Significance
Basal ganglia circuits are engaged in controlling psychomotor function. The circuits consist of striatonigral and striatopallidal pathways. The opposing but balanced activity of these two neural pathways is important for regulating movement-related functions. How the cell types of striatonigral and striatopallidal neurons are specified to construct these two pathways during development remains elusive. We found that the LIM homeodomain transcription factor Islet-1 (Isl1) was specifically expressed in striatonigral neurons during development. Genetic inactivation of Isl1 resulted in increased apoptosis, abnormal differentiation, and aberrant axonal projections of striatonigral neurons. These findings suggest that Isl1 plays a key role in specification of striatonigral neurons. Our study provides insights into genetic mechanisms by which basal ganglia circuits are built to function.
Keywords: homeobox gene, cell fate determination, axonal outgrowth, substantia nigra, globus pallidus
Abstract
Striatal projection neurons comprise two populations of striatonigral and striatopallidal neurons. These two neuronal populations play distinct roles in controlling movement-related functions in the basal ganglia circuits. An important issue is how striatal progenitors are developmentally specified into these two distinct neuronal populations. In the present study, we characterized the function of Islet-1 (Isl1), a LIM-homeodomain transcription factor, in striatal development. Genetic fate mapping showed that Isl1+ progeny specifically developed into a subpopulation of striatonigral neurons that transiently expressed Isl1. In Nestin-Cre;Isl1f/f KO mouse brain, differentiation of striatonigral neurons was defective, as evidenced by decreased expression of striatonigral-enriched genes, including substance P, prodynorphin, solute carrier family 35, member D3 (Slc35d3), and PlexinD1. Striatonigral axonal projections were also impaired, and abnormal apoptosis was observed in Isl1 KO striatum. It was of particular interest that striatopallidal-enriched genes, including dopamine D2 receptor (Drd2), proenkephalin, A2A adenosine receptor (A2aR) and G protein-coupled receptor 6 (Gpr6), were concomitantly up-regulated in Isl1 mutant striatum, suggesting derepression of striatopallidal genes in striatonigral neurons in the absence of Isl1. The suppression of striatopallidal genes by Isl1 was further examined by overexpression of Isl1 in the striatum of Drd2-EGFP transgenic mice using in utero electroporation. Ectopic Isl1 expression was sufficient to repress Drd2-EGFP signals in striatopallidal neurons. Taken together, our study suggests that Isl1 specifies the cell fate of striatonigral neurons not only by orchestrating survival, differentiation, and axonal projections of striatonigral neurons but also by suppressing striatopallidal-enriched genes. The dual action of developmental control by Isl1 in promoting appropriate striatonigral but repressing inappropriate striatopallidal genetic profiles may ensure sharpening of the striatonigral identity during development.
The striatum is the major input structure of basal ganglia circuits (1, 2). The striatum comprises medium-sized spiny projection neurons (MSNs) and aspiny interneurons. There are two populations of MSNs: striatonigral projection neurons of the direct pathway and striatopallidal projection neurons of the indirect pathway (1–3). These two neuronal populations are intermixed within the striatum without the laminar structure. The striatonigral neurons express dopamine D1 receptor (Drd1a) and substance P (SP), and they project their axons to the entopeduncular nucleus and the substantia nigra (SN). The striatopallidal neurons express dopamine D2 receptor (Drd2) and proenkephalin (Penk), and project their axons to the external segment of the globus pallidus (GP). Previous studies using FACS-array and translating ribosome affinity purification (TRAP) approaches have further characterized differential gene expression profiles in striatonigral and striatopallidal populations in postnatal striatum (4, 5).
At the functional level, direct and indirect pathways are functionally opposed in controlling movement in basal ganglia circuits. The imbalanced activity of these two pathways results in movement disorders, such as Parkinson disease (2, 3). Recent study further indicates that direct striatonigral and indirect striatopallidal pathways play distinct roles in control of reward and aversive behaviors, respectively (6). Despite the well-characterized functional differences between the striatonigral and striatopallidal pathways in basal ganglia circuits, the molecular specification of striatonigral and striatopallidal neurons during development remains largely elusive.
The LIM homeodomain protein Islet-1 (Isl1) was originally identified as a transcription factor that binds to the islet β cell-specific enhancer element in the insulin gene (7). Previous studies have indicated an important role of Isl1 in neuronal development. Null mutation of Isl1 results in cell death and/or defective differentiation of motor neurons and interneurons in the spinal cord (8, 9); bipolar, ganglionic, and amacrine cells in the retina (10–12); sensory nociceptive neurons in the dorsal root ganglia; and cholinergic neurons in the basal forebrain (13, 14). Isl1 is therefore engaged in genetic programs for controlling cell survival and differentiation of various cell types in the nervous system.
We and other groups have previously reported that Isl1 is transiently expressed in embryonic striatum and is down-regulated after birth, except in cholinergic interneurons (14, 15). In the present study, we first carried out a genetic fate mapping study. We found that Isl1+ progeny specifically developed into striatonigral projection neurons. Further studies by loss of function and gain of function identified a dual role for Isl1 in promoting striatonigral, but suppressing striatopallidal, developmental programs, suggesting an essential role of Isl1 in specification of the cell type of striatonigral neurons during development.
Results
Isl1 Cell Lineages Developed into Striatonigral Projection Neurons.
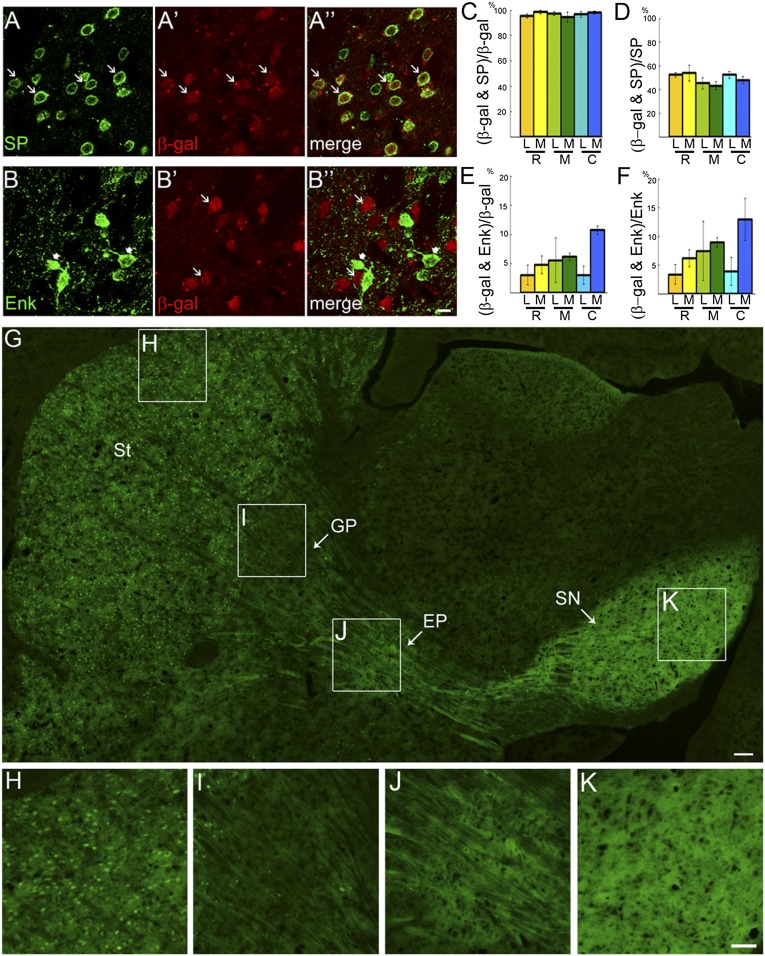
Isl1 is expressed in striatal cholinergic interneurons and is important for their survival and differentiation (14, 15). In addition to cholinergic interneurons, Isl1 is expressed in other striatal neurons in embryonic striatum (15). The down-regulation of Isl1 mRNA and protein in postnatal striatum prevented the cell types transiently expressing Isl1 to be identified in mature striatum (15) (Fig. S1). We therefore performed a cell lineage tracing study using a transgenic mouse approach. Isl1-Cre mice were intercrossed with Rosa26R or CAG-CAT-EGFP reporter mice (Figs. S2–S4). The lineage of Isl1-expressing cells could be traced by the β-gal reporter gene in Isl1-Cre;Rosa26R double-transgenic mouse brain. Results showed that many β-gal+ cells were distributed throughout the striatum at postnatal day (P) 25 (Figs. S3 and S4). Double immunostaining of β-gal and SP, a marker of striatonigral projection neurons (4), showed that ∼95% of β-gal+ cells coexpressed SP at rostral, middle, and caudal levels, whereas ∼50% of SP+ cells coexpressed β-gal from rostral to caudal levels (Fig. 1 A, C, and D). Thus, Isl1+ cell lineages comprised about half of the population of SP+ striatonigral neurons. Double immunostaining of β-gal and Met-enkephalin (Enk), a marker of striatopallidal projection neurons, showed that ∼5% of β-gal+ cells coexpressed Enk at rostral, middle, and caudal levels, whereas ∼7% of Enk+ cells coexpressed β-gal from rostral to caudal levels (Fig. 1 B, E, and F). Taken together, these data indicate that the majority of Isl1+ cell lineages developed into SP+ striatonigral neurons.
Fig. 1.
Majority of Isl1-expressing cells are striatonigral projection neurons. Double immunostaining of the striatonigral marker (SP) (A) and β-gal (A′) shows that more than 90% of β-gal+ cells coexpress SP [A′′ (arrows) and C], whereas ∼50% of SP+ cells coexpress β-gal+ (D) in P25 striatum of Isl1-Cre;Rosa26R brain at rostral (R, bregma 1.3), middle (M, bregma 0.7), and caudal (C, bregma 0.2) levels. The cell counts were performed in the lateral (L) and medial (M) striatum at each level. In contrast, double immunostaining of the striatopallidal marker Enk (B, arrowheads) and β-gal (B′, arrows) shows that less than 10% of β-gal+ cells coexpress Enk (B′′ and E) and less than 10% of Enk+ cells coexpress β-gal (F) in P25 striatum of Isl1-Cre;Rosa26R mice. (G–K) GFP-labeled striatal (St) neurons of the Isl1 cell lineage (H) selectively project their axons through the globus pallidus (GP) GP (I) to the entopeduncular nucleus (EP) (J) and substantia nigra (SN) SN (K) in a parasagittal section of P25 Isl1-Cre;CAG-CAT-EGFP brain. The boxed regions in G are shown at high magnification in H–K. Dense GFP+ fibers, presumably axonal terminals, are detected in EP (J) and SN (K). (Scale bars: A–B′′, 10 μm; G, 100 μm; H–K, 50 μm.)
To study the axonal projection pattern of Isl1+ cells, Isl1-Cre mice were intercrossed with the CAG-CAT-EGFP reporter mice (16). Many GFP+ cells were present in P25 striatum of Isl1-Cre;CAG-CAT-EGFP mice (Fig. 1 G and H). Extensive GFP+ axons were found to extend from the striatum via the GP to the SN of midbrain in parasagittal sections (Fig. 1 G–K). A high density of GFP+ neuropils was also found in the endopeduncular nucleus, the rodent equivalent of the globus pallidus internus (Gpi) that is targeted by striatonigral axons (Fig. 1J). Therefore, these results unequivocally indicate that Isl1 cell lineages developed into striatonigral neurons.
We also estimated recombination efficiency of Isl1-Cre activity by measuring the ratio of Isl1+;GFP+ cells/Isl1+ cells in embryonic day (E) 12.5 striatum of Isl1-Cre;CAG-CAT-EGFP mice. The Cre recombination efficiency was 87.3 ± 1.62% (Fig. S2). Therefore, the fact that Isl1-expressing cells constitute ∼50% of the striatonigral population is unlikely to be attributable to inefficient Isl1-Cre recombination.
Double staining of β-gal with striatal interneuron markers, including parvalbumin, neuronal NOS (nNOS), and calretinin, showed that none or, at most, a few cells coexpressed β-gal and these interneuron markers (Fig. S3 B–D). Double staining of β-gal and choline acetyltransferase (ChAT) showed that β-gal was colocalized in ChAT+ striatal interneurons (Fig. S3A), which was consistent with previous reports that Isl1 is expressed in striatal cholinergic interneurons (14, 15).
Isl1 Cell Lineages Were Present in Both Striosomal and Matrix Compartments of the Striatum.
Because Isl1 cell lineages comprised ∼50% of striatonigral neurons, we tested whether Isl1 cell lineages had a preference for developing into striosomal populations, which also project their axons to the SN (17–20). Double staining of β-gal and mu-opioid receptor (MOR1), a marker of striosomes, showed no preferential localization of β-gal+ cells within MOR1+ striosomes (Fig. S4A). Double staining of β-gal and calbindin, a marker of the matrix population, showed that β-gal+ cells were present in both calbindin-poor striosomes and calbindin-rich matrix (Fig. S4B). These findings indicate that Isl1 cell lineages contributed to the population of striatonigral neurons without compartment-specific preference.
Reduction of Striatonigral-Enriched Genes in Isl1 Mutant Striatum.
Given that Isl1 was transiently expressed in striatonigral neurons, we performed a loss-of-function study to investigate whether Isl1 might regulate neural development of striatonigral neurons. Previous studies have shown that Isl1 germ-line KO mice die at E10.5 owing to developmental defects in heart formation (8). We then intercrossed Isl1 conditional KO mice carrying a LoxP-flanked Isl1 null allele (Isl1f/f) with Nestin-Cre mice to generate neural-specific Isl1 KO mice. Because Nestin-Cre;Isl1f/f conditional KO (ncKO) mice died after birth, we then analyzed the phenotype of E18.5 mutant striatum in ncKO mice. Successful ablation of Isl1 and consequent loss of Isl1 protein in mutant brain was confirmed by Isl1 immunostaining, which demonstrated complete loss of Isl1 immunoreactivity in mutant striatum (Fig. S5).
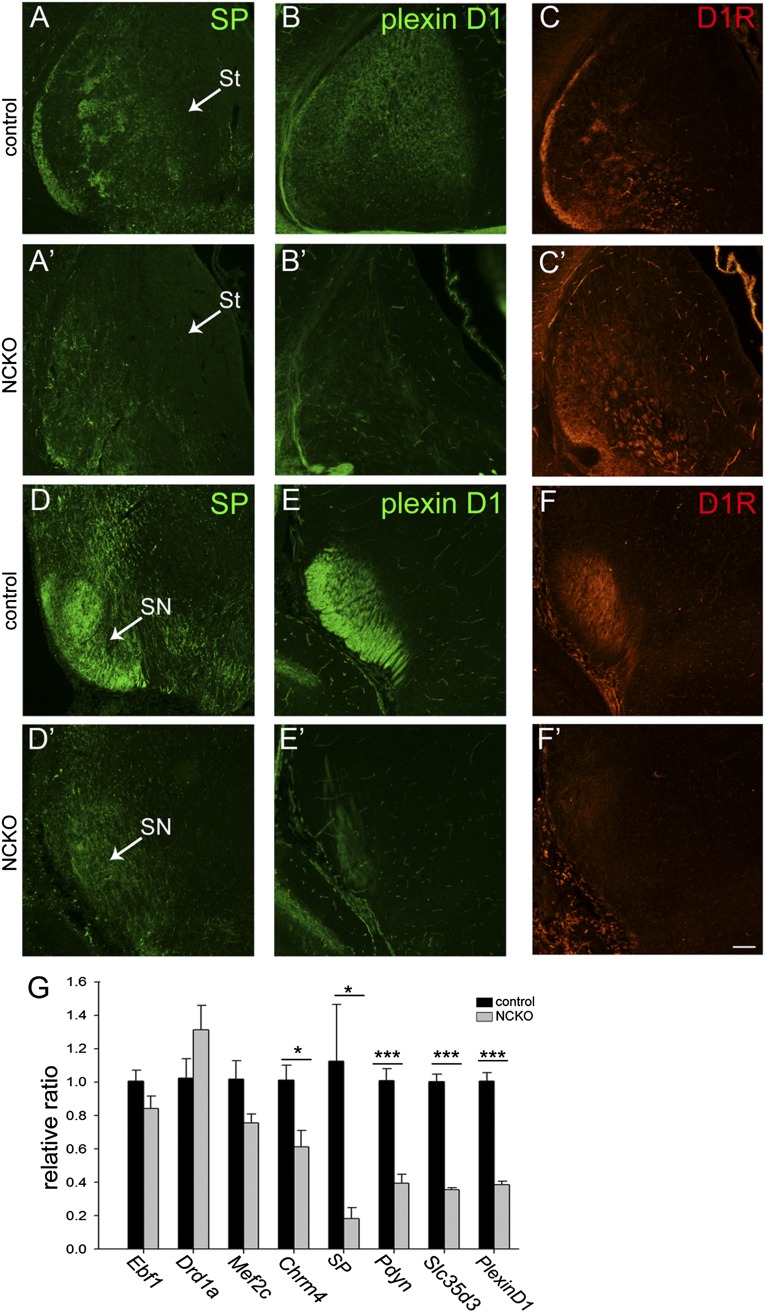
We investigated whether striatonigral-enriched genes were affected in Isl1 mutant striatum by examination of gene expression levels of SP, prodynorphin (Pdyn), Chrm4, Slc35d3, PlexinD1, Mef2c, Ebf1, and Drd1a, which are known to be preferentially expressed in striatonigral neurons (4). Quantitative RT-PCR (qRT-PCR) analyses showed significant reductions of SP (16.34 ± 5.9% of control, P < 0.05, n = 4), Pdyn (39.28 ± 8% of control, P < 0.001, n = 4), Slc35d3 (35.67 ± 4.0% of control, P < 0.001, n = 4), PlexinD1 (38.27 ± 2.8% of control, P < 0.001, n = 4), and Chrm4 (60.69 ± 18.8% of control, P < 0.05, n = 4) in ncKO striatum relative to controls (Fig. 2G). No significant change was noted for Ebf1 (84.16 ± 11.8% of control, P > 0.05, n = 4), Mef2c (77.97 ± 21.7% of control, P > 0.05, n = 4), and Drd1a (131.11 ± 24.3% of control, P > 0.05, n = 4) in mutant striatum relative to controls (Fig. 2G).
Fig. 2.
Down-regulation of striatonigral-enriched genes in Isl1 mutant brains at E18.5. SP immunoreactivity (A and A′) is significantly decreased in the striatum (St) (A′) and substantia nigra (SN) (D′) of Isl1 mutant mice. A similar reduction of plexinD1 immunoreactivity is found in the mutant St (B and B′) and SN (E and E′). D1R immunoreactivity is disorganized in the mutant St (C and C′) and dramatically decreased in SN (F and F′). (G) qRT-PCR analyses show a consistent reduction of striatonigral-enriched genes, including Chrm4, Slc35d3, PlexinD1, SP, and Pdyn, in Isl1 mutant St. A trend of a decrease in Mef2c is noted. n = 4. *P < 0.05; ***P < 0.001. (Scale bar: A–F′, 100 μm.)
Aberrant Expression Patterns of SP, PlexinD1, and Dopamine D1 Receptor in the Striatonigral Pathway of Isl1 Mutant Brain.
Down-regulation of striatonigral-enriched genes was further confirmed by immunostaining of SP and plexinD1. Expression of SP was patchy in control E18.5 striatum of Nestin-Cre;Isl1f/+ brain, particularly at the midcaudal level (Fig. 2A). SP+ axons projected from the control striatum via the GP, diencephalon/telencephalon boundary (DTB), and cerebral peduncle into SN (Fig. 2D). In Isl1 mutant striatum, SP immunoreactivity was dramatically reduced, with weak signals present in the ventrolateral striatum (Fig. 2A′). A few SP+ axons were found along the axonal trajectory routes to SN, such that only weak SP+ immunoreactivity was detected in SN (Fig. 2D′). Immunostaining of plexinD1, a striatonigral-enriched gene that marks striatofugal projections (21), showed that plexinD1 immunoreactivity was mainly present in the medial part of control E18.5 striatum (Fig. 2 B and E). In Isl1 mutant brain, plexinD1 immunoreactivity was dramatically reduced in the striatum and SN (Fig. 2 B′ and E′). Dopamine D1 receptor (D1R) immunostaining showed that instead of D1R+ patches in control E18.5 striatum at midcaudal levels, D1R immunoreactivity was concentrated in the ventrolateral mutant striatum without a patched pattern (Fig. 2 C and C′). Consistent with decreased SP+ and plexinD1+ axonal projections to SN, a significant reduction of D1R+ axons was also found in mutant SN (Fig. 2 F and F′).
Defective Striatonigral Projections in Isl1 Mutant Brain.
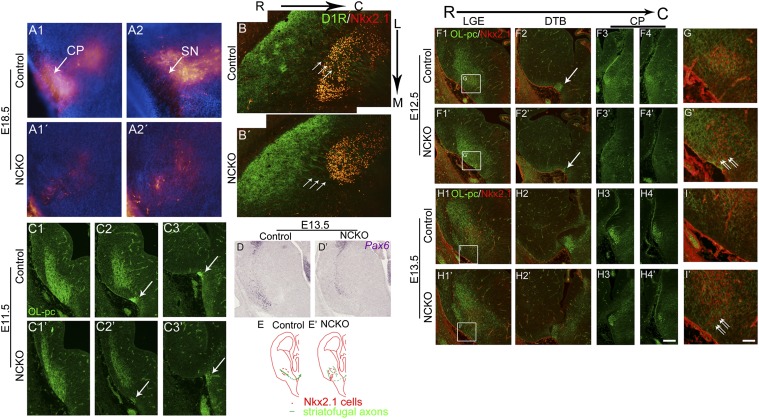
Concurrent decreases of SP+, plexinD1+, and D1R+ axons in the striatonigral pathway in Isl1 mutant brain suggested defective axonal outgrowth and/or navigation in Isl1 mutant brain. We therefore traced striatonigral axons by anterograde labeling of striatal axons with DiI at E18.5. Many DiI-labeled axons could be traced from the striatum via the DTB and cerebral peduncle into SN in control brain (Fig. 3 A1 and A2). By contrast, a significant reduction of DiI-labeled axons was found along the trajectory routes in the mutant brain. In mutant SN, only a few DiI-labeled axons were detected (Fig. 3 A1′ and A2′). These results clearly demonstrated that the axonal projections of striatonigral neurons were impaired in Isl1 mutant brain.
Fig. 3.
Defective striatofugal projections in developing Isl1 mutant brains. (A1–A2′) Anterograde labeling of striatofugal axons by placing DiI crystals in E18.5 rostral striatum. The descending DiI-labeled striatofugal axons project through cerebral peduncle (CP) (A1) into SN (A2) in control brain. In contrast, significant reduction of DiI-labeled striatofugal axons is observed in CP (A1′) and SN (A2′) in Isl1 mutant brain. (B) Striatal D1R+ axons enter Nkx2.1+ GP with a path at an angle of ∼45° (arrows) in control striatum as shown in horizontal sections of E18.5 brain. (B′) Routes of D1R+ axons shift to the plane parallel to the midline (arrows) when they enter the GP in Isl1 mutant brain. OL-pc+ striatofugal axons navigate from the striatal anlage (C1) through the ventral telencephalon to the DTB (C2 and C3, arrows) of E11.5 control brain. Reduction of OL-pc+ striatofugal axons is found at the DTB of Isl1 mutant brain (C2′ and C3′, arrows). (D and D′) Pax6+ cells, instead of lining the pallium/subpallium border, are located more laterally in E13.5 mutant telencephalon compared with those in control brain. (F1–G′) By E12.5, misrouted OL-pc+ axons are found in the regions around Nkx2.1+ GP or in the pial surface of the ventral mutant telencephalon in which ectopic Nkx2.1+ cells are detected (G and G′, double arrows). Reduction of OL-pc+ axons is also found in the path from the DTB to CP (F2 and F2′ (arrows) and F3–F4′). Similar results are found in E13.5 Isl1 mutant brain (H1–I′). The boxed regions in F1–H1′ are shown at high magnification in G and G′ and I and I′, respectively. (E and E′) Schematic drawings show the routes of striatofugal axons in E12.5–E13.5 control and mutant brains. C, caudal; L, lateral; M, medial; R, rostral. (Scale bars: A–D, F1–F4′, and H1–H4′, 100 μm; G–I′, 30 μm.)
Striatofugal axons are known to extend from the lateral ganglionic eminence (LGE) through the DTB into the cerebral peduncle as early as E13.5 (22). To examine the projection pattern of striatofugal axons at early developmental stages in Isl1 mutant brain, we analyzed the expression pattern of OL-protocadherin (OL-pc, protocadherin-10), a marker of striatofugal axons at early embryonic stages (22). In control E12.5 brain, OL-pc+ striatal axons extended through the Nkx2.1+ mantle zone of medial ganglionic eminence (MGE), passing through the DTB and then into the cerebral peduncle (Fig. 3 E–F4). In Isl1 mutant brain, however, OL-pc+ striatal axons did not converge through the Nkx2.1+ mantle zone. Instead, some extended aberrantly around the Nkx2.1− corridor cell region and the pial surface of the ventral telencephalon. Moreover, the density of OL-pc+ axonal bundles was significantly decreased in the DTB region (Fig. 3 E–F4′ and G′). Similar results were found in E13.5 mutant brains (Fig. 3 H1–H4′ and I′). Decreased OL-pc+ axonal projections were also observed in E11.5 mutant brain, in which weakly staining OL-pc+ axons were present in the ventrolateral ganglionic eminence (GE) and DTB (Fig. 3 C1–C3′). These findings indicated that defective striatofugal axonal projections occurred as early as E11.5 in Isl1 mutant brain.
Interestingly, the defective striatofugal projections in Isl1 mutant brain were similar to those of Pax6 mutant brain (22). We then examined Pax6 expression at E13.5 by in situ hybridization. We found that the number of Pax6+ cells was decreased in Isl1 mutant brains and, moreover, that the distribution of Pax6+ cells in the dorsal LGE (dLGE) along the pallium/subpallium borders was shifted laterally in ventral mutant telencephalon compared with control brain (Fig. 3 D and D′), suggesting that permissive areas along the trajectories of striatofugal axons were abnormal.
Later in development at E18.5, aberrant striatonigral projections over the mutant corridor region could be observed with D1R+ axons (Fig. 3B′). In horizontal sections of control striatum, D1R+ axons were observed in an oblique trajectory passing through the GP (Fig. 3B). In mutant striatum, trajectories of D1R+ axons, however, shifted more medially to enter the corridor region that was avoided in control striatum (Fig. 3B′).
Abnormal Death of Early-Born Cells in Isl1 Mutant Striatum.
Reductions in striatonigral markers and axonal projections raised the question as to whether abnormal cell death occurred in Isl1 mutant striatum. Consistent with this possibility, striatal areas were decreased by ∼30% in E18.5 mutant striatum at midcaudal and caudal levels (midcaudal level, control vs. mutant: 9.90 ± 0.66 × 105 vs. 7.14 ± 0.55 × 105, P < 0.001, n = 9; caudal level, control vs. mutant: 7.40 ± 0.76 × 105 vs. 5.64 ± 0.44 × 105, P < 0.01, n = 9; Fig. S6I). Immunostaining of activated caspase 3 (AC3) showed no significant change of AC3+ apoptotic cells in E12.5 mutant LGE (rostral level: control vs. mutant: 4.52 ± 0.35 vs. 4.69 ± 0.37, P > 0.05, n = 3; caudal level: 2.68 ± 0.31 vs. 4.83 ± 0.97, P > 0.05, n = 3; Fig. S6 A, A′, and D). In E13.5 mutant striatum, AC3+ apoptotic cells were increased by 1.98-fold and 1.67-fold at rostral and caudal levels, respectively (rostral: control vs. mutant: 1.69 ± 0.14 vs. 3.34 ± 0.22, P < 0.001, n = 3; caudal: 2.00 ± 0.26 vs. 3.34 ± 0.31, P < 0.05, n = 3; Fig. S6 B, B′, and E). By E15.5, AC3+ cells were also increased by 1.45-fold at the caudal level in Isl1 mutant striatum (rostral: control vs. mutant: 4.02 ± 0.24 vs. 5.17 ± 0.49, P > 0.05, n = 3; caudal: 5.53 ± 0.40 vs. 8.02 ± 0.57, P < 0.05, n = 3; Fig. S6 C, C′, and G). It was notable that AC3+ cells were predominantly present in the ventricular zone (VZ) and subventricular zone (SVZ) of E13.5 mutant striatum (Fig. S6F), suggesting that abnormal cell death occurred during early stages of differentiation.
Reduction of Early-Born Striatal Neurons in Isl1 Mutant Striatum.
To study further whether Isl1 mutation-induced apoptosis occurred in specific populations of developing striatal cells, we performed a pulse–chase study by pulse-labeling striatal cells with BrdU at E11.5, E12.5, or E15.5, and then quantifying the number of BrdU-labeled cells at E18.5 to assess for loss of labeled cells. Results demonstrated that BrdU+ cells labeled at E11.5 were reduced by ∼40% in E18.5 mutant striatum at midcaudal and caudal levels (midcaudal level, control vs. mutant: 123.8 ± 14.3 vs. 75.2 ± 8.7, P < 0.05, n = 3; caudal level, 104.7 ± 9.8 vs. 63.3 ± 8.4, P < 0.05, n = 3; Fig. S6 H, H′, and J). However, no significant difference in BrdU+ labeling at E12.5 or E15.5 was found between control and mutant striatum (Fig. S6 K and L).
A reduction in striatal area might also reflect altered cell proliferation in Isl1 mutant striatum. We assayed cell proliferation by immunostaining for phosphohistone 3 (pH3), a marker of mitotic cells. Results demonstrated no changes in pH3+ cell density in VZ of E12.5 and SVZ of E13.5 mutant LGE (E12.5 rostral: control vs. mutant: 0.056 ± 0.001 vs. 0.051 ± 0.002, P > 0.05, n = 3; caudal: 0.039 ± 0.005 vs. 0.040 ± 0.004, P > 0.05, n = 3; Fig. S7 A, A′, and C; E13.5 rostral: control vs. mutant: 1.82 ± 0.09 × 10−4 vs. 1.71 ± 0.13 × 10−4, P > 0.05, n = 3; caudal: 1.50 ± 0.11 × 10−4 vs. 1.56 ± 0.10 × 10−4, P > 0.05, n = 3; Fig. S7 B, B′, and F). Mild increases in pH3+ cell density were observed in E12.5 SVZ and E13.5 VZ at caudal levels (E12.5, control vs. mutant: 1.28 ± 0.01 × 10−4 vs. 1.48 ± 0.05 × 10−4, P < 0.05, n = 3; Fig. S7 A, A′, and D; E13.5, control vs. mutant: 3.97 ± 0.04 × 10−2 vs. 4.46 ± 0.06 × 10−2, P < 0.001, n = 3; Fig. S7 B, B′, and E).
Coordinated Increases of Striatopallidal-Enriched Genes in Isl1 Mutant Striatum.
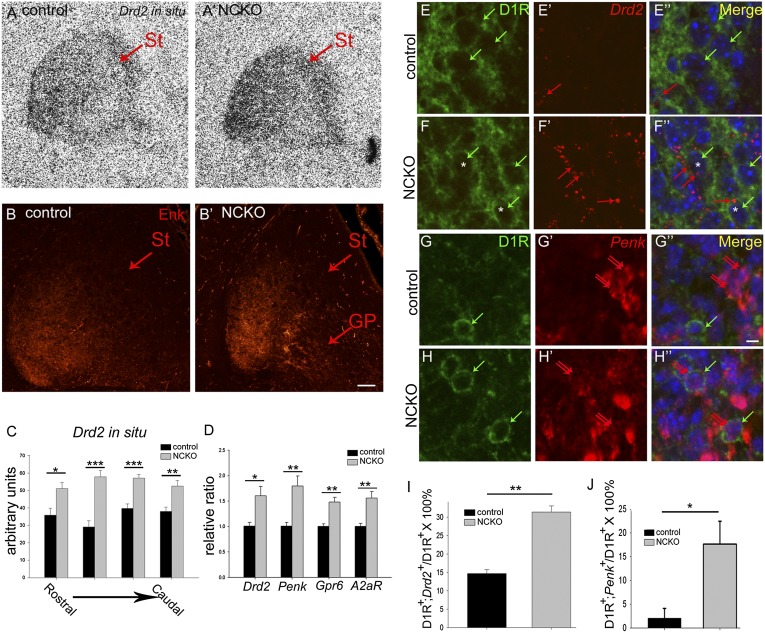
In addition to striatonigral neurons, we examined whether loss of Isl1 affected development of striatopallidal neurons. Remarkably, qRT-PCR analysis indicated that a repertoire of striatopallidal-enriched genes, including Drd2, Penk, adenosine receptor A2a (A2aR), and G protein-coupled receptor 6 (Gpr6) mRNAs (4), was significantly up-regulated in Isl1 mutant striatum (Drd2, 159.79 ± 25.68% of control, P < 0.05, n = 4; Penk, 178.91 ± 27.90% of control, P < 0.01, n = 4; A2aR, 155.13 ± 17.21% of control, P < 0.01, n = 4; Gpr6, 147.61 ± 10.31% of control, P < 0.01, n = 4; Fig. 4D). In situ hybridization analysis confirmed that Drd2 mRNA was increased by ∼1.5-fold in Isl1 mutant striatum throughout rostrocaudal levels (Fig. 4 A, A′, and C). Immunostaining of Met-Enk also demonstrated increased Enk protein expression in mutant striatum (Fig. 4 B and B′). These results revealed an unexpected role of Isl1 in suppressing striatopallidal-enriched genes during striatal development.
Fig. 4.
Up-regulation of striatopallidal-enriched genes in Isl1 mutant striatum at E18.5. (A, A′, and C) In situ hybridization shows that Drd2 mRNA expression is increased by ∼1.5-fold in mutant striatum from rostral to caudal levels. (B and B′) Enk immunoreactivity is increased in mutant striatum (St). (D) qRT-PCR analyses indicate a consistent up-regulation of striatopallidal-enriched genes, including Drd2, Penk, Gpr6, and A2aR in mutant St. (E–F′′ and I) D1R and Drd2 double immunostaining and in situ hybridization show that few cells coexpress D1R (cytoplasmic rings, green arrows) and Drd2 (red puncta, red arrows) in control St (E–E′′). Colocalization of D1R and Drd2 [F–F′′ (F and F′′, asterisks) and I] is significantly increased in mutant St as shown by D1R+ green cytoplasmic rings [E–F′′ (E, E′′, F, and F′′, green arrows)] containing Drd2+ red puncta [E–F′′ (E′, E′′, F′, and F′′, red arrows)]. (G–H′′ and J) Similar results are found in double labeling of D1R (G and H, green cytoplasmic rings) and Penk (G′, G′′, H′, and H′′, double red arrows). Cells coexpressing D1R and Penk are significantly increased in mutant striatum (G′′, H′′, and J). GP, globus pallidus; St, striatum. *P < 0.05; **P < 0.01; ***P < 0.001. Drd2 in situ hybridization, qPCR (n = 4); double labeling of D1R and Drd2 or Penk (n = 3). (Scale bars: A–B′, 50 μm; E–H′′, 5 μm.)
Given that Isl1 cell lineages develop into striatonigral neurons, the next question was whether Isl1 mutation-induced increases of striatopallidal genes occurred in striatonigral neurons. Because D1R, presumably still expressed by mutant striatonigral cells, remained in the ventrolateral part of Isl1 mutant striatum, it allowed us to use D1R as a marker for striatonigral neurons in mutant striatum. We performed D1R and Drd2 double immunostaining and in situ hybridization to see if D1R and Drd2 were colocalized in striatonigral neurons. In control striatum, a few D1R+ cells coexpressed Drd2 (D1R+;Drd2+cells/D1R+ cells × 100% = 14.67 ± 1.06%, n = 3; Fig. 4 E–E′′ and I). In contrast, D1R+ and Drd2+ double-labeled cells were increased by 2.14-fold in the mutant striatum (D1R+;Drd2+cells/D1R+ cells × 100% = 31.37 ± 1.70%, P < 0.01, n = 3; Fig. 4 E–F′′ and I). Similar results were found in double labeling of D1R and Penk, in which D1R+ and Penk+ double-labeled cells were significantly increased in mutant striatum (D1R+;Penk+ cells/D1R+ cells × 100%, control vs. mutant: 2.08 ± 2.08% vs. 17.70 ± 4.79%, P < 0.05, n = 3; Fig. 4 G–H′′ and J). These findings indicated that D1R-containing striatonigral and Drd2-containing striatopallidal neurons were predominantly distinct populations in control E18.5 striatum, and further suggested that loss of Isl1 led to derepression of striatopallidal Drd2 and Penk genes in striatonigral neurons.
Ectopic Isl1 Expression Suppressed Drd2 Expression in Striatopallidal Neurons.
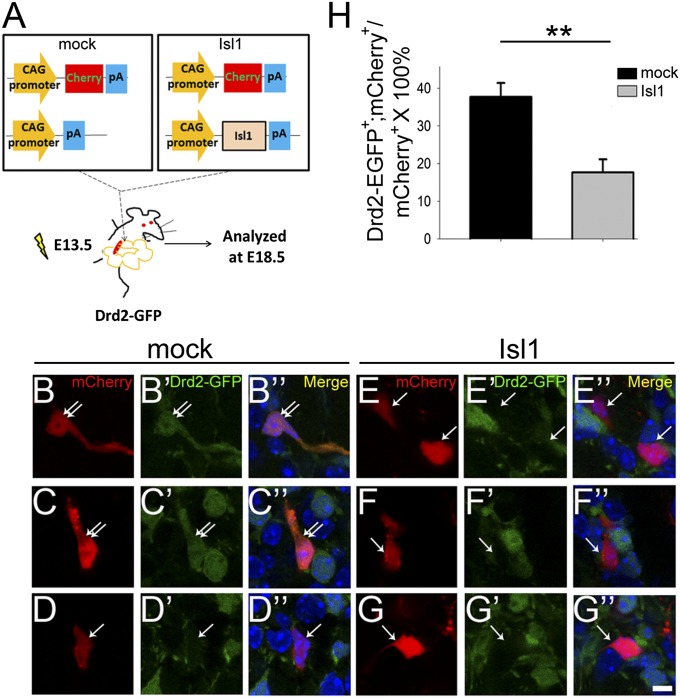
We then performed a gain-of-function study to see whether Isl1 could suppress the striatopallidal gene Drd2. A pdCALL2-myc-Isl1 expression plasmid was coelectroporated with pCAG-mCherry plasmid into E13.5 embryonic striatum by in utero electroporation. Most, if not all, mCherry+ cells coexpressed myc-Isl1 as demonstrated by double labeling of myc and mCherry (Fig. S8). Coelectroporation of pdCALL2-myc-Isl1 and pCAG-mCherry plasmids was then performed in E13.5 LGE of Drd2-EGFP BAC transgenic mice in which EGFP was driven by Drd2 promoter to be expressed in Drd2+ striatopallidal neurons. The electroporated brains were then analyzed at E18.5 (Fig. 5A). The ratio of Drd2-EGFP+;mCherry+ cells/mCherry+ cells was significantly decreased by 53% in the pdCALL2-myc-Isl1 group compared with the mock control group (mock vs. Isl1: 37.69 ± 3.71% vs. 17.64 ± 3.47%, P < 0.01, n = 4; Fig. 5 B–H). These results suggested that ectopic Isl1 expression was sufficient to repress Drd2 expression in striatopallidal neurons.
Fig. 5.
Overexpression of Isl1 is sufficient to suppress Drd2-EGFP expression. (A) Schematic drawings show coelectroporation of pCAG-mCherry and pdCALL2 or pdCALL2-myc-Isl1 plasmids into E13.5 striatum of Drd2-EGFP BAC transgenic mice by in utero electroporation. The brains were analyzed at E18.5 for colocalization of Drd2-EGFP and mCherry in striatal neurons. (B–D′′) mCherry+ cells coexpressing Drd2-EGFP (double arrows) are found in mock electroporated striatum. In Isl1 electroporated striatum, cells coexpressing Drd2-EGFP and mCherry were decreased by 53% (E–G′′) compared with mock control (H). The single arrows point to mCherry+ cells without EGFP. n = 4. **P < 0.01. (Scale bar: B–G′′, 5 μm.)
Discussion
In the present study, we have characterized cell types that transiently expressed Isl1 in striatum and further demonstrated the importance of Isl1 in striatal development. Isl1 was specifically expressed in ∼50% of striatonigral neurons during development. Conditional deletion of floxed Isl1 in neural progenitors by Nestin-Cre resulted in defective differentiation and axonal projection of striatonigral neurons. Moreover, striatopallidal genes were derepressed in striatonigral neurons, and overexpression of Isl1 was sufficient to suppress Drd2 expression. Therefore, Isl1 not only serves as a striatonigral marker in striatal neurogenesis but plays a pivotal role in promoting a striatonigral developmental program, concurrently suppressing striatopallidal genes to sharpen striatonigral cell identity.
Heterogeneous Populations of Striatonigral Projection Neurons.
We have previously shown transient Isl1 expression in the developing striatum, with the exception of cholinergic interneurons (15). Our present genetic cell lineage tracing study clearly indicated that striatonigral neurons were heterogeneous in terms of Isl1 expression. Isl1+ and Isl1− cell lineages contributed equally to SP+ striatonigral neuronal populations. There was no apparent preferential localization of Isl1+ neurons with respect to striosome/matrix compartments, nor did the axonal terminals of Isl1+ neurons show a specific topographical pattern in the SN. It will be of interest to determine whether there are any functional differences between these two populations of striatonigral neurons.
Isl1 Controls Striatonigral Axonal Projections.
Previous studies have shown that Isl1 regulates axonal growth and pathfinding in retinal ganglion cells and sensory nociceptive neurons (12, 13). In the present study, the importance of Isl1 in regulating striatonigral projections was demonstrated by decreased DiI-labeled and D1R+ striatonigral axons in Isl1 mutant brains at E18.5. Decreases in striatonigral projections was observed in mutant brains as early as E11.5–E12.5; at this stage, no significant increases in cell apoptosis were observed and no decreases in pH3+ mitotic cells were found in mutant brain.
Because Isl1+ cells were detected as early as E11.5 when OL-pc+ striatofugal axons appeared, it is likely that axons of Isl1+ cells constituted pioneer axons of the striatofugal pathway that innervated the SN. Defective striatonigral projections were already detected as early as E11.5–E12.5; at this stage, most OL-pc+ striatofugal mutant axons, instead of navigating through GP and the DTB to the cerebral peduncle, failed to converge through GP and were misrouted into the corridor cells area or the pial surface of ventral telencephalon. Because OL-pc is known to regulate outgrowth and navigation of striatofugal axons (22), Isl1 might regulate OL-pc to control striatofugal outgrowth. However, OL-pc expression did not appear to be altered in Isl1 ncKO striatum. It would be of interest to see whether defective trajectories of OL-pc+ striatofugal axons might be secondary to defective formation of the corridor cell region in which Isl1 is expressed (23).
The phenotype of defective striatofugal projections in Pax6 mutant brain is similar to that observed in Isl1 mutant striatum (22). Notably, Pax6+ cell numbers were decreased in Isl1 ncKO brains, and Pax6+ cells in dLGE along the pallium/subpallium boundaries were located more laterally in ventral mutant brains than in control brains. These findings suggested that Isl1 may non–cell-autonomously regulate the distribution of Pax6+ cells, which may, in turn, affect striatonigral projections. Another molecule known to be involved in striatonigral projection is Ebf1. In Ebf1 KO brain, striatonigral axons are able to project to SN but they are aberrantly distributed in SN (4). This phenotype is different from that in Isl1 mutant brain, because most striatonigral axons failed to reach the SN. Consistently, Ebf1 expression was not altered in Isl1 mutant striatum, suggesting that Isl1 and Ebf1 act in parallel to regulate axonal projections of striatonigral neurons.
Isl1 Regulates Cell Survival and Differentiation of Striatonigral Neurons.
Our study found that increased apoptotic AC3+ cells did not occur until E13.5 in LGE of striatal primordia. Many AC3+ cells were present in SVZ of the E13.5 germinal zone of ncKO brains, suggesting that a population of striatal cells died during early differentiation before sending out long-range striatofugal axons in SVZ of ncKO brains. Isl1 is clearly required for survival of this cell population. In fact, early-born BrdUE11.5 cells may constitute part of this cell population, because BrdUE11.5 cells were lost in E18.5 ncKO striatum. Nonetheless, there were other cell populations that could survive through E18.5 in Isl1 ncKO striatum, but the majority of them failed to develop into proper striatonigral neurons, and they had prominent defective striatonigral axonal projections. In E18.5 control striatum, D1R+ striosomes comprising early-born cells formed patches (24). Instead of forming patches, D1R+ cells in Isl1 ncKO striatum, presumably striatonigral cells, were aberrantly located in the ventrolateral striatum, but D1R+ axons were dramatically reduced in SN of mutant brains, suggesting that prenatal survival of striatonigral neurons may not depend on SN-derived factors, although it has been proposed that innervation of SN by striatonigral axons is required for survival of early-born striatonigral neurons during the cell death period in the first postnatal week of developing rat striatum (25). We cannot, however, rule out the possibility that there may be D1R− striatonigral cells with defective striatonigral projections that died in ncKO mutant striatum.
Isl1 Specifies the Cell Identity of Striatonigral Neurons.
In addition to controlling striatonigral axonal projections, Isl1 regulated the genetic profiles of striatonigral neurons, because a set of striatonigral-enriched genes, including SP, Pdyn, Slc35d3, Chrm4, and PlexinD1, was coordinately decreased in Isl1 ncKO stratum. Such coordinated regulation of striatonigral-enriched genes by Isl1 implicates a central role of Isl1 in specifying the cell fate of striatonigral neurons. The importance of Isl1 in specification of striatonigral neurons is further evidenced by the concomitant up-regulation of a set of striatopallidal-enriched genes, including Drd2, Penk, Gpr6, and A2aR in mutant striatum. Double labeling of D1R and Drd2 or Penk showed increases of D1R+/Drd2+ and D1R+/Penk+ double-labeled cells in Isl1 ncKO striatum, suggesting derepression of striatopallidal genes in striatonigral neurons without Isl1. Overexpression of Isl1 in Drd2-EGFP BAC transgenic striatum further supported this hypothesis, because Isl1 overexpression was sufficient to suppress Drd2-EGFP signals. Given that Isl1 was expressed in striatonigral neurons, the derepression of striatopallidal-enriched genes as a result of loss of Isl1 suggested that Isl1 normally suppresses striatopallidal-enriched genes. Taken together, the function of Isl1 was to specify the identity of striatonigral cells by promoting striatonigral-enriched genes and suppressing striatopallidal-enriched genes.
Regarding the molecular mechanisms by which Isl1 specifies cell types, Isl1 is known to specify different cell types by interacting with different transcription regulators (26). A recent study indicates that coexpression of Ngn2, Isl1, and Lhx3 is able to specify mouse ES cells into spinal motor neurons (27). Interestingly, replacing Lhx3 with Phox2a leads to specification of cranial, rather than spinal, motor neurons. The different cell fate specification by Isl1–Lhx3 and Isl1–Phox2a complexes is determined by binding of these two complexes to distinct genomic locations (27). It is plausible that Isl1 may interact with other transcriptional regulators to specify striatonigral cells.
It has been reported that a gene expression program common to the spinal cord and hindbrain, including Lhx1, Lhx2, Lbxcor1, Olig1, and Olig2, is derepressed in developing sensory neurons of Isl1 KO mice (13). Prolonged, rather than transient, expression of basic helix-loop-helix transcription factors, including Neurog1, Neurod1, Neurod4, and Neurod6, is also found in developing sensory neurons of Isl1 KO mice. Conceivably, Isl1 may interact with different cofactors to activate or inhibit different target genes. Along this line, one may speculate that Isl1, by interacting with different transcriptional regulators, may differentially promote striatonigral genetic programs and repress striatopallidal genetic programs.
Given the selective expression of Isl1 and its key role in development of striatonigral cell lineages, it is essential to know the upstream factors that regulate Isl1 expression. Isl1 is downstream of the Gsx2 and Dlx1/2 genetic cascades that control patterning and specification of striatal progenitors (28, 29). Isl1 is also inducible by sonic hedgehog in the process of differentiation of basal telencephalic cells from ES cells (30). It is yet unknown how Isl1 is programmed to be selectively expressed in striatonigral cell lineages. It is likely that combined actions of different signaling molecules may direct specific expression of Isl1 in striatonigral cell lineages, which will require further study in the future.
Materials and Methods
Animals.
Isl1-Cre (31), Isl1f/f (13), and Nestin-Cre (32) (kindly provided by C. Lai, Scripps Research Institute, La Jolla, CA); CAG-CAT-EGFP (16) (kindly provided by M. Colbert, Cincinnati Children’s Hospital, Cincinnati, OH); Rosa26R [R26R; kindly provided by T.-F. Tsai, National Yang-Ming University (NYMU)]; and Drd2-EGFP BAC transgenic mice [stock no. 000230-UNC; Mutant Mouse Regional Resource Center (MMRRC)] were housed in a specific pathogen-free room with 12-h light/dark cycle in the animal center at NYMU. Transgenic mice were maintained by back-crossing with C57/BL6J strain mice, except that Drd2-EGFP mice were back-crossed with FVB/N strain mice. To obtain Isl1 conditional KO mice, Nestin-Cre;Isl1f/+ mice were intercrossed with Isl1f/f mice. The animal protocols were approved by the Institutional Animal Care and Use Committee of NYMU.
Genotyping.
The genotyping of the genetically modified mice was performed by PCR with tail genomic DNA. The mouse tails were collected at 3–4 wk after birth. Following incubation in the lysis buffer (10 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% SDS, 100 μg/mL protease K) at 55 °C overnight, the lysates were treated with RNase A for 30 min at 37 °C, followed by protein precipitation solution (Promega), to remove RNA and proteins. The genomic DNA was extracted by isopropanol (1:1 ratio) and dissolved in Tris⋅EDTA buffer for at least 2 h at 50 °C. PCR genotyping primers for transgenic mice are listed as follows: Drd2-GFP-5′ (5′-CCTAC GGCGT GCAGT GCTTC AGC-3′) and Drd2-GFP-3′ (5′-CGGCG AGCTG CACGC TGCGT CCTC-3′); Isl1-5′ (5′-GGTCT CTGGA ACATC CCACA T-3′) and Isl1-3′ (5′-CTGTT CCTAC TCCCC ATTCA CT-3′); Isl1-Cre-5′ (5′-ACAGC AACTA TTTGC CACCT AGCC-3′) and Isl1-Cre-3′ (5′-TCCCT GAACA TGTCC ATCAG GT-3′); Nes-cre-5′ (5′-GCTAA ACATG CTTCA TCGTC G G-3′) and Nes-cre-3′ (5′-GATCT CCGGT ATTGA AACTC CAGC-3′); CAT-5′ (5′-CTGCT AACCA TGTTC ATGCC-3′) and CAT-3′ (5′- GGTAC ATTGA GCAAC TGACT G -3′); and R26R common 5′ (5′-AAAGT CGCTC TGAGT TGTTA T-3′), R26R WT 3′ (5′-GGAGC GGGAG AAATG GATAT G-3′), and R26R mutant 3′ (5′-GCGAA GAGTT TGTCC TCAAC C-3′). In general, the program of PCR amplification is similar for each with different annealing temperatures: 95 °C for 3 min followed by 35 cycles at 95 °C for 30 s and different annealing temperatures for each genotype for 30 s, 72 °C for 40 s, and for an additional 5 min at 72 °C to complete the reaction. The annealing temperatures are as follows: 68 °C for Drd2-GFP, 58 °C for Nestin-Cre, 60 °C for Isl1 and Isl1-Cre, 65 °C for R26R, and 66 °C for CAG. The sizes of PCR products are follows: 300 bp for Drd2-GFP allele, 550 bp for Nes-Cre allele, 500 bp for Isl1 floxed allele, 450 bp for Isl1 WT allele, 350 bp for Isl1-Cre allele, 300 bp for CAG allele, 500 bp for R26R WT allele, and 250 bp for R26R mutant allele.
qRT-PCR.
Striatal tissue was dissected from E18.5 forebrain. Total RNA was prepared by guanidinium thiocyanate-phenol-chloroform extraction. Two micrograms of RNA was primed with oligo(dT) (Roche) and reverse-transcribed into cDNAs using SuperScript reverse transcriptase (Invitrogen) following the manufacturer’s instructions. The DNA sequences of each primers are listed as follows: Pdyn forward, 5′-GGACA GGAGA GGAAG CAGAC-3′, reverse, 5′-AGAAG CAGGA AACTA CACAG ACC-3′; SP forward, 5′-AAGCC TCAGC AGTTC TTTGG-3′, reverse, 5′-GTGCG TTCAG GGGTT TATTT AC-3′; Drd1a forward, 5′-AGATC GGGCA TTTGG AGAG-3′, reverse, 5′-GGATG CTGCC TCTTC TTCTG-3′; Chrm4 forward, 5′-AGTGG GCAGT GTTCC TCTCT AC-3′, reverse, 5′-GTGCT TGCTT GTGTC ATTCG-3′; Ebf1 forward, 5′-AGGTT GGATT CTGCT ACGAA AGTT-3′, reverse, 5′-TGATT CCTCT TAAAA AGGCC TGA-3′; Slc35d3 forward, 5′-CCTGC CCATG TACGT AGTCT TCAA-3′, reverse, 5′-CGATC AGGAT ACAGG CCACG AATA-3′; Mef2c forward, 5′-GGATG AGCGT AACAG ACAGG T-3′, reverse, 5′-ATCAG TGCAA TCTCA CAGTC G-3′; Gpr6 forward, 5′-CTTAG CAGCC ACCAG AAAGG-3′, reverse, 5′-TGGCT ACCCA CCACA CAATA-3′; Drd2 forward, 5′-CTCCT CCATC GTCTC GTTCT AC-3′, reverse, 5′-CTTGA GTGGT GTCTT CAGGT TG-3′; Penk forward, 5′-AGAAG CGAAC GGAGG AGAGA-3′, reverse, 5′-TTCAG CAGAT CGGAG GAGTT G-3′; A2aR forward, 5′-TCAGC CTCTT GGCTA TTGCC-3′, reverse, 5′-CTCAA ACAGA CAGGT CACCC G-3′; and GAPDH forward, 5′-CGTGG AGTCT ACTGG TGTCT TC-3′, reverse, 5′-TGCAT TGCTG ACAAT CTTGA G-3′. The real-time PCR reactions using SYBR green dye-based detection were performed by the NYMU Genome Research Center using an Applied Biosystems 7900HT Fast Real-Time PCR System.
Administration of BrdU.
Timed-pregnant mice were i.p. injected with BrdU (100 mg/kg) at E11.5, E12.5, or E15.5. The embryos were harvested at E18.5. The day of plug positivity was defined as E0.5.
Preparation of Brain Tissue.
Littermates of P25 and 2-mo-old mice were used. The mice were anesthetized by i.p. injection of sodium pentobarbital and then transcardially perfused with ice-cold 4% (wt/vol) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.4). The brains were postfixed in the same fixative overnight and then cryoprotected by 30% (wt/vol) sucrose in 0.1 M PB for 3 d at 4 °C. The brains were sectioned at 20 μm. The sections were stored at 4 °C for histological analyses. For harvesting embryonic brains, pregnant mice were deeply anesthetized with 3% (wt/vol) sodium pentobarbital before abdominotomy. Embryos were removed from the uterus and kept in ice-cold PBS, and the brains were removed and immersed for fixation with 4% PFA for at least 20 h and then cryoprotected by 30% sucrose in 0.1 M PB for 2 d. Brains were cut at 12–18 μm using a cryostat (Leica).
Immunohistochemistry.
Immunostaining was performed as previously described (17). Antigen retrieval was performed for Nkx2.1, Isl1 immunostaining [3A4, 4D5 antibodies; 10 mM citric acid buffer (pH 6.0) at 95 °C for 15 min] and BrdU immunostaining [1 N of HCl/0.1 M PB at 45 °C for 30 min and 0.1 M borated buffer (pH 8.6) for 10 min]. Sections were washed with 0.1 M PBS twice for 5 min, 0.1% Triton X-100 in 0.1 M PBS for 5 min, and 3% (vol/vol) H2O2/10% (vol/vol) methanol in 0.1 M PBS for 5 min. Sections were incubated with the following primary antibodies overnight at room temperature: polyclonal rabbit anti-AC3 antibody (1:1,000; Cell Signaling), monoclonal mouse anti–β-gal antibody (1:1,000; Boehringer Mannheim), monoclonal rat anti-BrdU antibody (1:500; Accurate), monoclonal mouse anticalbindin antibody (1:1,000; Swant), monoclonal mouse anticalretinin antibody (1:1,000; Chemicon), polyclonal goat anti-ChAT antibody (1:200; Chemicom), polyclonal goat anti-D1R antibody (1:2,000; Frontier Institute), polyclonal rabbit anti-Isl1 antibody (1:2,000; kindly provided by S. Pfaff, Salk Institute, San Diego, CA), monoclonal 3A4 or 4D5 mouse anti-Isl1 antibody (1:100; Developmental Studies Hybridoma Bank), polyclonal rabbit anti–Met-Enk antibody (1:5,000; ImmunoStar), polyclonal chicken anti-c-myc antibody (1:1,000; Novus), monoclonal mouse anti-Nkx2.1 antibody (1:200; NeoMarkers), monoclonal mouse antineuronal NOS antibody (1:2,000; Sigma), polyclonal rabbit anti-MOR1 antibody (1:2,000; ImmunoStar), monoclonal rat anti–OL-pc antibody (1:1,000) (22), monoclonal mouse antiparvalbumin antibody (1:1,000; Sigma), polyclonal goat anti-plexinD1 antibody (1:1,000; R&D Systems), polyclonal rabbit anti-pH3 antibody (1:1,000; Millipore), and polyclonal rabbit anti-SP antibody (1:3,000; Eugene Tech, Inc.). After incubation with primary antibodies, the sections were incubated with appropriate secondary antibodies (1:500) for 1 h: biotinylated goat anti-rabbit antibody, horse anti-mouse antibody, rabbit anti-goat antibody and rabbit anti-rat antibody. The sections were then processed with avidin-biotin based immunostaining (Vectastain Elite ABC kit; Vector Laboratories). Avidin-FITC (Vector Laboratories) or a tyramide amplification system (TSA; PerkinElmer) was used to detect signals. Immunostained sections were counterstained with DAPI (Molecular Probes).
X-Gal Staining.
X-gal staining was performed as previously described (33). After X-gal staining, sections were immunostained as described above.
Axonal Tracing.
After fixation in 4% PFA, E18.5 brains were sectioned using a vibrotome at 100 μm. Small crystals of DiI (Invitrogen) were placed in the anterior part of striatum with a fine glass pipette. The DiI-labeled brain slices were stored in 4% PFA in the dark for ∼2 mo at 37 °C. The DiI-labeled brains were sectioned sectioned at 100 μm and counterstained with DAPI.
In Situ Hybridization.
The brain sections of WT and littermate mutant mice were mounted on the same slides to ensure they were processed under the same conditions. The rat Drd2L riboprobe (128–1,372 bp) was kindly provided by K. Kobayashi (Fukushima Medical University, Fukushima, Japan). Mouse Pax6 (NM_013627, 1,158–2,042 bp; Allen Brain Atlas) and Penk (NM_001002927, 312–1,127 bp; Allen Brain Atlas) were cloned by PCR and then subcloned into pGEM-T-easy vector (Promega). In situ hybridization was performed with 35S-UTP–labeled or digoxigenin-labeled probes as previously described (34).
Plasmid Construction.
The rat Isl1 cDNA was subcloned from the pCS2-MT-myc-Isl1 plasmid (kindly provided by T. Jessell, Columbia University, New York, NY) into the pdCALL2 plasmid that was derived from the pCCALL2 plasmid (kindly provided by A. Nagy, Mount Sinai Hospital, Toronto, ON, Canada). The pCAG-mCherry plasmid was kindly provided by S.-J. Chou (Academia Sinica, Taipai, Taiwan).
In Utero Electroporation.
The pCAG-mCherry, pdCALL2, and pdCALL2-myc-Isl1 plasmids were prepared using an endotoxin-free kit (Qiagene). The pCAG-mCherry plasmid was mixed with the pdCALL2 or pdCALL2-myc-Isl1 plasmids at a ratio of 3:1 (vol/vol). Mixed plasmids (1.2 μL at 2 μg/μL) were injected into the lateral ventricle of E13.5 forebrains of Drd2-EGFP mouse embryos, followed by electroporation (33 V, 30 ms, four pulses; BTX ECM 830). The electroporated brains were harvested at E15.5 or E18.5 for histological analysis.
Image Analyses, Quantification, and Statistics.
Microscopic analysis was performed using a fluorescence microscope (Olympus BX51) and a confocal microscope (Zeiss LSM700). Brightness adjustments for the whole field of photomicrographs were made, and final plates were composed using Adobe Photoshop CS (Adobe Systems). Quantification of cell number and area was performed with the aid of ImageJ software (National Institutes of Health). The number of pH3+ cells was normalized with the length of VZ or the area of SVZ in the germinal zone. The Student t test was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank Drs. S.-J. Chou, M. Colbert, T. Jessell, K. Kobayashi, C. Lai, A. Nagy, S. Pfaff, and T.-F. Tsai for providing reagents and transgenic mice and H.-F. Wang for help in the initial stage of the project. This work was supported by National Science Council Grants NSC99-2311-B-010-005-MY3, NSC101-2321-B-010-021, NSC102-2321-B-010-018, and NSC102-2911-I-010-506 and by an Aiming for Top University Grant, Ministry of Education, Taiwan.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319138111/-/DCSupplemental.
References
- 1.Graybiel AM. Neurotransmitters and neuromodulators in the basal ganglia. Trends Neurosci. 1990;13(7):244–254. doi: 10.1016/0166-2236(90)90104-i. [DOI] [PubMed] [Google Scholar]
- 2.Crittenden JR, Graybiel AM. Basal Ganglia disorders associated with imbalances in the striatal striosome and matrix compartments. Front Neuroanat. 2011;5:59. doi: 10.3389/fnana.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 5.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S. Distinct roles of synaptic transmission in direct and indirect striatal pathways to reward and aversive behavior. Neuron. 2010;66(6):896–907. doi: 10.1016/j.neuron.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344(6269):879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- 8.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 9.Liang X, et al. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci. 2011;47(3):215–222. doi: 10.1016/j.mcn.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007;503(1):182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci USA. 2008;105(19):6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135(11):1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, et al. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci. 2008;28(13):3291–3297. doi: 10.1523/JNEUROSCI.5730-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H-F, Liu F-C. Developmental restriction of the LIM homeodomain transcription factor Islet-1 expression to cholinergic neurons in the rat striatum. Neuroscience. 2001;103(4):999–1016. doi: 10.1016/s0306-4522(00)00590-x. [DOI] [PubMed] [Google Scholar]
- 16.Burns KA, et al. Nestin-CreER mice reveal DNA synthesis by nonapoptotic neurons following cerebral ischemia hypoxia. Cereb Cortex. 2007;17(11):2585–2592. doi: 10.1093/cercor/bhl164. [DOI] [PubMed] [Google Scholar]
- 17.Jiménez-Castellanos J, Graybiel AM. Compartmental origins of striatal efferent projections in the cat. Neuroscience. 1989;32(2):297–321. doi: 10.1016/0306-4522(89)90080-8. [DOI] [PubMed] [Google Scholar]
- 18.Gerfen CR. The neostriatal mosaic. I. Compartmental organization of projections from the striatum to the substantia nigra in the rat. J Comp Neurol. 1985;236(4):454–476. doi: 10.1002/cne.902360404. [DOI] [PubMed] [Google Scholar]
- 19.Fujiyama F, et al. Exclusive and common targets of neostriatofugal projections of rat striosome neurons: A single neuron-tracing study using a viral vector. Eur J Neurosci. 2011;33(4):668–677. doi: 10.1111/j.1460-9568.2010.07564.x. [DOI] [PubMed] [Google Scholar]
- 20.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74(5):858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Chauvet S, et al. Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron. 2007;56(5):807–822. doi: 10.1016/j.neuron.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 2007;10(9):1151–1159. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- 23.López-Bendito G, et al. Tangential neuronal migration controls axon guidance: A role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125(1):127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caille I, Dumartin B, Le Moine C, Begueret J, Bloch B. Ontogeny of the D1 dopamine receptor in the rat striatonigral system: An immunohistochemical study. Eur J Neurosci. 1995;7(4):714–722. doi: 10.1111/j.1460-9568.1995.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 25.Fishell G, van der Kooy D. Pattern formation in the striatum: Neurons with early projections to the substantia nigra survive the cell death period. J Comp Neurol. 1991;312(1):33–42. doi: 10.1002/cne.903120104. [DOI] [PubMed] [Google Scholar]
- 26.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 27.Mazzoni EO, et al. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci. 2013;16(9):1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long JE, et al. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512(4):556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K. Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron. 2009;63(4):451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K, et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8(3):288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 31.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 33.Liao WL, et al. Modular patterning of structure and function of the striatum by retinoid receptor signaling. Proc Natl Acad Sci USA. 2008;105(18):6765–6770. doi: 10.1073/pnas.0802109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CW, et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci USA. 2004;101(8):2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.