Significance
Apaf-1 interacting protein (APIP) inhibits two main types of programmed cell death: apoptosis and pyroptosis. In addition, APIP is a 5-methylthioribulose-1-phosphate dehydratase (MtnB) in the methionine salvage pathway. We verified its enzymatic activity directly through an enzyme assay and determined its high-resolution structure. Furthermore, we explored the relationship between two distinct functions of APIP/MtnB, cell death inhibition and methionine salvage, and determined that it functions as a cell death inhibitor independently of its MtnB enzyme activity for apoptosis, but dependently for caspase-1–induced pyroptosis. Our results provide groundwork for studies of the role of APIP/MtnB in development of cancers and inflammatory diseases.
Keywords: squamous carcinoma, systemic inflammatory response syndrome
Abstract
APIP, Apaf-1 interacting protein, has been known to inhibit two main types of programmed cell death, apoptosis and pyroptosis, and was recently found to be associated with cancers and inflammatory diseases. Distinct from its inhibitory role in cell death, APIP was also shown to act as a 5-methylthioribulose-1-phosphate dehydratase, or MtnB, in the methionine salvage pathway. Here we report the structural and enzymatic characterization of human APIP as an MtnB enzyme with a Km of 9.32 μM and a Vmax of 1.39 μmol min−1 mg−1. The crystal structure was determined at 2.0-Å resolution, revealing an overall fold similar to members of the zinc-dependent class II aldolase family. APIP/MtnB exists as a tetramer in solution and exhibits an assembly with C4 symmetry in the crystal lattice. The pocket-shaped active site is located at the end of a long cleft between two adjacent subunits. We propose an enzymatic reaction mechanism involving Glu139* as a catalytic acid/base, as supported by enzymatic assay, substrate-docking study, and sequence conservation analysis. We explored the relationship between two distinct functions of APIP/MtnB, cell death inhibition, and methionine salvage, by measuring the ability of enzymatic mutants to inhibit cell death, and determined that APIP/MtnB functions as a cell death inhibitor independently of its MtnB enzyme activity for apoptosis induced by either hypoxia or etoposide, but dependently for caspase-1-induced pyroptosis. Our results establish the structural and biochemical groundwork for future mechanistic studies of the role of APIP/MtnB in modulating cell death and inflammation and in the development of related diseases.
The programmed death of dangerous cells, either infected or transformed, has critical importance for the survival of the multicellular organism and therefore is also of great medical relevance. APIP, Apaf-1 interacting protein, was initially identified as an inhibitor of apoptotic cell death induced by hypoxia/ischemia and cytotoxic drugs (1). Recently APIP was also shown to inhibit pyroptosis, an inflammatory form of cell death, induced by Salmonella infection (2). Thus, APIP has been implicated in two major types of programmed cell death: apoptosis and pyroptosis. In apoptosis, APIP inhibits the mitochondrial pathway involving caspase-9 but not the receptor pathway involving caspase-8 (1, 3). In pyroptosis, APIP’s inhibitory function was recently revealed in a functional genetic screen for the SNP associated with increased caspase-1–mediated cell death in response to Salmonella infection (2) and subsequently confirmed by cell viability assays (2, 4). Intriguingly, other SNPs near APIP were found in patients suffering from systemic inflammatory response syndrome (2), which further implicates APIP in inflammation.
Distinct from its inhibitory role in the programmed cell death, APIP was recently shown to act as an enzyme in the methionine salvage pathway (2, 4). The amino acid sequence of human APIP exhibits 23–26% identity to the previously characterized Bacillus and yeast 5-methylthioribulose-1-phosphate dehydratase (MtnB) (4). The methionine salvage pathway converts MTA (5-methylthioadenosine) to methionine through six enzymatic reaction steps, and MtnB is the third enzyme in the pathway and catalyzes the dehydration of MTRu-1-P (5-methylthioribulose-1-phosphate) to DK-MTP-1-P (2,3-diketo-5-methylthiopentyl-1-phosphate) (Fig. 1B) (4, 5). In the absence of methionine, cells supplemented with MTA exhibit decreased viability when APIP expression is reduced (2, 4). These studies indicate that APIP is an MtnB enzyme in the methionine salvage pathway.
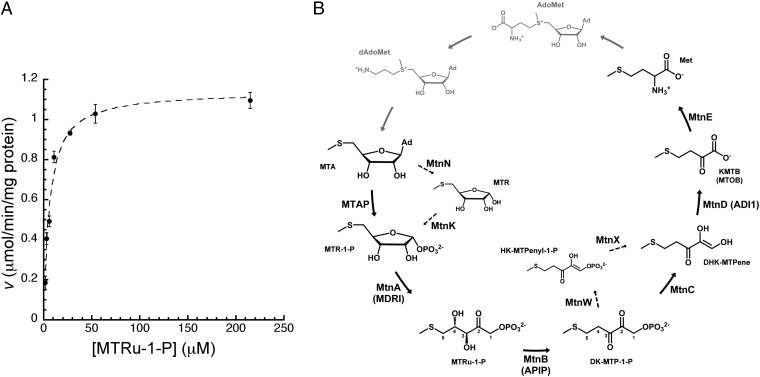
Fig. 1.
APIP as an MtnB enzyme in the methionine salvage pathway. (A) Initial reaction rate was plotted at seven different concentrations of the substrate MTRu-1-P for Michaelis-Menten kinetic analysis. Data represent mean values with SE from three independent measurements. (B) Methionine salvage pathway characterized in Homo sapiens and Saccharomyces cerevisiae converts MTA to methionine (Met) through the common six enzymatic reactions. Dashed line represents B. subtilis methionine salvage reaction steps distinct from H. sapiens and S. cerevisiae. Gray colored enzymatic steps and metabolites represent biochemical links that are not conceptually part of the methionine salvage pathway. AdoMet, S-adenosyl-l-methionine; dAdoMet, decarboxylated AdoMet; DHK-MTPene, 1,2-dihydroxy-3-keto-5-methylthiopentene; HK-MTPenyl-1-P, 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate; Met, l-methionine; MTOB, 4-methylthio-2-oxobutyrate; MTR, 5-methylthioribose.
The methionine salvage pathway is found in all organisms, from bacteria to plants and animals (6). The role of this pathway is to recycle MTA, which is a by-product of the polyamine synthetic process, back to methionine (Fig. 1B). The methionine salvage pathway is beneficial as a means of recycling the sulfur present in MTA because assimilation of sulfur is thermodynamically costly (6). The metabolic importance of the pathway is underscored in humans because methionine is one of the essential amino acids needed to be provided through the diet, in which it is one of the most limiting amino acids (6). Recently, the methionine salvage pathway attracted medical interest because it was implicated in cell death and inflammation and diseases associated with these processes. For example, metabolites such as MTA and 2-keto-4-methylthiobutyrate (KMTB) have effects of apoptosis induction (6–9). MTA was also shown to induce caspase-1–dependent pyroptosis in the inflammatory response to bacterial infection (2). In addition, the 5-methylthioadenosine phosphorylase (MTAP, which catalyzes the first step) is a tumor suppressor implicated in a various human cancers (6, 10), and aci-reducton dioxygenase 1 (ADI1, also called MtnD, which catalyzes the fifth step) has a similar role in prostate cancer (11, 12). Human APIP/MtnB, which is the focus of the present study, is another example of a methionine salvage enzyme that is implicated in cell death and inflammation. APIP/MtnB was recently reported to be up-regulated in squamous carcinoma cells from tongue and larynx (13) and down-regulated in the cells and tumors of non–small-cell lung carcinoma (14). In addition, APIP/MtnB is implicated in inflammatory conditions that likely involve caspase-1–dependent pyroptosis, such as systemic inflammatory response syndrome (2).
Studies of APIP/MtnB to date have focused mainly on its functional role either in cell death or in methionine salvage. To gain a better understanding of APIP/MtnB at a molecular and biochemical level, we carried out a structural and biochemical characterization in this study. The MtnB enzyme activity of APIP was verified by an in vitro enzyme assay. In addition, the crystal structure was determined at 2.0-Å resolution, which revealed details of the active site architecture and led to a proposed catalytic mechanism. Furthermore, we explored the relationship between two distinct functions of APIP/MtnB, cell death inhibition, and methionine salvage, by testing its enzymatic mutants derived from the crystal structure for their ability to inhibit two main types of programmed cell death: pyroptosis and apoptosis.
Results and Discussion
APIP Is an MtnB Enzyme in the Methionine Salvage Pathway.
APIP was predicted to be an MtnB enzyme in the methionine salvage pathway by a recent bioinformatic analysis of the primary sequence (4), which was supported by cell viability assays from two independent studies, as noted above (2, 4). We tested the MtnB enzyme activity of human APIP in an in vitro enzyme assay. The preparation of MTRu-1-P, the substrate of MtnB, was not successful because of the difficulties in purification after synthesis. So MTRu-1-P was indirectly supplied to the assay system by addition of MtnA enzyme, which precedes the MtnB reaction in the methionine salvage pathway, and its substrate, MTR-1-P (methylthioribose-1-phosphate) (Fig. 1B). The concentration of MTRu-1-P was calculated from the equilibration constant between MTR-1-P and MTRu-1-P. More details are described in Materials and Methods. Product formation was measured by monitoring the increase in absorbance at 280 nm for the reaction coupled to Bacillus 2,3-diketo-5-methylthiopentyl-1-phosphate enolase (MtnW), which follows the MtnB enzyme in the bacterial methionine salvage pathway (Fig. 1B). MtnB enzyme activity of APIP was evident, with Vmax of 1.39 μmol min−1 mg−1 and Km of 9.32 µM deduced from the reaction rates measured at seven different substrate concentrations (Fig. 1A). The observed MtnB enzyme activity for APIP is consistent with recent reports that reduced APIP expression caused decreased viability in cells supplemented with MTA as a source of methionine and that the overexpression of APIP decreased cellular MTA levels (2, 4). The results from our enzymatic assay indicate that human APIP is the MtnB enzyme responsible for the dehydration step converting MTRu-1-P to DK-MTP-1-P in the methionine salvage pathway (Fig. 1).
Overall Structure of APIP/MtnB.
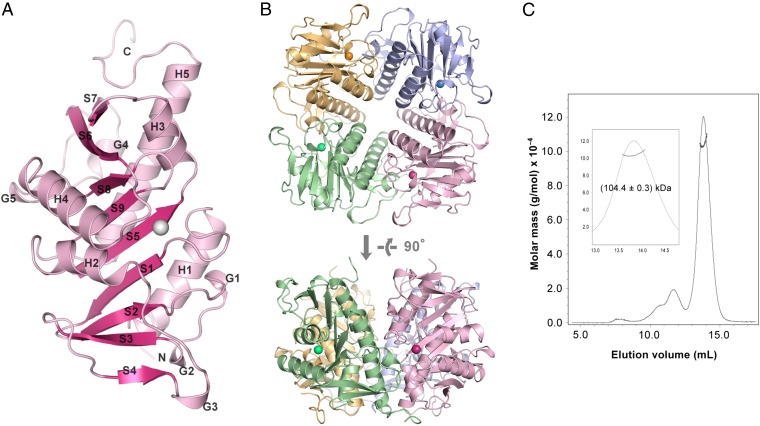
The crystal structure of APIP/MtnB was determined by the multiwavelength anomalous diffraction method using the bromide ion as an anomalous scatterer for phasing. The structure was refined to 18.3% of Rwork and 22.0% of Rfree at 2.0-Å resolution, and the details of the crystallographic analysis are summarized in Table S1. The asymmetric unit contains four APIP/MtnB molecules forming a tetramer with C4 symmetry (Fig. 2B). The final refined model contains residues 20–242 and one zinc ion for each APIP/MtnB molecule. The monomer consists of a central nine-stranded β-sheet (S1–S9) flanked on both sides by five α-helices (H1–H5) and four 310 helices (G1–G4) (Fig. 2A). The overall fold shows close similarity to members of the class II aldolase family, including FucA [l-fuculose 1-phosphate aldolase; Protein Data Bank (PDB) ID 1FUA], RhuA (l-rhamnulose-1-phosphate aldolase; PDB ID 1OJR), and RibE (l-ribulose-5-phosphate 4-epimerase; PDB ID 1JD1) (15–17), as analyzed by structural alignment using the DALI server giving Z-scores of 18.6–23.1 for 179–197 aligned residues (18). In particular, the strand order and direction, 6↓-7↑-8↑-9↓-5↑-1↓-2↑-3↓-4↑, of the central β-sheet is conserved between APIP/MtnB and the aforementioned class II aldolase family members, and noticeable structural variations reside mainly in the surrounding helices.
Fig. 2.
Overall fold of APIP/MtnB and its tetrameric assembly. (A) The monomeric APIP/MtnB structure is sketched in ribbon representation. An active site zinc ion is shown in a gray ball. (B) Tetramer assembly of APIP/MtnB in C4 symmetry is observed in an asymmetric unit of the crystal lattice. Zinc ions are represented as balls colored similarly as the corresponding subunits embracing them. (C) SEC-MALS shows that APIP/MtnB exists as a tetramer in solution.
APIP/MtnB, similar to other members of the class II aldolase family, forms a tetramer of C4 symmetry in the crystal lattice. We investigated whether APIP/MtnB exists as a tetramer in solution by measuring the mass using multiangle light scattering (MALS) coupled with size-exclusion chromatography. The measured mass was 104.4 kDa, which is exactly four times the monomeric molecular mass of 26.1 kDa, indicating that APIP/MtnB exists as a tetramer in solution (Fig. 2C). The interface between adjacent subunits covers 2,114–2,865 Å2, which is in the range of a relatively large protein–protein interface area (19). The interface on one subunit is located at the edge of the central β-sheet and involves three loop regions (L[H1-S1], L[S7-H4], L[S8-S9]), helix H3, and the 310 helix G1. The interface on the other subunit is composed mainly of a helix H5, part of helix H1, and the 310 helix G4.
Active Site Architecture and Catalytic Mechanism.
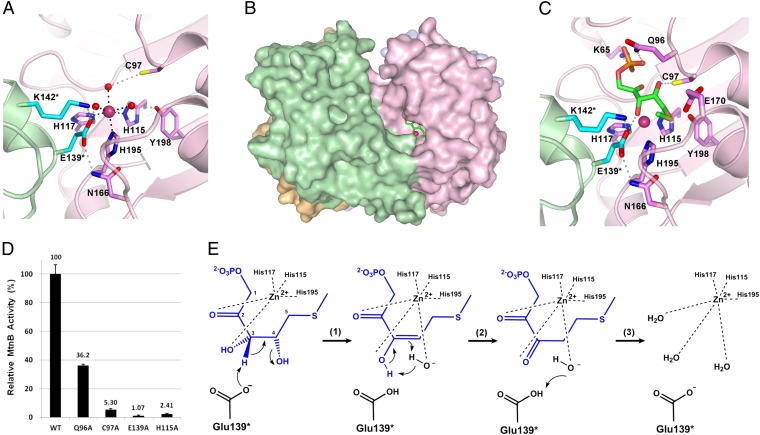
During the structure refinement, a strong spherical electron density was found near the imidazole groups of three histidine residues (His115, His117, and His195). The density is connected to the NE2 atoms of the three imidazole rings of these histidine residues even at a 2.5 σ contour level of an omit map. We assigned this density to a zinc ion in analogy to the other class II aldolase family members, which are all zinc-dependent (15–17). To confirm the presence of the zinc ion in APIP/MtnB, we performed an X-ray fluorescence spectroscopy analysis on the APIP/MtnB crystal. The wavelength scan around the K edge of zinc atom clearly showed a peak at 9,671.1 eV (Fig. S1A). Moreover, the data set collected at the wavelength of the peak, 1.28201 Å, further confirmed the presence of the zinc ion: its anomalous difference map showed a strong density at the position of the modeled zinc ion in the active site at the contouring level of 5 σ cutoff (Fig. S1B). Because zinc ions had not been added during sample preparation, these results suggest that APIP/MtnB acquired the zinc ion inside the cell on culturing and bound to it tightly. A pocket-shaped active site, defined by the presence of the catalytic zinc ion, is located at one end of a long cleft along the subunit interface (Fig. 3B). The zinc ion is octahedrally coordinated by six ligands: three histidine residues (His115, His117, and His195) and three water molecules (Fig. 3A).
Fig. 3.
Active site architecture and the catalytic mechanism. (A) Structural features of the active site are represented. The features contributed by the one subunit are colored in pink, and the adjacent subunit in green. Hydrogen bond is represented as a dotted line in gray, and the coordination between zinc and ligand in black. A pink ball is for the zinc ion, and red balls for water molecules. Asterisk denotes the residues contributed from the adjacent subunit. (B) Surface is represented with the docked substrate, MTRu-1-P, from the same view as Fig. 2B, Lower. (C) Substrate binding mode is sketched from the docking model with the same representing scheme and color code as in A and B. (D) Data represent mean relative specific activities of mutants with SE of three independent measurements. The wild type’s specific activity is set to 100%. (E) Proposed catalytic mechanism, starting from the substrate-bound state (Left), which corresponds to the docking model. (Right) Active site observed in the crystal structure.
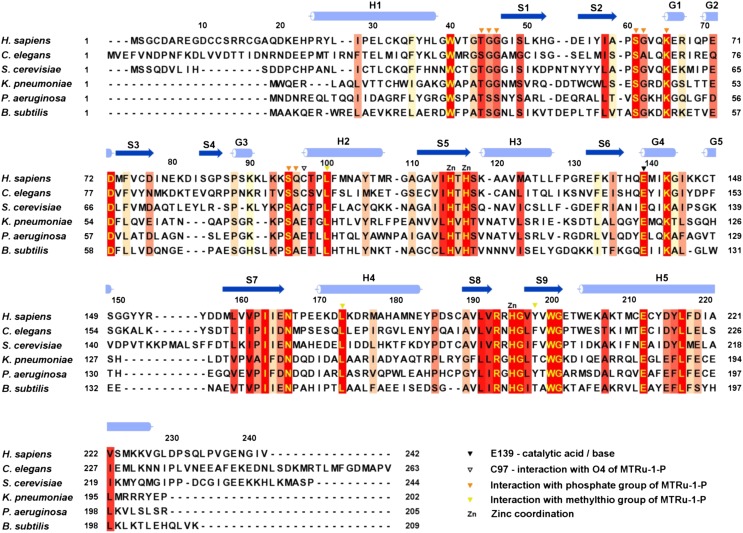
Near the zinc ion, the surface of the active site pocket is formed by several residues, including Cys97, Tyr198, Glu139*, Lys142*, and Asn166. Of these, Cys97, Glu139*, and Tyr198 form hydrogen bonds with the three water molecules that coordinate the zinc ion (an asterisk indicates that the residue is contributed by the neighboring subunit) (Fig. 3A). Notably, Glu139* is firmly positioned by the hydrogen bonds with Lys142* and Asn166, and these three residues are absolutely conserved from bacteria to humans, which strongly implies that Glu139* plays a critical role in catalysis (Figs. 3A and 4). This is further discussed below with a proposed catalytic mechanism.
Fig. 4.
Alignment of APIP/MtnB sequences. The sequences are retrieved from UniProtKB database (http://www.uniprot.org).
Structural information about the substrate-binding mode can provide insight into the catalytic mechanism and the related roles of the active site residues. To obtain this information for APIP/MtnB, we deduced a substrate-docked model using AutoDock4 (20) because determining the structure of the substrate-bound state was hampered by difficulties in preparation of the substrate, MTRu-1-P. The top solution, with an estimated free energy of binding of −4.98 kcal/mol, shows a reasonable substrate-bound structure as assessed by comparison with previously reported crystal structures of other class II aldolase family members, FucA and RhuA, in complex with their common substrate analog, PGH (phosphoglycolohydroxamate) (PDB IDs 4FUA and 1GT7) (15, 16). The overall binding of MTRu-1-P to APIP/MtnB is very similar to PGH bound to FucA and RhuA. The phosphate group of MTRu-1-P is bound deep within the pocket, and two oxygen atoms, O2 and O3, coordinate to the zinc ion, in a manner similar to PGH-bound structures of FucA and RhuA (Fig. 3C) (15, 16). More importantly for the catalytic mechanism, O3 and O4 of MTRu-1-P are hydrogen-bonded to Glu139* and Cys97, respectively (Fig. 3C), as discussed further below.
The substrate-docked model led us to propose an enzyme reaction mechanism involving Glu139* as a catalytic acid/base (Fig. 3E). The leftmost sketch in Fig. 3E is a simplified presentation of the active site occupied by the substrate, MTRu-1-P, shown in Fig. 3C. In the first step of the proposed mechanism, Glu139* abstracts a proton from C3 of MTRu-1-P, and the hydroxide is removed from C4 to coordinate the zinc ion, resulting in enol formation. The second step is a thermodynamic slide from enol to keto by tautomerization, which results in the di-keto product DK-MTP-1-P. In the third step, Glu139* donates a proton to the hydroxide ion, and the product DK-MTP-1-P diffuses out of the active site. As water molecules come in, the active site returns to the state observed in the crystal structure, with three water molecules coordinating the zinc ion together with three histidine residues to form the octahedral coordination geometry (Fig. 3A and the rightmost sketch of Fig. 3E). It is important to note that Glu139* is precisely positioned toward C3 of MTRu-1-P through hydrogen bonds with Lys142* and Asn166 in the substrate-docked model, which explains the absolute conservation of these three residues from bacteria to eukaryotes (Figs. 3C and 4 and Fig. S2). In addition, the mutation of Glu139* abolished the enzyme activity almost completely, which further suggests that Glu139* plays a critical role in catalysis (Fig. 3D).
Inhibition of Cell Death by APIP/MtnB and Its Enzymatic Function.
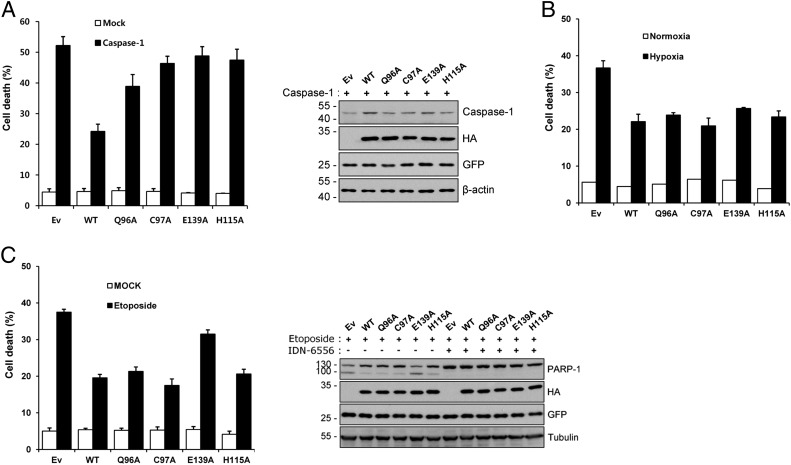
As noted above, APIP/MtnB was first identified in our previous studies as an inhibitor of caspase-9–dependent apoptosis induced by ischemic/hypoxic injury or by cytotoxic agents such as etoposide and cisplatin (1, 3). Recently it was reported that APIP/MtnB inhibits not only the caspase-9–dependent apoptosis but also the caspase-1–dependent pyroptosis (2). We investigated whether the inhibition of two main types of programmed cell death by APIP/MtnB is associated with its MtnB enzymatic activity by testing the enzymatic mutants for the ability of cell death inhibition. For pyroptosis assay, the death of HeLa cell is induced by caspase-1 overexpression, and the APIP/MtnB enzymatic mutants were evaluated for the ability of cell death inhibition. The Q96A mutant inhibited the cell death in a degree approximately half that of the wild type, and the other three mutants, C97A, E139A, and H115A, lost almost all ability of cell death inhibition (Fig. 5A). These results correlate well with the enzymatic assay in which the mutants lost the enzymatic activity to similar degrees; the Q96A mutant retained 36.2% of the enzymatic activity of the wild type, and the other three mutants, C97A, E139A, and H115A, exhibited less than 6% (Fig. 3D). In particular, the mutation of Glu139, a catalytic acid/base, caused the largest defect commonly in both functions of cell death inhibition and MtnB enzyme catalysis (Figs. 5A and 3D). These results suggest that the APIP/MtnB inhibits the cell death induced by caspase-1 overexpression in a manner dependent upon its MtnB enzyme activity.
Fig. 5.
Evaluation of APIP/MtnB enzymatic mutants for the inhibition of cell death. (A) Caspase-1–induced cell death. (Left) HeLa cells were transiently cotransfected with pEGFP and the indicated constructs for 18 h. After staining with 0.5 µg/mL ethidium homodimer (EtHD), cell death rates were measured by counting the number of both GFP- and EtHD-positive cells among total GFP-positive cells after staining with EtHD. Ev, empty vector. Values indicate mean ± SD (n = 3). (Right) Whole-cell extracts were prepared and subjected to Western blotting using the indicated antibodies. (B) Hypoxia-induced cell death. HeLa cells were transiently cotransfected with pEGFP and the indicated constructs for 24 h and then exposed to a hypoxic condition of 1% O2 for 48 h. Cell death rates were measured by trypan blue exclusion assay; bars represent mean ± SD (n = 3). (C) Etoposide-induced cell death. (Left) HeLa cells were transiently cotransfected with pEGFP and the indicated constructs for 24 h and then treated with 40 μM etoposide for 36 h. After staining with 0.5 μg/mL EtHD, cell death rates were measured by counting the number of both GFP- and EtHD-positive cells among total GFP-positive cells. Bars indicate mean ± SD (n = 3). (Right) Cells were treated the same as described above in the presence or absence of 10 μM IDN-6556. Western blotting was performed with the indicated antibodies. PARP-1, poly(ADP ribose) polymerase-1.
For caspase-9–dependent apoptosis assay, we tested the APIP/MtnB mutants in both hypoxic and cytotoxic conditions in which APIP/MtnB was previously shown as an inhibitor of apoptosis (1, 3). In the hypoxia-induced cell death assay, all four mutants inhibited the death of HeLa cells to a similar degree as the wild type (Fig. 5B). Given that these mutants exhibited decreased MtnB enzyme activity (partial for Q96A and almost complete for the other three mutants; Fig. 3D), these results suggest that the inhibition of hypoxic cell death by APIP/MtnB is not dependent upon its MtnB enzyme activity. In the etoposide-induced cell death assay, three of the mutants, Q96A, C97A, and H115A, showed similar levels of cell death inhibition as the wild type, and E139A exhibited a significantly lower level (Fig. 5C). The important point of these results is that the degrees of inhibition of etoposide-induced cell death by the wild type and the mutants do not correlate with their MtnB enzyme activities (Fig. 3D). For example, the C97A, H115A, and E139A mutants lost almost all MtnB enzyme activity (Fig. 3D). However, C97A and H115A mutants show a similar level of cell death inhibition as the wild type, and the E139A mutant shows a significantly lower level (Figs. 3D and 5C). The lack of correlation implies that the inhibition of etoposide-induced cell death by APIP/MtnB is not dependent upon its MtnB enzyme, same as for the hypoxia-induced cell death. These results are consistent with the recent study by Ko et al. (2), in which overexpression of the C97A mutant resulted in a dominant negative phenotype that rendered cells unable to use MTA as their source of methionine, but still showed the retained ability to inhibit the cell death induced by caspase-9 overexpression (2).
Cell Death and Methionine Salvage Pathway.
Given that APIP is an MtnB enzyme as well as a cell death inhibitor, it leads to the question as to how these two distinct functions are related to each other. As shown above, the inhibition of pyroptosis by APIP/MtnB is dependent on its MtnB enzymatic function. In the recent study by Ko et al. (2), the exogenous addition of MTA, the starting substrate for the methionine salvage pathway, resulted in an enhancement of pyroptosis, as did decreased APIP expression, implying that the inhibition of pyroptosis by APIP/MtnB must be mediated by MTA. This mechanism may explain why the APIP/MtnB enzymatic mutants failed to inhibit caspase-1–induced cell death; overexpression of the mutants caused the defect in methionine salvage pathway and the accumulation of MTA, which resulted in their failure to inhibit the pyroptosis. The proinflammatory state has an increased demand for methionine to support leukocyte proliferation and synthesis of acute phase proteins and polyamines (2, 21), thus it was proposed that this demand explains the link between pyroptosis and MTA (2).
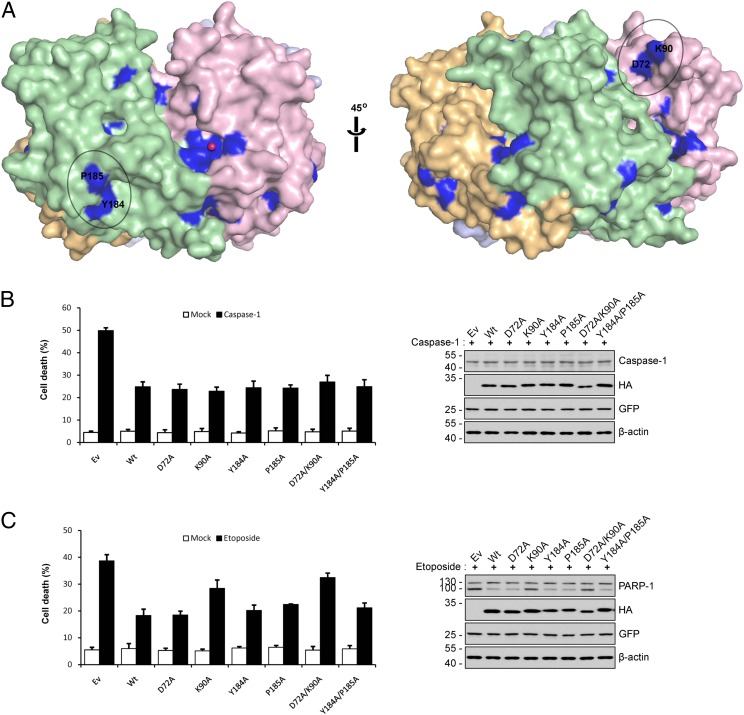
In contrast to pyroptosis, the antiapoptotic function of APIP/MtnB is not dependent upon its MtnB activity, as shown here and by Ko et al. (2) in different experimental conditions. What then is the mechanism of apoptosis inhibition by APIP/MtnB? We showed in previous studies that APIP/MtnB suppresses hypoxic/ischemic cell death by two different mechanisms (1, 3). APIP/MtnB competes with procaspase-9 for binding to Apaf-1 and thereby inhibits the activation of caspase-9 (1). In addition, APIP/MtnB activates AKT and ERK1/2, which inactivate caspase-9 by phosphorylation (3). These mechanisms provide a possible explanation for why APIP’s role in the inhibition of apoptosis is independent of its MtnB enzyme activity, as shown for the cell death induced by hypoxia or etoposide in the present study and by caspase-9 overexpression in the recent study by Ko et al. (2). Furthermore, the antiapoptotic function of APIP/MtnB may involve interaction with the proteins affecting apoptosis, such as Apaf-1 or proteins in AKT and ERK1/2 signaling pathways. In this regard, it is tempting to speculate that some conserved surface residues may be responsible for such interactions. Thus, we identified conserved surface residues by aligning APIP/MtnB sequences of four different multicellular model organisms—human, fruit fly, plant, and yeast (Fig. S3)—and by locating them on the molecular surface (Fig. 6A). Even though most of the conserved residues are located in the active site and tetrameric interface as expected, two small conserved surface patches were found distantly from the active site and the interface. Specifically, one patch is composed of Asp72 and Lys90, and the other of Tyr184 and Pro185. To find out which residue among them may play a possible role in protein–protein interaction for apoptosis inhibition, we examined their mutational effects on APIP/MtnB’s function inhibiting cell death (Fig. 6). We prepared four single mutants (D72A, K90A, Y184A, and P185A) and two double mutants (D72A/K90A and Y184A/P185A) and tested the mutants for the ability to inhibit pyroptosis and apoptosis in the same experimental settings used for the enzymatic mutants of Fig. 5 A and C, respectively. All of the tested surface mutants showed a similar level of pyroptosis inhibition as the wild type, which perhaps is because the surface residues are located distantly from the active site so that their mutation could not affect the catalytic activity. These results are consistent with the notion that pyroptosis inhibition by APIP is dependent upon its MtnB enzyme activity. For apoptosis inhibition, two mutants, K90A and D72A/K90A, showed significant loss of apoptosis inhibition activity, and all of the other mutants, in contrast, showed a similar level of apoptosis inhibition as the wild type. Considering that the single mutant of D72A retains a similar level of apoptosis inhibition activity as the wild type, the mutation of Lys90 seems to cause the loss of apoptosis inhibition activity observed for the double mutant D72A/K90A as for the single mutant K90A. These results may imply that Lys90 plays a role in apoptosis inhibition, presumably through protein–protein interaction in the apoptosis signaling pathways mentioned above.
Fig. 6.
Evaluation of APIP/MtnB surface mutants for the inhibition of cell death. (A) The conserved residues on the molecular surface are colored in blue as in Fig. S3. Outside the active site and the tetramric interface, two conserved patches are found: one patch consists of D72 and K90, and the other of Y184 and P185. (B) Caspase-1–induced cell death. (Left) HeLa cells were transiently cotransfected with pEGFP and the indicated constructs for 18 h. After staining with 0.5 µg/mL EtHD, cell death rates were measured by counting the number of both GFP- and EtHD-positive cells among total GFP-positive cells after staining with EtHD. Values indicate mean ± SD (n = 3). (Right) Whole-cell extract were prepared and subjected to Western blotting using the indicated antibodies. (C) Etoposide-induced cell death. (Left) HeLa cells were transiently cotransfected with pEGFP and the indicated constructs for 24 h, and then treated with 40 μM etoposide for 36 h. After staining with 0.5 μg/mL EtHD, cell death rates were measured by counting the number of both GFP- and EtHD-positive cells among total GFP-positive cells. Bars indicate mean ± SD (n = 3). (Right) Whole-cell extracts were prepared and subjected to Western blotting using the indicated antibodies.
The role of the methionine salvage pathway in metabolism is to recycle the sulfur-containing byproduct of the polyamine synthesis and convert it into one of the essential amino acids. Apart from its metabolic importance, there is a strong medical interest in the pathway stemming from findings that implicate the component enzymes (MTAP, MtnD/ADI1, MtnA/MRD1) and metabolites (MTA and KMTB/MTOB) in cancers and inflammatory diseases (2, 6, 9–11, 22). Human APIP/MtnB is another example of a methionine salvage enzyme that is implicated in these diseases. The involvement of APIP/MtnB in cell death and inflammation may indicate that it plays a role in the establishment of these related diseases; recent clinical studies reported that APIP expression is up-regulated in squamous carcinoma cells from tongue and larynx (13) or down-regulated in the cells and tumors of non–small-cell lung carcinoma (14). In addition, APIP is associated with inflammatory diseases, such as systemic inflammatory response syndrome and cystic fibrosis (2, 23). Thus, future studies of APIP/MtnB should aim to elucidate the detailed molecular mechanism of how APIP/MtnB modulates cell death and inflammation and contributes to the development of related diseases. Our current structural and biochemical characterization of APIP/MtnB lays the groundwork for those future studies.
Materials and Methods
Structure Determination.
Several constructs of APIP/MtnB were tested for expression in an Escherichia coli system, and then the best behaving soluble construct encompassing residues 20–242 was chosen for the crystallographic study. Details for the preparation of recombinant APIP/MtnB protein and the crystal samples were reported previously (24). Structure was determined at 2.4-Å resolution by multiwavelength anomalous diffraction (MAD) method using Br as an anomalous scatterer. For NaBr soaking, the native crystal of APIP/MtnB was transferred to the cryoprotecting solution containing 0.1 M NaBr in addition to the ingredients described previously (24) and then was put therein for 30 s before flash-cooling. MAD data were collected at three wavelengths of the peak, the inflection, and the high-energy remote. The SOLVE/RESOLVE software package was used for the location of anomalous scatterers, phase calculation, density modification, and initial model building (25). Subsequent manual model rebuilding and refinement was performed iteratively against the data at 2.0-Å resolution by using Coot (26), Refmac (24), and CNS (27). A Ramachandran plot for the final model was generated by Dynarama in Coot (26). Statistics for phasing and structure refinement are summarized in Table S1.
Site-Directed Mutagenesis.
Mutants of APIP/MtnB were generated by the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Expression and purification of the mutants were done following the same protocol as for the wild type (24).
Size Exclusion Chromatography Combined with MALS.
Size exclusion chromatography–MALS (SEC-MALS) measurement was performed on the AKTA basic system (GE Healthcare) connected to a miniDAWN TREOS (Wyatt Technology). A Superdex-200 HR 10/300 GL column (GE Healthcare) was used with the buffer of 20 mM Tris·HCl (pH 8.0), 0.1 mM TCEP, 100 mM NaCl, and 5% (vol/vol) glycerol at a flow rate of 0.5 mL/min. BSA (NEB) was used as reference to determine the detector delay volume and normalization coefficients.
Enzyme Assay.
Michaelis-Menten kinetics.
MtnB enzyme activity of APIP/MtnB was assayed by measuring the increase of absorbance at 280 nm for HK-MTPenyl-1-P produced in the reaction coupled with Bacillus MtnW. The substrate MTRu-1-P was prepared beforehand in 100 μL of the reaction mixture containing 50 mM Tris·HCl (pH 7.5), 1 mM MgCl2, 28 μg of Bacillus methylthioribose-1-phosphate isormerase (MtnA), and seven different concentrations of MTR-1-P. The exact MTRu-1-P concentration in the reaction mixture was calculated from the equilibrium constant between MTRu-1-P and MTR-1-P ([MTRu-1-P]/[MTR-1-P] = 6.0). MTR-1-P was synthesized as previously described (28, 29). The reaction was started by adding the limiting amount (0.3 μg in 0.2 μL) of the APIP/MtnB protein to the reaction mixture and was monitored at 280 nm with an HP 8453 UV-Visible Spectrophotometer (Hewlett Packard). The concentration of the product, HK-MTPenyl-1-P, was calculated from its molecular extinction coefficient, 9.5 × 103 M−1 cm−1 at 280 nm. Three independent measurements were performed.
Relative activity of mutants to the wild type.
The overall experimental procedure is the same as above, except that 0.72 μg of APIP/MtnB protein sample is added into the mixture for the start of the reaction. MTR-1-P was synthesized as previously described (30). The reaction was monitored at 280 nm with a DU 800 Spectrophotometer (Beckman Coulter). Three independent measurements were performed.
Preparation of the methionine salvage pathway enzymes.
Each gene for Bacillus subtilis MtnK, MtnA, MtnW, or human MTAP was amplified from the genomic DNA (ATCC) or the cDNA (Korea Human GenBank) by PCR and inserted into pET16b vector (Novagen) or its derivative for the bacterial expression in an N-terminally His-tagged form. Each protein is overexpressed in the Rosetta2 (DE3) strain of E. coli (Novagen), transformed with the corresponding expression vector, and purified through a His-tag affinity column and desalting column (GE Healthcare).
Docking in Silico.
A 2D structure of the substrate, MTRu-1-P, was drawn, and its 3D coordinates were produced in PDB format by the PRODRG Server (31). The coordinate were then converted into PDBQT format with united atom types and partial charges for each atom type by AutoDock4 (20). The protein model for docking was prepared by the removal of water molecules from the final refined coordinates of APIP. Then the polar hydrogen atoms were added and Kollman charges were assigned for all atoms in the model (20). A grid map of 50 Å × 50 Å × 50 Å with 0.503 Å grid spacing was generated around the active site zinc atom. The Lamarckian genetic algorithm was applied, and 30 individual runs were performed for a 150 population size with a 250,000 maximum number of energy evaluations (20).
Cell Death Assay.
Hypoxia induction, etoposide treatment, and cell viability measurement were done mainly in the same way as reported previously (1, 3). HeLa cells were transiently cotransfected with pEGFP and pcDNA3-HA-APIP/MtnB. For hypoxic cell death, the trypan blue exclusion assay was used for cell death rate measurement. In the case of caspase-1–induced or etoposide-induced cell death, the cell death rates were measured by counting the number of both GFP- and EtHD-positive cells.
Preparation of Figures for Structure and Sequence Alignment.
Structural figures were generated from the coordinates of the crystal structure and the substrate-docked model by using the PyMOL Molecular Graphics System (Schrödinger). Sequence alignment was performed using ClustalX2 (32) and edited by JalView (33).
Supplementary Material
Acknowledgments
We thank the staff of beamlines 5C and 7A of Pohang Accelerator Laboratory, Korea, and beamlines NW12 and 17A of Photon Factory, Japan, for help with data collection, and Dr. Hyun Kyu Song of Korea University for generously sharing the miniDAWN TREOS system. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by Ministry of Education, Science and Technology Grant 2009-0074396 (to J.K.Y.), Global Research Laboratory Grant NRF-2010-00341 (to Y.-K.J.), and by Grant A092006 of the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4M6R).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308768111/-/DCSupplemental.
References
- 1.Cho DH, et al. Induced inhibition of ischemic/hypoxic injury by APIP, a novel Apaf-1-interacting protein. J Biol Chem. 2004;279(38):39942–39950. doi: 10.1074/jbc.M405747200. [DOI] [PubMed] [Google Scholar]
- 2.Ko DC, et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc Natl Acad Sci USA. 2012;109(35):E2343–E2352. doi: 10.1073/pnas.1206701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho DH, et al. Suppression of hypoxic cell death by APIP-induced sustained activation of AKT and ERK1/2. Oncogene. 2007;26(19):2809–2814. doi: 10.1038/sj.onc.1210080. [DOI] [PubMed] [Google Scholar]
- 4.Mary C, et al. Functional identification of APIP as human mtnB, a key enzyme in the methionine salvage pathway. PLoS ONE. 2012;7(12):e52877. doi: 10.1371/journal.pone.0052877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirkov I, Norbeck J, Gustafsson L, Albers E. A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. FEBS J. 2008;275(16):4111–4120. doi: 10.1111/j.1742-4658.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- 6.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life. 2009;61(12):1132–1142. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- 7.Tang B, Kadariya Y, Murphy ME, Kruger WD. The methionine salvage pathway compound 4-methylthio-2-oxobutanate causes apoptosis independent of down-regulation of ornithine decarboxylase. Biochem Pharmacol. 2006;72(7):806–815. doi: 10.1016/j.bcp.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Avila MA, García-Trevijano ER, Lu SC, Corrales FJ, Mato JM. Methylthioadenosine. Int J Biochem Cell Biol. 2004;36(11):2125–2130. doi: 10.1016/j.biocel.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Li TW, et al. S-Adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol Pharmacol. 2009;76(1):192–200. doi: 10.1124/mol.108.054411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadariya Y, et al. Mice heterozygous for germ-line mutations in methylthioadenosine phosphorylase (MTAP) die prematurely of T-cell lymphoma. Cancer Res. 2009;69(14):5961–5969. doi: 10.1158/0008-5472.CAN-09-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oram SW, et al. Expression and function of the human androgen-responsive gene ADI1 in prostate cancer. Neoplasia. 2007;9(8):643–651. doi: 10.1593/neo.07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oram S, et al. Identification and characterization of an androgen-responsive gene encoding an aci-reductone dioxygenase-like protein in the rat prostate. Endocrinology. 2004;145(4):1933–1942. doi: 10.1210/en.2003-0947. [DOI] [PubMed] [Google Scholar]
- 13.Järvinen AK, et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer. 2008;47(6):500–509. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- 14.Moravcikova E, et al. Down-regulated expression of apoptosis-associated genes APIP and UACA in non-small cell lung carcinoma. Int J Oncol. 2012;40(6):2111–2121. doi: 10.3892/ijo.2012.1397. [DOI] [PubMed] [Google Scholar]
- 15.Dreyer MK, Schulz GE. Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure. J Mol Biol. 1996;259(3):458–466. doi: 10.1006/jmbi.1996.0332. [DOI] [PubMed] [Google Scholar]
- 16.Kroemer M, Merkel I, Schulz GE. Structure and catalytic mechanism of L-rhamnulose-1-phosphate aldolase. Biochemistry. 2003;42(36):10560–10568. doi: 10.1021/bi0349266. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, et al. The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization. Biochemistry. 2001;40(49):14763–14771. doi: 10.1021/bi0112513. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol. 1996;266:653–662. doi: 10.1016/s0076-6879(96)66041-8. [DOI] [PubMed] [Google Scholar]
- 19.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285(5):2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 20.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimble RF, Grimble GK. Immunonutrition: role of sulfur amino acids, related amino acids, and polyamines. Nutrition. 1998;14(7-8):605–610. doi: 10.1016/s0899-9007(98)80041-5. [DOI] [PubMed] [Google Scholar]
- 22.Kabuyama Y, et al. A mediator of Rho-dependent invasion moonlights as a methionine salvage enzyme. Mol Cell Proteomics. 2009;8(10):2308–2320. doi: 10.1074/mcp.M900178-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang W, Yang JK. Crystallization and preliminary X-ray crystallographic analysis of human Apaf-1-interacting protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 12):1518–1520. doi: 10.1107/S1744309112042832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Ashida H, Saito Y, Kojima C, Yokota A. Enzymatic characterization of 5-methylthioribulose-1-phosphate dehydratase of the methionine salvage pathway in Bacillus subtilis. Biosci Biotechnol Biochem. 2008;72(4):959–967. doi: 10.1271/bbb.70651. [DOI] [PubMed] [Google Scholar]
- 29.Ashida H, et al. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science. 2003;302(5643):286–290. doi: 10.1126/science.1086997. [DOI] [PubMed] [Google Scholar]
- 30.Della Ragione F, Cartenì-Farina M, Gragnaniello V, Schettino MI, Zappia V. Purification and characterization of 5′-deoxy-5′-methylthioadenosine phosphorylase from human placenta. J Biol Chem. 1986;261(26):12324–12329. [PubMed] [Google Scholar]
- 31.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 32.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 33.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.