Significance
Plant survival in a highly varied environment requires flexibility in gene regulation. To capture dynamics of mRNA translation at the genome scale, we precisely mapped individual ribosomes to mRNAs of whole seedlings under control and low-oxygen conditions. The results demonstrate nearly 100-fold variation in the efficiency of translation of individual mRNAs under both conditions and provide unique insights into posttranscriptional and translational regulation modulated by low-energy stress in Arabidopsis thaliana.
Keywords: ribosome profiling, uORF, alternative splicing, long intergenic noncoding RNA, translational efficiency
Abstract
Translational regulation contributes to plasticity in metabolism and growth that enables plants to survive in a dynamic environment. Here, we used the precise mapping of ribosome footprints (RFs) on mRNAs to investigate translational regulation under control and sublethal hypoxia stress conditions in seedlings of Arabidopsis thaliana. Ribosomes were obtained by differential centrifugation or immunopurification and were digested with RNase I to generate footprint fragments that were deep-sequenced. Comparison of RF number and position on genic regions with fragmented total and polysomal mRNA illuminated numerous aspects of posttranscriptional and translational control under both growth conditions. When seedlings were oxygen-deprived, the frequency of ribosomes at the start codon was reduced, consistent with a global decline in initiation of translation. Hypoxia–up-regulated gene transcripts increased in polysome complexes during the stress, but the number of ribosomes per transcript relative to normoxic conditions was not enhanced. On the other hand, many mRNAs with limited change in steady-state abundance had significantly fewer ribosomes but with an overall similar distribution under hypoxia, consistent with restriction of initiation rather than elongation of translation. RF profiling also exposed the inhibitory effect of upstream ORFs on the translation of downstream protein-coding regions under normoxia, which was further modulated by hypoxia. The data document translation of alternatively spliced mRNAs and expose ribosome association with some noncoding RNAs. Altogether, we present an experimental approach that illuminates prevalent and nuanced regulation of protein synthesis under optimal and energy-limiting conditions.
Gene expression in eukaryotes is modulated at multiple levels from chromatin organization through protein production. This includes the highly energy-demanding process of protein synthesis. A typical way to assess translation is to isolate transcripts in association with one or more ribosomes (i.e., polysomes) by differential centrifugation. To facilitate the capture of ribosome-associated mRNA, we developed a method for translating ribosome affinity immunopurification (TRAP) incorporating FLAG-tagged ribosomal protein L18 (RPL18) into functional ribosomes in the model plant Arabidopsis thaliana (1). Two advantages of TRAP are that it reduces contamination of polysome preparations with mRNA–ribonucleoprotein (mRNP) complexes of similar density and can be used to obtain mRNAs from subpopulations of cells of a multicellular organ or tissue in plants (2, 3), as well as in mammals and insects (4, 5).
Comparative profiling of the transcriptome (total mRNA) and translatome (polysomal mRNA) of Arabidopsis seedlings and rosette leaves has revealed that the proportion of the mRNA from an individual gene undergoing translation, its translational status, ranges from >10% to <95% under normal growth conditions (6, 7). The translational status of individual transcripts is regulated by diverse environmental stimuli, including carbon availability (8, 9), cold (10, 11), dehydration (6, 10, 12), excess cadmium (13), heat (14), hypoxia (7, 15), pathogens (16), photomorphogenic illumination (17), reillumination (18), salinity (10), singlet oxygen (19), symbionts (20), and unanticipated darkness (21). Translation is also modulated by regulatory molecules, including auxin (22, 23), gibberellins (24), and polyamines (25). Most of the recent studies documenting dynamics in mRNA translation were performed using microarrays, but the application of high-throughput mRNA sequencing (mRNA-seq) to evaluate translatomes has provided greater detail of RNAs associated with ribosomes (3, 26). In yeast, the translatome is recognized as an accessible proxy of de novo protein synthesis (27).
As an extension of translatome analyses, it would be advantageous to quantify the number of individual ribosomes per transcription unit relative to transcript abundance, because an mRNA can engage one to many ribosomes. It would also be desirable to know the local density of the ribosomes along the message because this may provide additional information on the synthesis of the encoded protein. This is because ribosomes may stall due to a low abundance of transfer RNA (tRNA), constraints of folding or targeting of the nascent protein, or regulation of termination or binding of a microRNA (miRNA)–Argonaute complex. Also, one or more short upstream ORFs (uORFs) can inhibit the translation of the main ORF (mORF). In such cases, translatome data may overestimate the amount of synthesized protein. Hence, the mapping of the number and distribution of individual ribosomes along a protein-coding region (i.e., ORF) can provide a more accurate estimate of translational activity of an mRNA.
Ribosome profiling is the quantitative genome-wide mapping of regions of mRNA protected from nuclease digestion by ribosomes. This short-read RNA sequencing methodology, developed using yeast, revealed that cycloheximide-treated ribosomes protect ∼28-nt regions [ribosome footprints (RFs)] within protein-coding ORFs (28). The mapping of RFs to the yeast genome identified rate-limiting steps of translation of individual mRNAs under reduced carbon availability and during spore formation (28, 29). In mouse cells, the coupling of RF profiling with inhibitors of the initiation phase of translation expanded the predicted proteome (30) and was used to identify the subset of transcripts regulated during translation by mammalian target of rapamycin complex 1 (31). RF profiling was also used to evaluate miRNAs associated with transcripts engaged in translation, confirming a relationship between translational inhibition and turnover of miRNA targets in HeLa cells (32). RF profiling using tiling arrays was recently extended to maize chloroplasts, resolving several unanswered questions regarding organellar translation (33). RF profiling might also be applied to immunopurified (TRAPed) ribosomes and used to explore dynamics of translation in response to stimuli in plants.
Our previous work demonstrated that whole seedlings of Arabidopsis exposed to low-oxygen stress (hypoxia/anoxia) limit the translation of ∼70% of cytosolic mRNAs as an energy conservation mechanism that is rapidly reversed by reoxygenation (15). To understand the regulation of translation in response to hypoxia better, we performed RF profiling using mRNA–ribosome complexes obtained by differential centrifugation or immunopurification. The number of ribosomes that mapped to each gene transcript was compared to the transcript steady-state (total) abundance as an estimate of translational efficiency. Ribosome distribution along the mRNA was also examined. Our goal was to determine if translation was differentially regulated during the initiation, elongation, or termination phase. RF profiling exposed ∼100-fold variation in the efficiency of translation of individual mRNAs under both conditions and confirmed that hypoxia primarily limits the process of initiation. The results uncovered other intriguing aspects of posttranscriptional and translational control that regulate the nascent proteome under both optimal and low-oxygen conditions.
Results and Discussion
Isolation of RF Fragments by Centrifugation or Immunopurification.
The digestion of mRNA–ribosome complexes with nuclease produces ca. 30-nt regions of the transcript that are physically protected by the footprint of a ribosome. These fragments can be isolated, converted into cDNA, deep-sequenced, and mapped back to the genome to provide spatial and quantitative information on mRNA translation. Here, we tailored RF profiling to polysomes isolated by centrifugation or immunopurification (i.e., TRAP) from whole seedlings of Arabidopsis (Fig. 1A and Fig. S1). For both methods, we used a genotype that expresses FLAG-tagged RPL18 under the quasiconstitutive 35S cauliflower mosaic virus promoter. In the first method, polysomes were collected from detergent-treated cell extracts by sucrose gradient centrifugation, followed by RNase I digestion that reduced most polysomes to 80S monosomes (Fig. 1B), which were reisolated from sucrose gradients. In the second method, FLAG-tagged ribosome–mRNA complexes were TRAPed by immunoprecipitation and then digested with RNase I. The ribosomes obtained by either approach include nucleolar preribosomes. Because immunopurification does not rely on ultracentrifugation, the ribosomes isolated by TRAP should not be contaminated with nonribosomal mRNPs that cosediment with ribosomes. Following purification of the ca. 30-nt RF fragments, libraries were constructed for strand-specific (directional) high-throughput sequencing. Because the footprints were contaminated with some abundant rRNA fragments also generated by RNase I digestion, the library construction included a subtractive hybridization step. Our goal was to monitor differential regulation of mRNA translation in response to low-oxygen stress. Therefore, the analysis was performed with seedlings that were maintained under nonstress conditions (normoxia) or exposed to a short nonlethal period of hypoxia. The same samples were used for isolation of total and TRAPed polysomal poly(A)+ mRNA that was also directionally sequenced.
Fig. 1.
RF profiling analysis of control (normoxic) and hypoxic seedlings exposes regulation of translational initiation. (A) Overview of experimental strategy. (B) Absorbance (A254 nm) of sucrose density gradient fractionated ribosomes from RNase I- or mock-treated (control) cell extracts. (C) Representative sucrose density gradient profiles of ribosomes from normoxic and 2-h hypoxic seedlings. (D) Size distribution and relative abundance of RF reads and total and polysomal mRNA processed in parallel. (E) Fraction of 30-nt reads at the first (A), second (U), and third (G) nucleotide positions, respectively, of the start codon [average and SD for nonstress samples (RF#1 and RF#2)]. (F) Comparison of RF rpkM values per protein-coding ORF for all genes with >128 reads from one representative biological replicate. (G) Coverage values of 30-nt reads within and at the ends of the ORFs of genes. Inferred ribosome position relative to the acceptor (A), peptidyl (P), and exit (E) sites of the ribosome with the start codon in the P site and the stop codon in the A site. Bars show RFs, and lines show total (T) and polysomal (P) mRNA reads for the two conditions. Reads are mapped at the first nucleotide of the RF fragment. Data in D–G represent reads mapped uniquely to the genome with two or fewer mismatches. D, F, and G are data from RF#1. Figs. S2 and S3 provide corresponding analyses of other datasets.
High Ribosome Occupancy at the Start Codon of the Coding Sequence.
We obtained over 94 million RF sequence reads in our ribosome sequencing (Ribo-seq) libraries for each of two independent biological replicate samples of gradient-purified ribosomes (referred to as the RF#1 and RF#2 datasets) and over 58 million RF reads for immunopurified ribosomes (referred to as the TRAP-RF dataset). In each case, normoxia and hypoxia datasets were produced. The duplicated total and polysomal mRNA-seq libraries yielded 22–68 million reads per library. These short sequences were mapped to the A. thaliana Col-0 genome, allowing no more than two mismatches and only unique alignments (Dataset S1A). The vast majority of the RF dataset reads mapped to gene transcript ORFs (78.38% normoxia, 72.53% hypoxia), with far fewer reads mapping to 5′ UTRs (9.11% normoxia, 8.58% hypoxia), introns (4.55% normoxia, 5.01% hypoxia), and 3′ UTRs (7.24% normoxia, 12.58% hypoxia). Values for the TRAP-RF dataset were very similar.
In our hands, the RF preparation method was robust because the read counts per ORF in the replicated RF libraries were highly similar (r ≥ 0.90, Pearson correlation) (Fig. S2 A and B). The RFs obtained by the two methods of ribosome isolation were generally consistent (r = 0.86 normoxia, r = 0.66 hypoxia) (Fig. S2 C and D). A lack of congruency was expected because high-speed centrifugation yields all extracted ribosomes, whereas TRAP isolates tagged ribosome complexes from the cells that express the 35S promoter (2). We found the modal RF size was 30 nt in Arabidopsis seedlings (Fig. 1D), which is 2 nt larger than the footprint reported for yeast (28). In both the RF and TRAP-RF datasets, the average footprint density in the UTRs was ∼10-fold more than reported for yeast (28), although the level was quite variable among genes (Fig. S3A).
To survey ribosome positioning across mRNAs, the footprints were mapped along the annotated 5′ UTRs, ORF, and 3′ UTRs, and the distribution was compared with the reads obtained by mRNA-seq for total and polysomal mRNAs (Fig. 1G and Figs. S3 B and C and S4). Regardless of the method of RF isolation, the read coverage (ribosome occupancy) was principally within transcribed regions and read size was similar in the 5′ UTR, ORF, and 3′ UTR (Fig. S4). At this global level, RF coverage was the highest from position −19 to the annotated start codon (Fig. 1G and Figs. S3 B and C and S5). This demonstrates that an Arabidopsis ribosome with an AUG at the peptidyl (P) site of the ribosome protects 16–18 nt 5′ and 12–14 nt 3′ of the start codon. This footprint was 4–6 nt larger 5′ to the AUG than that of yeast and mammalian ribosomes (28, 32). As in those studies, our data support the long-standing notion that start codon recognition and ribosome subunit joining are the primary rate-limiting processes of translation (Fig. S5 A–C). Termination was also rate-limiting because read coverage increased ∼20 nt 5′ of the stop codon. This peak corresponds to the trailing edge of the ribosome with the stop codon in the acceptor (A) site of the ribosome (Fig. S5 D–F). There was a >50% decline in footprints in the first 30 nt of the 3′ UTR, regardless of the treatment condition or ribosome isolation method. The use of a genome browser to display RF density on individual genes revealed gene-to-gene variation in 3′ UTR reads, which raises the possibility that some nuclease protection may be due to RNA binding proteins or structured RNA regions.
To look for evidence of 3-nt periodicity in RFs at the start codon as reported for yeast (28), we focused on the start codon (AUG) of annotated ORFs. The frequency of footprints with a 5′ end at the adenine nucleotide of this triplet was ∼75%, regardless of the condition or method of ribosome isolation (Fig. 1E and Fig. S2 E–J). By contrast, the 5′ termini of total and polysomal mRNA-seq reads at this triplet were more equally distributed between the three nucleotides, consistent with the mechanical fragmentation of the mRNA samples during library construction. These data suggest that RF mapping of high read coverage can provide codon-level resolution in plants.
Hypoxia Reduces Ribosome Protection Associated with Initiation and Increases Protection Associated with Termination.
In this study, 2 h of hypoxia caused a 47 ± 4% reduction in polysomes concomitant with an increase in monosomes and ribosomal subunits (Fig. 1C), consistent with earlier findings (15). The range in RF read number per gene (Fig. 1F and Fig. S3 D and E) was highly similar under normoxic and hypoxic conditions. However, hypoxia reduced the frequency of ribosomes in the initiation and stop codon regions (Fig. 1G and Figs. S3 B and C and S5). This was observed with both methods of ribosome isolation. The presence of fewer footprints at the AUG codon strongly supports the conclusion that hypoxia inhibits initiation. On the other hand, scarcer footprints in the stop codon region could reflect fewer translating ribosomes or increased ribosome runoff without termination during the stress. We noted higher footprint coverage in the 3′ UTR under hypoxia in both RF datasets but not in the TRAP-RF dataset (Figs. S3 B and C and S5 D–F). The reason for this difference is unclear but could reflect mRNP contamination in the ribosomes obtained by the conventional ultracentrifugation protocol.
Hypoxia Selectively Alters Translational Efficiency of mRNAs.
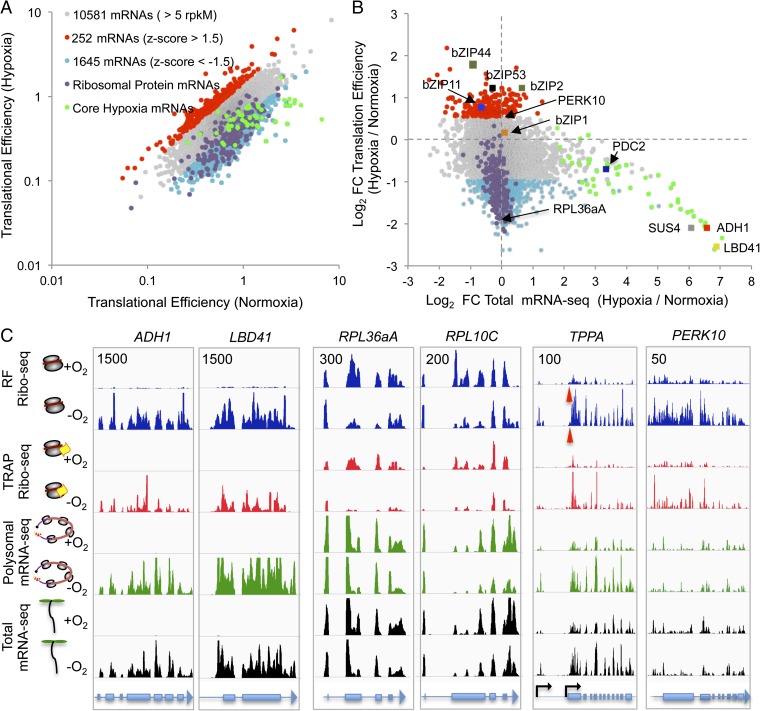
To gain insight into the efficiency of translation at the gene-specific level, we compared the number of RF and mRNA-seq reads that mapped to individual ORFs. Here, the biologically replicated datasets were combined. We then identified all ORFs with a sufficient number of RF and mRNA-seq reads in all normoxic and hypoxic samples [10,581 genes with more than 5 reads per kilobase per million reads (rpkM)]. Translational efficiency was calculated as the relative number of footprint reads to mRNA-seq reads (28). This revealed an ∼100-fold variation in translational efficiency among genes under both conditions (Fig. 2A and Dataset S1B) and identified 252 (2.4%) transcripts with increased and 1,645 (15.6%) transcripts with decreased translational efficiency in response to hypoxia (|z-score| >1.5). The TRAP-RF dataset yielded a similar perspective on hypoxia-regulated translation (Fig. S6 A and B and Dataset S1D), although a higher percentage of the genes that passed the >5-rpkM filter displayed increased [671 (6.2%)] or decreased [1,117 (10.3%)] translational efficiency.
Fig. 2.
Translational efficiency of individual mRNAs is perturbed by hypoxia. (A) Comparison of translational efficiency under normoxia and hypoxia (reads mapped to ORFs with ≥5 rpkM). Values are given for the number of genes evaluated and genes with conditionally altered translational efficiency. (B) Change in mRNA abundance and translational efficiency in response to hypoxia for the same genes as in A. (C) Coverage of RF and TRAP-RF, polysomal mRNA, and total mRNA reads on selected genes. ADH1 (ALCOHOL DEHYDROGENASE1; AT1G77120) and LBD41 (LATERAL ORGAN BOUNDARY DOMAIN41; AT3G02550) are core hypoxia-responsive genes (2). RPL36aA (AT3G23390) and RPL10C (AT1G66580) encode ribosomal proteins. TPPA (TREHALOSE PHOSPHATE PHOSPHATASE A; AT5G51460) has two transcription start sites with use of the 5′ site more prevalent under hypoxia. The red arrowheads indicate a region present only in the shorter transcript. The black arrows indicate the two transcription start sites. PERK10 (PROLINE-RICH EXTENSIN-LIKE RECEPTOR KINASE 10; AT1G26150) shows higher ribosome occupancy under hypoxia. The maximum read value of the scale used for each gene is indicated at the upper left.
Looking more deeply, we found that 32% (n = 76) of the mRNAs with significantly elevated translational efficiency under hypoxia possessed one or more predicted uORF of ≥60 nt (≥20 amino acids) 5′ to the mORF compared with 6% of the mRNAs with reduced translational efficiency in the RF dataset. As reported previously (7, 15), many mRNAs displayed reduced translation in the absence of a change in mRNA accumulation during hypoxia. The translationally repressed mRNAs were highly enriched for proteins associated with translation [Gene Ontology (GO):0006412; adjusted P value = 6.84E-30], including 138 of 219 cytosolic ribosomal proteins in the RF dataset (Fig. 2A and Dataset S1C). The TRAP-RF data showed the same phenomena. This indicates that hypoxia influences uORF translation and reconfirms that hypoxia reduces ribosomal protein mRNA translation, as seen in response to several other energy-limiting conditions (6, 8, 12, 18, 21).
Dynamics in mRNA accumulation and translation were globally assessed by comparing changes in translational efficiency with those of total RNA accumulation in response to hypoxia (Fig. 2B). For the 1,897 mRNAs with a significant increase or decrease in translational efficiency, only 45 increased and 14 decreased in total message abundance [edgeR/generalized linear model (GLM) |log2 fold change| >1; false discovery rate (FDR) of <0.05]. Corresponding values in the TRAP-RF dataset were 29 mRNAs with increased abundance and 1 mRNA with decreased abundance of the 1,888 transcripts with significantly altered translational efficiency (Fig. S6B). These data demonstrate that altered rates of initiation, elongation, or termination, which are the factors that determine translational efficiency, are not generally coordinated with changes in message abundance in response to hypoxia.
Our study further clarified the coordinated increase in stress protein mRNAs in the transcriptome and translatome (7, 15). Comparison of the fold change in transcript abundance with the fold change in translational efficiency (RF dataset) revealed that the strongly up-regulated transcripts (core hypoxia-responsive mRNAs) (2) had similar or lower translational efficiency during hypoxia (Fig. 2B). The same analysis with the TRAP-RF dataset indicated that core hypoxia-responsive mRNAs effectively recruited ribosomes in proportion during the stress (i.e., maintained the prestress translational efficiency). These data demonstrate that the stress-induced mRNAs are effectively translated, but not at a higher efficiency.
Evaluation of the differentially translated mRNAs with a genome browser confirmed regulation of RF coverage under hypoxia. ALCOHOL DEHYDROGENASE1 (ADH1) and LATERAL ORGAN BOUNDARY DOMAIN41 (LBD41) mRNAs are representative of highly hypoxia-induced mRNAs with proportional increases in RFs as a consequence of their induction and successful ribosome recruitment (Fig. 2C). These hypoxia-responsive mRNAs can be contrasted to those encoding ribosomal protein (RP) genes, as illustrated by RPL36aA and RPL10C. Two other examples of regulation are shown in Fig. 2C. TPPA, encoding a trehalose 6-phosphate phosphatase, illustrates variation in the 5′ leader due to alternative transcription start sites that could conditionally influence translation status, as seen in yeast (34). PROLINE-RICH EXTENSIN-LIKE RECEPTOR KINASE 10 (PERK10), encoding a proline-rich protein, exemplifies increased ribosome occupancy during hypoxia in the absence of a marked change in transcript abundance. These views illustrate the diverse landscape of ribosome-protected fragments that can be evaluated with single-ribosome resolution in our dataset.
Hypoxia Alters Ribosome Occupancy on Upstream ORF-Containing mRNAs.
Genes with short uORFs 5′ to the mORF are prevalent in plants, with an estimated 20% of Arabidopsis protein-coding genes possessing a uORF (35–37), This includes about 30 gene families with one or more uORFs encoding an evolutionarily conserved peptide ORF (CPuORF). It is thought that uORFs act as a barrier to translation of the mORF, because leaky scanning past the AUG of the uORF or reinitiation at the AUG of mORF is either limited or highly regulated (38). To evaluate the function of this mRNA feature, we identified 2,020 genes with one or more ≥60-nt uORF. These were examined at a global level by using the longest uORF of each transcript to generate RF plots with the RF and TRAP-RF datasets (Fig. 3A and Fig. S7A). This exposed the enrichment of RFs at the start and stop codons of the uORF (Fig. 3A). The footprints at the uORF start codon were less evident in the TRAP dataset (Fig. S7B), possibly because TRAP relies on a tagged 60S subunit and would not capture an mRNA engaged with a single 43S preinitiation complex. By narrowing our analysis to 51 genes with uORFs that inhibit mORF translation based on visual inspection in a genome browser (Dataset S1B), the enrichment of RF termini 16–18 nt 5′ of the AUG was more evident in both RF and TRAP-RF datasets (Fig. S7 C and D). This was particularly clear under normoxia. Taken together, these data provide strong evidence that uORFs are translated in Arabidopsis.
Fig. 3.
Hypoxia alters translation of uORFs relative to mORFs on multicistronic mRNAs. (A) Coverage values of 30-nt reads at the annotated start codon region, uORF coding sequence, and stop codon region of the longest uORF identified computationally (n = 2,020 with uORF ≥60 nt). The plot is displayed as in Fig. 1G. (B) RF coverage (RF#1) on S1 class bZIP transcription factor genes with multiple uORFs. The scale maximum read value is the same as in Fig. 2C. (C) Tabulation of change in ratio of RFs on the mORF relative to the uORF under normoxia and hypoxia. (D) Comparison of RF (RF#1 and TRAP-RF) and total mRNA-seq reads on SUPPRESSOR OF ACAULIS 51 (SAC51).
Next, we addressed whether uORFs may act as a barrier to mORF translation. To investigate this possibility, we inspected individual uORF-containing genes, focusing first on the S1 class basic leucine zipper (bZIP) transcription factors, which have more than three uORFs. uORF2 of bZIP11 is the longest of the four uORFs on this transcript and restricts the translation of the mORF in leaves fed sucrose (9). We found that the log2 ratio of RFs that map to the bZIP11 mORF relative to uORF2 fell from −3.84 to −2.03 (FDR = 6.22E-09, normoxia; FDR = 1.97E-03, hypoxia) (Fig. 3 B and C). There was pronounced inhibition of translation of the transcription factor ORF under normoxia, which was lessened by hypoxia. We also found many RFs just 5′ of the uORF2 stop codon under normoxia. This is consistent with the proposal that translocation is inhibited as the ribosome approaches the end of uORF2, due to specific amino acids in the uORF2 peptide (9). The increase in ribosomes translating the bZIP11 coding sequence during hypoxia was accompanied by a greater number of footprints in the intercistronic region between the uORF and mORF. Four other S1 class bZIP mRNAs with multiple uORFs displayed a uORF barrier with RFs enhanced near the stop codon of the longest uORF, which was lessened by hypoxia (Fig. 3 B and C).
The transcript encoding the transcription factor SUPPRESSOR OF ACAULIS 51 (SAC51) provided another example of translational inhibition by a uORF that was dampened during hypoxia (Fig. 3D). In this case, RF coverage was highest near to the start and stop codons of the first (CPuORF38) and third (CPuORF40) uORFs in normoxic seedlings and increased markedly in the mORF during hypoxia. However, a net increase in SAC51 synthesis during the stress is unlikely because of a decline in the steady-state abundance of the transcript. Many other mRNAs with CPuORFs displayed elevated translational efficiency in response to hypoxia in both the RF and TRAP-RF datasets (Fig. S6 C–E).
The increase in ribosome occupancy in the intercistronic and mORF regions of uORF mRNAs under hypoxia could result from reduced recognition of the AUG of the uORF by the scanning preinitiation complex (i.e., leaky scanning to downstream AUGs) or enhanced reinitiation after termination of uORF translation. Increased mORF translation due to leaky scanning seems more likely because reinitiation is promoted by target of rapamycin kinase (23), which is typically inactive under energy-limiting conditions. These data provide genome-scale confirmation that uORFs capture ribosomes, and thereby limit the production of protein from the downstream mORF. They also show that an energy crisis can reduce the effectiveness of the uORF barrier to mORF translation.
RF Profiling Reveals Regulated Alternative Splicing.
Alternative splicing is displayed by over 60% of intron-containing genes in plants, but little is known about the translation of alternatively spliced mRNAs (39). We found that ∼5% of the RFs mapped to introns (Fig. S3A and Dataset S1A). Because growth conditions can influence the selection of splice sites and intron retention (40–42), we questioned whether ribosome profiling could be used to demonstrate translation of distinct mRNA isoforms. We therefore sought intron retention events in genes that were not differentially expressed based on ORF reads in total RNA but had >10 reads on at least one intron under both normoxia and hypoxia. This yielded ∼130 examples of significantly altered intron retention (Fig. 4A and Dataset S1G). Hypoxia increased intron retention events in a number of mRNAs encoding the splicing machinery. For example, there was enhanced retention of intron 6 of SMALL NUCLEAR RNP 70K (U1-70K) (Fig. 4B), which causes an in-frame termination codon leading to the production of a truncated and nonfunctional protein (43). This was also seen for mRNAs encoding the splicing factors SR30, SR34, SR35, and SR45 (Fig. S8A) and proteins associated with the circadian clock (Fig. 4 A and B, Fig. S8B, and Dataset S1G). In the case of GLYCINE-RICH RNA-BINDING PROTEIN (GRP7)/COLD CIRCADIAN RHYTHM AND RNA-BINDING PROTEIN 2(CCR2), encoding the slave oscillator, enhanced retention of intron 1 was accompanied by significantly reduced translational efficiency under hypoxia (log2-fold change = −1.40, z-score of 2.37; Fig. 4B). The transcript isoform containing intron 1 has a premature termination codon and is a target of nonsense-mediated decay (44). Regulated accumulation and translation of alternatively spliced GLYCINE-RICH RNA-BINDING PROTEIN 8 (GRP8) mRNA was also evident (Fig. S8B). The dataset includes examples of stress-regulated intron skipping. The really interesting new gene (RING) E3 ligase gene XB3 ORTHOLOG 5 IN ARABIDOPSIS THALIANA (XBAT35; AT3G23280) produces transcripts that possess or lack exon 8, which encodes a nuclear localization signal (45). Exon 8 skipping prevailed under hypoxia (Fig. S8B). These results illustrate that RF mapping can capture dynamically regulated variations in splicing and provide insight into the consequences on protein production.
Fig. 4.
RF profiling reveals conditional alternative splicing. (A) Fold change (FC) in response to hypoxia of RF read number on coding sequences compared with introns. Only introns with ≥10 reads under both treatments are represented. Genes in red had significantly changed RFs on introns (|Fold Change| >2.0, FDR of ≤0.1). Genes discussed in the main text are denoted. (B) RF coverage on selected genes. U1-70K, SMALL NUCLEAR RIBONUCLEOPARTICLE 70K (AT3G50670); GRP7, GLYCINE-RICH RNA-BINDING PROTEIN (AT2G21660). dsRNA read data are from Zheng et al. (46). Gene transcript (blue diagrams) and protein predictions (red diagrams) are shown. The red arrowhead indicates a retained intron.
In the process of evaluating intron retention events, we observed some islands of high RF coverage, such as within intron 1 of EU.K.ARYOTIC INITIATION FACTOR 4A (eIF4A) (Fig. S8B). Reports of secondary structures in protein-depleted RNA fractions obtained from Arabidopsis (46, 47) led us to consider that some fragments in the Ribo-seq libraries are derived from regions of ribosome-associated transcripts that are RNase I-resistant due to dsRNA formation. In fact, the reads in intron 1 of eIF4A coincided with a structured region of the mRNA predicted by Zheng et al. (46). In Arabidopsis, high dsRNA read counts were reported for relatively few ORFs (Dataset S1B), and their presence was poorly correlated with translational efficiency (r2 < 0.01). In genomes with high GC content, it may be important to consider dsRNA reads in Ribo-seq data.
Evidence of Noncoding RNA Translation.
Plants produce over 30,000 putative noncoding RNAs (ncRNAs) that are 5′-capped and 3′-polyadenylated (48, 49). These include natural antisense transcripts (NATs), long intergenic/intronic ncRNAs (lincRNAs), and RNAs encoding ORFs of only 30–100 amino acids [small ORFs (sORFs)] (50). Some of these molecules function in chromatin modification or as miRNA mimics, whereas others are involved in ribosome biogenesis [i.e., small nucleolar RNAs (snoRNAs)]. Here, we used existing compendia of ncRNAs (49, 50) to seek ones that may be ribosome-associated. First, we identified ncRNAs with mRNA-seq reads (>5 rpkM) in both normoxic and hypoxic samples. Only 0.7% (n = 210) of the reported ncRNAs passed this filter. We then calculated the translational efficiency of these RNAs and found the values to be generally similar to those of mRNAs (Fig. S9A). By visual inspection of the read data with a genome browser, we categorized most of the 210 ncRNAs into one of four groups (Fig. 5 and Dataset S1H).
Fig. 5.
RF profiling exposes noncoding mRNAs associated with ribosomes. (A) RF coverage on a lincRNA (At5NC0011780) encoding an uncharacterized structured RNA and a snoRNA, a small ORF RNA (sORF0629/AT1G31935), and a lincRNA with no ORF (AT3G64540). (B) Strand-specific RFs (individual reads) on a NAT encoding a small ORF (seedGroup6045) and a NAT (AT2G05812) overlapping an active protein-coding gene (AT2G05810). Red reads are oriented from left to right (5′ to 3′), and blue reads are oriented from right to left (5′ to 3′). dsRNA read data are from Zheng et al. (46). Blue boxes represent the RNA, and red boxes show the position of ORFs.
Group 1 ncRNAs had few polysomal mRNA-seq reads relative to total mRNA-seq and RF reads (n = 23) but were highly structured (46) (Dataset S1H). As an example, lincRNA AtNC0011780 is polycistronic, encoding a nonannotated structured RNA and a snoRNA (AT5G10572.1) (Fig. 5A). A subsequent survey of RFs on annotated snoRNAs confirmed their prevalence in Ribo-seq reads and depletion in polysomal mRNA-seq reads. snoRNAs are derived from mono- and polycistronic poly(A)+ RNAs that guide cleavage and modification of pre-rRNA within the nucleolus during ribosome biogenesis (51). The paucity of snoRNAs in the polysomal RNA fraction, which was purified from TRAPed ribosomes by poly(A)+ selection, is consistent with 3′ processing of these RNAs (Fig. S9B). We conclude that the group I ncRNAs are structured RNAs, which copurify with ribosomes or preribosomes.
We detected three other groups of ncRNAs with RF reads. Group 2 had read coverage consistent with initiation at an AUG codon, elongation, and termination [n = 18 (i.e., sORF0629, 86 aa)] (Fig. 5A). This group included NATs that apparently encode proteins (i.e., SeedGroup6045 NAT, 132 aa) (Fig. 5B) and pre-miR414 (Fig. S9B). We confirmed RF coverage on an 88-aa ORF of pre-miR414, supporting earlier suggestions that this transcript encodes a functional ORF rather than a miRNA precursor (52). Group 3 ncRNAs was composed of 42 active NATs (i.e., mRNA-seq reads) (Fig. 5B). The directional construction of the libraries allowed us to confirm that both the NAT and the opposing protein-encoding ORF can be occupied by ribosomes in some cases. However, only a subset of the NATs had an ORF encoding more than 30 aa. Most were similar to a group of 39 lincRNAs that had limited coding capacity but extensive RF coverage [group 4 (i.e., At3g63540)] (Fig. 5A). These ncRNAs were unstructured and encoded very short ORFs (<30 aa). They could be RNAs specifying short biologically active peptides, as recently identified in mammalian cells (53). This enigmatic cohort of ribosome-associated ncRNAs will require further study to determine whether they are translated or function in a manner that involves association with ribosomes.
Conclusions
This analysis illustrates the value of ribosome profiling in the investigation of posttranscriptional regulation of nuclear gene expression in plants. We show that ribosome profiling can be accomplished with RFs obtained by TRAP, a method that limits contamination by cosedimenting mRNPs and facilitates cell type-specific ribosome isolation. The comparison of RFs obtained by centrifugation or TRAP from seedlings grown under optimal conditions with those temporarily deprived of oxygen revealed translational control at initiation and termination. We identified examples of transcription start site variation, alternative splicing, uORF barriers, and regulated elongation. The data reconfirmed that hypoxia-induced mRNAs encoding proteins relevant to stress survival are elevated in polysomes during the stress but clarified that this was not associated with the capacity to recruit more ribosomes per transcript. On the other hand, translational efficiency was reduced for a large cohort of mRNAs, including many that encode components of the energy-consuming translational machinery. We hypothesize that these translationally repressed mRNAs become sequestered into stress granule-like complexes similar to those described in animals (54). Taken together, our data demonstrate that hypoxia-induced synthesis of proteins such as ADH1 is due to transcriptional induction (55), successful translational initiation, and avoidance of sequestration.
A value of RF profiling is the insight it provides into the nascent proteome. Our study confirmed at the global scale that uORFs function to limit the number of ribosomes on mORFs under control growth conditions. Interestingly, a dampening of ribosome occupancy on uORFs usually coincided with a rise in the proportion of ribosomes on the downstream mORF during hypoxia. However, this increase in ribosomes on a mORF does not ensure higher protein production because mRNA abundance also needs to be considered. Our investigation also demonstrated differential regulation of splicing that is likely to influence mRNA stability and protein isoform production. Future ribosome profiling analyses in plants might use specific inhibitors of translation and translation factor mutants to clarify other aspects of translational control, such as the use of non-AUG start sites. With the demonstration of miRNAs in immunopurified polysomes from Medicago (20) and a peripheral endoplasmic reticulum-associated protein required for miRNA-mediated translational repression in Arabidopsis (56), RF profiling may prove useful in the investigation of the relationship between miRNAs and the translation or cleavage of their targets, as in animals (32, 57). However, this may require biochemical and genetic strategies to enrich for ribosomes containing miRNAs and their targets and will merit validation. The finding of some ncRNAs in immunopurified ribosomes raises the possibility that these unusual transcripts can encode short peptides or carry out functions coupled to translation. Thus, in addition to improved prediction of the nascent proteome, ribosome profiling promises to help unravel the intertwined posttranscriptional mechanisms that contribute to the plasticity of plants.
Materials and Methods
Plant Growth and Treatment.
The transgenic A. thaliana 35S:HF-RPL18B line 12-2-4 (ecotype Col-0) was used in this study (1). Seedlings were grown on solid media [0.43% (wt/vol) Murashige and Skoog salts, 1% (wt/vol) sucrose] at 23 °C under a 16-h light (80 μE⋅m−2⋅s−1) and 8-h dark cycle for 7 d. Hypoxia was imposed as described previously (15) for 2 h under low light (7 μE⋅m−2⋅s−1). Control samples (normoxic) were held in air under the same light and humidity conditions. Whole seedlings were harvested directly into liquid nitrogen and stored at −80 °C.
Preparation of Total and Immunopurified Polysomal RNA and Ribosome-Protected Fragments.
Detailed procedures are given in SI Materials and Methods. TRAP was used to isolate mRNA–ribosome complexes for polysomal mRNA (58). Total RNA was obtained from the same cell homogenate. RNA was purified by use of TRIzol reagent (Invitrogen). Two methods were used to obtain the ribosome–mRNA complexes used to prepare RF fragments: differential centrifugation and TRAP (Fig. S1). In the first method, the polysome pellet (P170) was resuspended in 300 μL of RNase I digestion buffer [RNID; 20 mM Tris⋅HCl (pH 8.0), 140 mM KCl, 35 mM MgCl2, 50 μg/mL cycloheximide, 50 μg/mL chloramphenicol], and 10 units of RNase I (Ambion) was added per 100 OD A260 units of ribosomes. Digestion was for 2 h at room temperature. The resultant monosomes were obtained by centrifugation through a 20–60% (wt/vol) sucrose gradient (58). In the second method, TRAP was performed as described (58), with the exception that wash buffer was replaced by RNID. In-gel RNase I digestion was performed by resuspension of the washed EZview red anti-FLAG affinity gel (Sigma) in 400 μL of RNID with the addition of 250 units of RNase I and incubation for 2 h at room temperature. RFs were extracted by use of TRIzol reagent.
Construction of Libraries and Sequencing.
Detailed procedures are given in SI Materials and Methods. For RF and TRAP-RF Ribo-seq libraries, the oligo-dT tailing and size selection method described by Ingolia et al. (28, 59) was used with some modifications for plants. Two abundant rRNA fragments were removed by subtractive hybridization with biotinylated oligos using Dynabeads MyOne Streptavidin C1 (Invitrogen). PCR amplification of libraries was performed for 15 cycles, and the products were separated by gel electrophoresis to obtain ∼120- to 125-bp fragments. Ribo-seq libraries were not multiplexed. For the total and polysomal mRNA-seq libraries, poly(A)+ mRNA was obtained using Oligo-dT DynaBead (Invitrogen) purification and subjected to partial alkaline hydrolysis (28). Randomly fragmented mRNAs were size-selected (25–35 nt) and subjected to 3′-end dephosphorylation and 5′-end phosphorylation. Libraries were constructed by use of a NEXTflex Small RNA Sequencing Kit (Bioo Scientific) and NEXTflex Small RNA Barcodes (Set A; Bioo Scientific). The mRNA-seq libraries were quadruplexed. Libraries were quality-controlled by evaluation of RF size and quantity on an Agilent 2100 Bioanalyzer. The mRNA-seq libraries were combined in equal molar amounts for library multiplexing. High-throughput sequencing was performed by use of a HiSeq2000 (Illumina) sequencer, and FASTQ files were obtained. Details on all bioinformatic procedures and analyses are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Nicholas Ingolia for advice on the ribosome profiling method and Reed Sorenson and Maureen Hummel for stimulating discussions throughout this project. This research was funded by National Science Foundation Grants IOS-0750811 (to J.B.-S.) and MCB-1021969 (to J.B.-S. and T.G.). J.B. was funded by Marie Curie European Economic Community Fellowship PIOF-GA-2012-327954.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50597).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317811111/-/DCSupplemental.
References
- 1.Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138(2):624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(44):18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas A, et al. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS ONE. 2012;7(7):e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaguchi R, Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005;33(3):955–965. doi: 10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 2005;96(4):647–660. doi: 10.1093/aob/mci217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolaï M, et al. Large-scale analysis of mRNA translation states during sucrose starvation in arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol. 2006;141(2):663–673. doi: 10.1104/pp.106.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmani F, et al. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150(3):1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SH, et al. Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol. 2012;159(3):1111–1124. doi: 10.1104/pp.112.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juntawong P, Sorenson R, Bailey-Serres J. Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 2013;74(6):1016–1028. doi: 10.1111/tpj.12187. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi R, Girke T, Bray EA, Bailey-Serres J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 2004;38(5):823–839. doi: 10.1111/j.1365-313X.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- 13.Sormani R, et al. Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol. 2011;52(2):436–447. doi: 10.1093/pcp/pcr001. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura H, et al. A computational and experimental approach reveals that the 5′-proximal region of the 5′-UTR has a Cis-regulatory signature responsible for heat stress-regulated mRNA translation in Arabidopsis. Plant Cell Physiol. 2013;54(4):474–483. doi: 10.1093/pcp/pcs189. [DOI] [PubMed] [Google Scholar]
- 15.Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008;56(5):743–755. doi: 10.1111/j.1365-313X.2008.03642.x. [DOI] [PubMed] [Google Scholar]
- 16.Moeller JR, et al. Differential accumulation of host mRNAs on polyribosomes during obligate pathogen-plant interactions. Mol Biosyst. 2012;8(8):2153–2165. doi: 10.1039/c2mb25014d. [DOI] [PubMed] [Google Scholar]
- 17.Liu MJ, Wu SH, Chen HM, Wu SH. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piques M, et al. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol. 2009;5:314. doi: 10.1038/msb.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khandal D, et al. Singlet oxygen-dependent translational control in the tigrina-d.12 mutant of barley. Proc Natl Acad Sci USA. 2009;106(31):13112–13117. doi: 10.1073/pnas.0903522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynoso MA, Blanco FA, Bailey-Serres J, Crespi M, Zanetti ME. Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 2012;(73):289–301. doi: 10.1111/tpj.12033. [DOI] [PubMed] [Google Scholar]
- 21.Juntawong P, Bailey-Serres J. Dynamic Light Regulation of Translation Status in Arabidopsis thaliana. Front Plant Sci. 2012;3:66. doi: 10.3389/fpls.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA. 2012;109(48):19537–19544. doi: 10.1073/pnas.1214774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schepetilnikov M, et al. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32(8):1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro DM, Araújo WL, Fernie AR, Schippers JH, Mueller-Roeber B. Translatome and metabolome effects triggered by gibberellins during rosette growth in Arabidopsis. J Exp Bot. 2012;63(7):2769–2786. doi: 10.1093/jxb/err463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010;38(2):353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moghe GD, et al. Characteristics and significance of intergenic polyadenylated RNA transcription in Arabidopsis. Plant Physiol. 2013;161(1):210–224. doi: 10.1104/pp.112.205245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100(7):3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335(6068):552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zoschke R, Watkins KP, Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25(6):2265–2275. doi: 10.1105/tpc.113.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rojas-Duran MF, Gilbert WV. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA. 2012;18(12):2299–2305. doi: 10.1261/rna.035865.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayden CA, Jorgensen RA. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen RA, Dorantes-Acosta AE. Conserved Peptide Upstream Open Reading Frames are Associated with Regulatory Genes in Angiosperms. Front Plant Sci. 2012;3:191. doi: 10.3389/fpls.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaughn JN, Ellingson SR, Mignone F, Arnim Av. Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA. 2012;18(3):368–384. doi: 10.1261/rna.031179.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy B, von Arnim AG. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book. 2013;11:e0165. doi: 10.1199/tab.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22(6):1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20(1):45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalyna M, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40(6):2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leviatan N, Alkan N, Leshkowitz D, Fluhr R. Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS ONE. 2013;8(6):e66511. doi: 10.1371/journal.pone.0066511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golovkin M, Reddy AS. Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell. 1996;8(8):1421–1435. doi: 10.1105/tpc.8.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36(22):6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho SD, Saraiva R, Maia TM, Abreu IA, Duque P. XBAT35, a novel Arabidopsis RING E3 ligase exhibiting dual targeting of its splice isoforms, is involved in ethylene-mediated regulation of apical hook curvature. Mol Plant. 2012;5(6):1295–1309. doi: 10.1093/mp/sss048. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Q, et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6(9):e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F, et al. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell. 2012;24(11):4346–4359. doi: 10.1105/tpc.112.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ben Amor B, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19(1):57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24(11):4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada K, et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA. 2013;110(6):2395–2400. doi: 10.1073/pnas.1213958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown JW, Echeverria M, Qu LH. Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci. 2003;8(1):42–49. doi: 10.1016/s1360-1385(02)00007-9. [DOI] [PubMed] [Google Scholar]
- 52.Xie Z, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138(4):2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9(1):59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey-Serres J, Sorenson R, Juntawong P. Getting the message across: Cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 2009;14(8):443–453. doi: 10.1016/j.tplants.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Gibbs DJ, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature. 2011;479(7373):415–418. doi: 10.1038/nature10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153(3):562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336(6078):233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- 59.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.