, Liu CJ, Chen YS, Lo JF, Yu CC. Elimination of head and neck cancer initiating cells through targeting glucose regulated protein78 signaling. Mol Cancer. 2010;9:283. doi: 10.1186/1476-4598-9-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL, et al. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J Clin Invest. 2003;112:575–587. doi: 10.1172/JCI17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xing PX, Hu XF, Pietersz GA, Hosick HL, McKenzie IF. Cripto: a novel target for antibody-based cancer immunotherapy. Cancer Res. 2004;64:4018–4023. doi: 10.1158/0008-5472.CAN-03-3888. [DOI] [PubMed] [Google Scholar]
- 80.Hu XF, Li J, Yang E, Vandervalk S, Xing PX. Anti-Cripto Mab inhibit tumour growth and overcome MDR in a human leukaemia MDR cell line by inhibition of Akt and activation of JNK/SAPK and bad death pathways. Br J Cancer. 2007;96:918–927. doi: 10.1038/sj.bjc.6603641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parisi S, D'Andrea D, Lago CT, Adamson ED, Persico MG, Minchiotti G. Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. J Cell Biol. 2003;163:303–314. doi: 10.1083/jcb.200303010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parish CL, Parisi S, Persico MG, Arenas E, Minchiotti G. Cripto as a target for improving embryonic stem cell-based therapy in Parkinson's disease. Stem Cells. 2005;23:471–476. doi: 10.1634/stemcells.2004-0294. [DOI] [PubMed] [Google Scholar]
- 83.Lonardo E, Parish CL, Ponticelli S, Marasco D, Ribeiro D, Ruvo M, De Falco S, Arenas E, Minchiotti G. A small synthetic cripto blocking peptide improves neural induction, dopaminergic differentiation, and functional integration of mouse embryonic stem cells in a rat model of Parkinson's disease. Stem Cells. 2010;28:1326–1337. doi: 10.1002/stem.458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.