Significance
Proper insulin signaling is necessary for regulating glucose metabolism and is often deregulated in cancer. Insulin activation of the phosphoinositide-3 kinase (PI3K) pathway is negatively regulated by the tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN). We previously described a novel inhibitor of PTEN activity, phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 (P-REX2). Here, we show that P-REX2 inhibition of PTEN occurs through the pleckstrin homology (PH) domain and requires PTEN C-terminal tail phosphorylation. Furthermore, loss of Prex2 results in reduced insulin signaling and increased Pten activity, and mice lacking P-rex2 display impaired glucose uptake and insulin resistance. This reveals a crucial role for the PH domain of P-REX2 in the regulation of glucose metabolism through the inhibition of PTEN.
Keywords: metabolism, diabetes
Abstract
Insulin activation of phosphoinositide 3-kinase (PI3K) signaling regulates glucose homeostasis through the production of phosphatidylinositol 3,4,5-trisphosphate (PIP3). The dual-specificity phosphatase and tensin homolog deleted on chromosome 10 (PTEN) blocks PI3K signaling by dephosphorylating PIP3, and is inhibited through its interaction with phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 (P-REX2). The mechanism of inhibition and its physiological significance are not known. Here, we report that P-REX2 interacts with PTEN via two interfaces. The pleckstrin homology (PH) domain of P-REX2 inhibits PTEN by interacting with the catalytic region of PTEN, and the inositol polyphosphate 4-phosphatase domain of P-REX2 provides high-affinity binding to the postsynaptic density-95/Discs large/zona occludens-1-binding domain of PTEN. P-REX2 inhibition of PTEN requires C-terminal phosphorylation of PTEN to release the P-REX2 PH domain from its neighboring diffuse B-cell lymphoma homology domain. Consistent with its function as a PTEN inhibitor, deletion of Prex2 in fibroblasts and mice results in increased Pten activity and decreased insulin signaling in liver and adipose tissue. Prex2 deletion also leads to reduced glucose uptake and insulin resistance. In human adipose tissue, P-REX2 protein expression is decreased and PTEN activity is increased in insulin-resistant human subjects. Taken together, these results indicate a functional role for P-REX2 PH-domain–mediated inhibition of PTEN in regulating insulin sensitivity and glucose homeostasis and suggest that loss of P-REX2 expression may cause insulin resistance.
Phosphatases are essential for the regulation of many signal transduction pathways, and altered phosphatase activity disrupts various cellular processes. Phosphatases are divided into two families, the serine (Ser)/threonine (Thr) phosphatases and the tyrosine (Tyr) phosphatases, which include the subfamily of dual-specificity phosphatases (1). Serine/threonine phosphatases are predominantly regulated by the formation of inhibitor complexes (2). Direct phosphorylation of both phosphatases and their inhibitors has also been implicated in serine/threonine phosphatase regulation (2). Protein tyrosine phosphatases (PTPs) are mainly regulated by reversible oxidation of the catalytic pocket (3). However, phosphorylation has also been implicated in their regulation (4).
The dual-specificity phosphatase and tensin homolog deleted from chromosome 10 (PTEN) was discovered through the mapping of homozygous deletions in cancer (5, 6). PTEN has the conserved PTP catalytic motif within its phosphatase domain (PD) and a C2 domain, both of which are required to dephosphorylate its primary substrate, phosphatidylinositol 3,4,5-trisphosphate (PIP3). This generates phosphatidylinositol 4,5-bisphosphate. thereby inhibiting PIP3-mediated recruitment and activation of the serine/threonine kinase AKT (7–9). Beyond these domains, the C-terminal tail of PTEN is phosphorylated at Ser-366, Ser-370, Ser-380, Thr-382, Thr-383, and Ser-385. High stoichiometry phosphorylation has been reported at Ser-370, -380. and -385 in vivo (10, 11). Furthermore, incorporation of 32P into PTEN during orthophosphate labeling in vivo is substantially reduced when the cluster of Ser-380, Thr-382, and Thr-383 are mutated to alanines. Phosphorylation at this cluster of three residues has been implicated in the regulation of PTEN stability and phosphatase activity (12). C-terminal tail phosphorylation is also required for the formation of an intramolecular interaction that occurs between the tail and the catalytic region of PTEN, which inhibits PTEN membrane recruitment and PIP3 access (13). In addition, the C terminus of PTEN contains a postsynaptic density-95/Discs large/zona occludens-1 (PDZ)-binding domain (PDZ-BD), providing a binding site for several PDZ-domain–containing proteins (14).
P-REX2A and P-REX2B were identified by two different groups through database searches for proteins homologous to P-REX1 (15, 16). P-REX2A is a guanine nucleotide exchange factor (GEF) for the small GTPase RAC and responds to both PIP3 and the beta-gamma subunits of G proteins (15). The structural domains of P-REX2A include the catalytic DHPH (Diffuse B-cell lymphoma homology and pleckstrin homology) domain tandem, two DEP (Disheveled, EGL-10, and pleckstrin homology) domains, two PDZ domains, and a C-terminal inositol polyphosphate-4 phosphatase (IP4P) domain. P-REX2B, a splice variant of P-REX2, lacks the C-terminal phosphatase domain. P-REX2 plays an important role in endothelial cell RAC1 activation and migration, as well as Purkinje cell dendrite morphology in the cerebellum (17, 18). Recently, we reported that P-REX2 interacts with PTEN to inhibit its phosphatase activity in a noncompetitive manner, thereby activating the phosphoinositide-3 kinase (PI3K) signaling pathway in cells (19). Here, we examine the mechanism by which P-REX2 inhibits PTEN and uncover that the PH domain of P-REX2 is responsible for inhibiting PTEN phosphatase activity. PH-domain–mediated inhibition is highly regulated by both the DH domain of P-REX2 and PTEN C-terminal tail phosphorylation. Furthermore, P-REX2 inhibition of PTEN plays a physiological role in the regulation of insulin-stimulated PI3K signaling and glucose metabolism.
Results
P-REX2 Inhibition of PTEN Is Regulated by Phosphorylation of the C-Terminal Tail of PTEN.
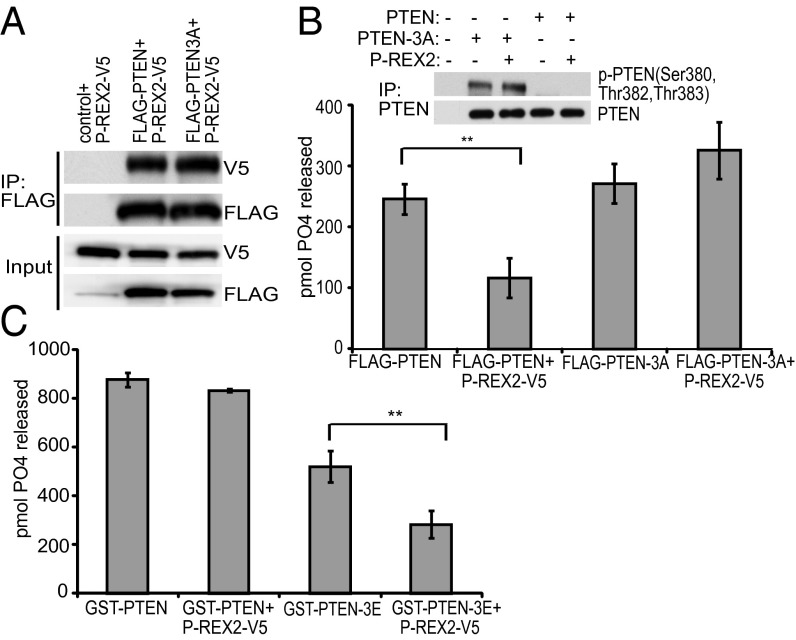
Due to the presence of critical phosphorylation sites located on the C-terminal tail of PTEN, we were interested in determining if phosphorylation affects P-REX2 binding. To this end, we transfected a FLAG-tagged PTEN mutant containing unphosphorylatable alanine substitutions at the phosphorylated cluster containing amino acids Ser-380, Thr-382, and Thr-383 (FLAG-PTEN-3A) or wild-type FLAG-PTEN into HEK293 cells with control vector or P-REX2-V5. The FLAG-PTEN/P-REX2-V5 complex was immunoprecipitated with anti-FLAG antibody. P-REX2 bound equally well to PTEN and PTEN-3A (Fig. 1A), suggesting that tail phosphorylation at these sites does not affect the physical interaction between P-REX2 and PTEN. To investigate whether phosphorylation of the tail regulates P-REX2-mediated PTEN inhibition, we transfected FLAG-PTEN or FLAG-PTEN-3A with or without P-REX2 into U87 cells, a PTEN-deficient glioblastoma cell line, and immunoprecipitated protein complexes with anti-PTEN antibody. PTEN phosphatase assays were performed by incubating immunoprecipitated PTEN with 20 µM di-C8-d-myo-PIP3 with a reaction endpoint of 30 min, and we observed that P-REX2 inhibited PTEN phosphatase activity by 53%, but did not inhibit PTEN-3A (Fig. 1B). Similar levels of PTEN inhibition were also observed after incubation with PIP3 for 15 min (Fig. S1A). C-terminal phosphorylated PTEN and total PTEN were present at equal levels in all samples, implying that P-REX2 expression does not interfere with PTEN C-terminal tail phosphorylation or immunoprecipitation (Fig. 1B).
Fig. 1.
P-REX2 inhibition of PTEN is regulated by PTEN tail phosphorylation. (A) HEK293 cell lysates coexpressing P-REX2 along with empty vector, FLAG-PTEN, or FLAG-PTEN-3A were incubated with FLAG-M2 antibody conjugated to agarose beads. Immunoprecipitated protein complexes were analyzed by immunoblotting. (B) U87 cell lysates coexpressing FLAG-PTEN or FLAG-PTEN-3A with empty vector or P-REX2-V5 were incubated with a rabbit monoclonal PTEN antibody conjugated to agarose beads. Phosphatase assays were performed on immunoprecipitated protein complexes by incubating protein-bound beads with 20 µM soluble di-C8-d-myo-PIP3 at 37 °C for 30 min. Levels of free phosphate (PO4) released are shown. ±SD, n = 3. Representative levels of immunoprecipitated PTEN are shown. (C) In vitro phosphatase assays were performed in triplicate by incubating equimolar amounts of purified GST-PTEN WT or GST-PTEN-3E and purified P-REX2-V5 in the presence of PIP3 at 37 °C for 30 min. Levels of free phosphate are shown from a representative experiment repeated three times. ±SD, **P < 0.01.
To verify that PTEN tail phosphorylation is required for P-REX2 inhibition, we performed phosphatase assays on purified bacterially produced, and therefore unphosphorylated, recombinant GST-PTEN and a partial phosphorylation mimicking mutant containing glutamic acid substitutions at amino acids Ser-380, Thr-382, and Thr-383 (GST-PTEN-3E) in the presence of P-REX2. In accordance with our previous results, P-REX2-V5 inhibited GST-PTEN-3E phosphatase activity by 46%, but did not inhibit GST-PTEN at a reaction endpoint of 30 min (Fig. 1C). GST-PTEN-3E was also sensitive to inhibition by P-REX2 after 15 min and 5 min of incubation with PIP3 (Fig. S1A). Adding decreasing molar amounts of P-REX2 to the reaction resulted in stoichiometric inhibition of PTEN without altering PTEN solubility (Fig. S1 B and C). Pull-down experiments using lysates from HEK293 cells overexpressing P-REX2 showed that P-REX2 bound with high stoichiometry to both GST-PTEN and GST-PTEN-3E (Fig. S1D). Therefore, although P-REX2 interacts with high stoichiometry to both phosphorylated and unphosphoryated PTEN, it can recognize the phosphorylation status of PTEN and inhibit only when the C-terminal tail of PTEN is phosphorylated.
To examine if P-REX2 affects PTEN activity by altering its C-terminal tail phosphorylation, we overexpressed PTEN alone or in combination with P-REX2 in U87 cells. Membrane and cytosolic fractions were isolated from starved and insulin stimulated cells, and PTEN C-terminal phosphorylation was analyzed by Western blot. P-REX2 expression did not appear to increase the phosphorylation of Ser-380 and Thr-382 and Thr-383 in either the membrane or cytosolic fraction (Fig. S1E).
P-REX2 Binds to Multiple Sites on PTEN.
Because the PTEN tail regulates P-REX2 inhibition of PTEN as well as P-REX2 binding (19), we sought to understand the P-REX2-PTEN tail interaction in more detail. We transfected HEK293 cells with P-REX2 along with a series of N-terminal FLAG-tagged PTEN constructs containing stepwise deletions in the tail of PTEN, including a two-amino-acid deletion of the PDZ-BD, in HEK293 cells (Fig S2A). Coimmunoprecipitation experiments revealed that a two-amino-acid deletion at the C terminus of the PDZ-BD [FLAG-PTEN(1–401)] led to a major reduction of P-REX2 binding (Fig. S3A). P-REX2, however, still bound to FLAG-PDC2, a mutant containing the phosphatase domain and C2 domain (amino acids 1–352, Fig S3B), validating that P-REX2 binds to the phosphatase-C2 region of PTEN. Overall, these results demonstrate that P-REX2 binds to the PDZ-BD and also interacts with a region of PTEN located outside of the PTEN tail.
The Inositol Polyphosphate 4-Phosphatase Domain of P-REX2 Docks to the PDZ-BD of PTEN.
To determine how P-REX2 interacts with the PDZ-BD, we performed pull-down experiments with GST-TAIL, which only contains the C-terminal tail region (amino acids 353–403), or GST-PDC2 (Fig S2 A and B). We found that P-REX2ΔDHPH, a mutant lacking the DHPH domain, had strong affinity for the PTEN tail but not the phosphatase or C2 domains (Fig. S3C). To confirm this interaction, P-REX2ΔDHPH was expressed in HEK293 cells with FLAG-tagged C2TAIL or C2TAIL402STOP and complexes were immunoprecipitated with anti-FLAG antibody. P-REX2ΔDHPH bound to C2TAIL, and this interaction was dependent on the PDZ-BD of PTEN (Fig. S3D).
To further map this interaction, we performed immunoprecipitation experiments using the P-REX2 C-terminal IP4P or the DEP-PDZ domain tandem (DEPPDZ) with C2TAIL or C2TAIL402STOP. Surprisingly, we found that it was the IP4P domain that interacted with the PDZ-BD domain (Fig. S3E). We validated this result by showing that P-REX2ΔIP4P, a P-REX2 mutant lacking the inositol phosphatase domain, coimmunoprecipitated equally with C2TAIL and C2TAIL402STOP (Fig. S3F). Furthermore, binding of P-REX2ΔIP4P to PTEN was reduced compared with full-length P-REX2, likely due to the lack of a PDZ-BD interaction.
The PH Domain of P-REX2 Interacts with the Phosphatase and C2 Domains of PTEN.
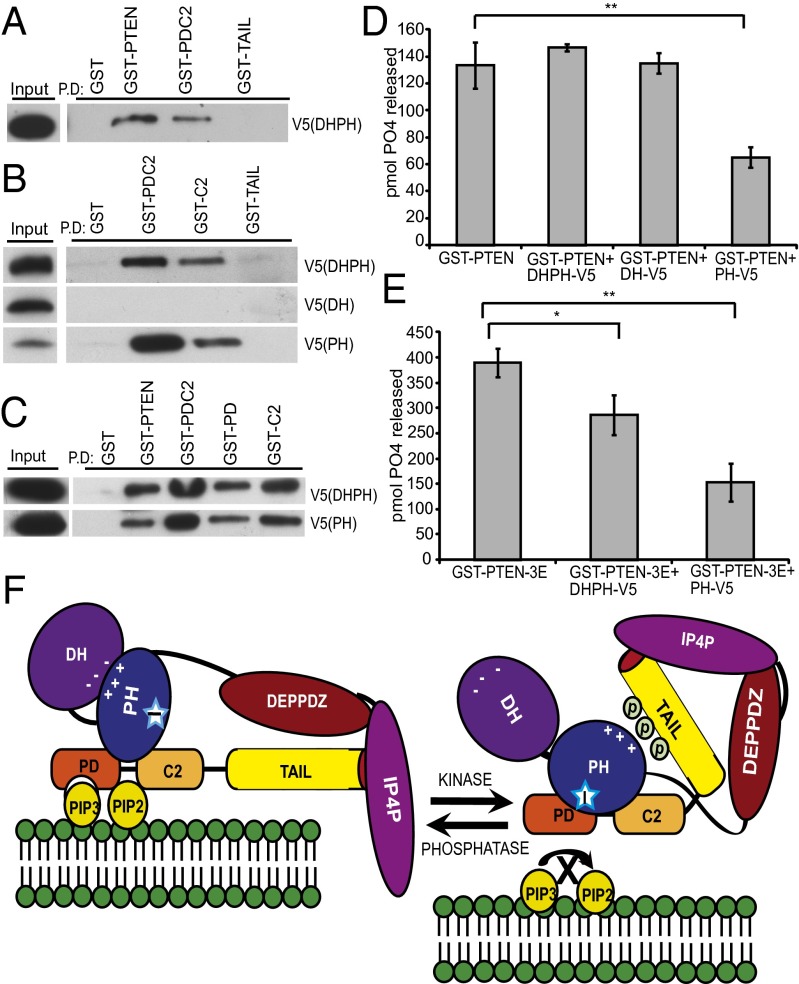
Prior work showed that the GST-PTEN could bind to in vitro translated DHPH (19). To further investigate this, we performed pull-down experiments using GST-PTEN deletion mutants. DHPH-V5 expressed in HEK293 cells bound to GST-PTEN and GST-PDC2, but not to GST-TAIL (Fig. 2A), demonstrating that it binds in a different manner from the IP4P domain. Further analysis revealed that in vitro translated DHPH and PH domains bound to GST-PDC2 and GST-C2 (C2 domain), whereas the DH domain did not bind to any region of PTEN (Fig. 2B). These results suggest that the PH domain alone is necessary for P-REX2 binding to the phosphatase and C2 domains of PTEN. We confirmed this interaction by coimmunoprecipitation of PH-V5 or DHPH-V5 with FLAG-PDC2 expressed in cells (Fig. S4 A and B). To analyze PH domain binding in more detail, we performed pull-down assays with GST-PD and GST-C2. The DHPH and PH domains bound strongly to both GST-PD and GST-C2 (Fig. 2C). Therefore, the PH domain interfaces with both the phosphatase and C2 domains, which together make up the minimal catalytic region of PTEN.
Fig. 2.
P-REX2 PH domain interacts with and inhibits the catalytic domain of PTEN. (A) HEK293 cell lysates coexpressing DHPH-V5 were incubated with recombinant GST-PTEN, GST-PDC2, GST-TAIL, or GST control Sepharose beads. Pulled-down protein complexes were analyzed by immunoblotting. (B and C) PH-V5, DH-V5, or DHPH-V5 were in vitro translated and 10 µL of protein lysate was incubated with GST control, GST-PTEN, GST-PDC2, GST-PD, or GST-C2 Sepharose beads. Pulled-down protein complexes were analyzed by immunoblotting. (D and E) In vitro phosphatase assays were performed in triplicate by incubating equimolar amounts of GST-PTEN WT or GST-PTEN mutants with purified V5-tagged P-REX2 domains as indicated in the presence of PIP3. Levels of free phosphate are shown from representative experiments repeated at least twice. ±SD, **P < 0.05 and *P < 0.01. (F) Model of PTEN inhibition by P-REX2. The DH domain of P-REX2 restricts PH domain inhibition of PTEN phosphatase activity when the C-terminal tail of PTEN is unphosphorylated. However, phosphorylation of the tail is “read” by the DH domain, switching the ionic interaction to the phospho-tail and unleashing the PH domain inhibitory region (stars) of P-REX2 through interaction with the phosphatase and C2 domains of PTEN on its nonlipid-binding surface.
The PH Domain of P-REX2 Constitutively Inhibits PTEN Phosphatase Activity in the Absence of the DH Domain.
Because the PH domain binds to the phosphatase and C2 domains of PTEN, we asked whether it could inhibit PTEN without C-terminal tail regulation. We performed phosphatase assays with recombinant GST-PTEN protein and purified V5-tagged P-REX2 domains and found that PH-V5 inhibited GST-PTEN phosphatase activity by 51%, whereas the DH domain and surprisingly, the DHPH domain did not (Fig. 2D). This indicates that the PH domain can inhibit PTEN regardless of its phosphorylation state. We suspected that phosphorylation of the PTEN tail may be regulating DHPH-domain–mediated PTEN inhibition, and therefore performed phosphatase assays using the GST-PTEN-3E mutant. DHPH-V5 and PH-V5 were capable of inhibiting GST-PTEN-3E by 27% and 61%, respectively (Fig. 2E). Our results suggest that the DH domain of P-REX2 blocks PH domain inhibition of PTEN, and also “reads” PTEN tail phosphorylation, which unleashes the PH domain and allows for PTEN inhibition (Fig. 2F).
The PH Domain of P-REX2 Inhibits PTEN Activity in Cells.
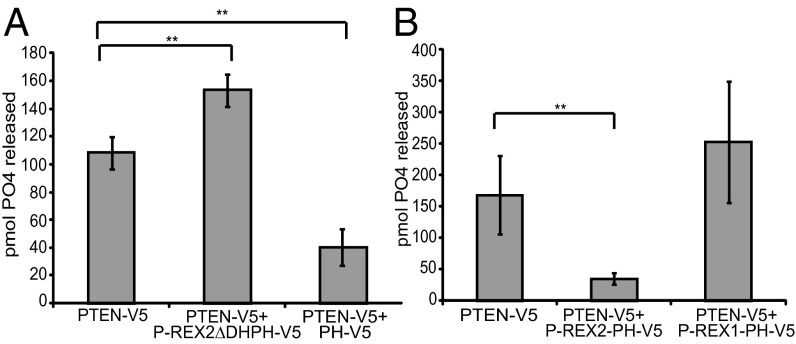
To determine if PH domain inhibition of PTEN could be measured from cell extracts, we performed phosphatase assays on immunoprecipitated PTEN-V5 expressed in HEK293 cells with PH-V5 or P-REX2ΔDHPH-V5. We used a shared V5 tag to immunoprecipitate protein complexes due to the weak interaction between PTEN and the PH domain. The PH domain successfully inhibited eukaryotic PTEN activity by 63%, whereas P-REX2ΔDHPH led to a 40% increase in PTEN activity (Fig. 3A and Fig. S4C). These results demonstrate that the PH domain can inhibit PTEN produced from mammalian cells, and P-REX2ΔDHPH may act as a dominant negative mutant by blocking the formation of the wild-type inhibitory complex.
Fig. 3.
The PH domain of P-REX2, but not P-REX1, inhibits PTEN produced in mammalian cells. (A) Lysates from HEK293 cells coexpressing PTEN-V5 with empty vector, PH-V5, or P-REX2ΔDHPH-V5 were incubated with V5-agarose beads. Phosphatase assays were then performed on the immunoprecipitated protein. Levels of free phosphate are shown. ±SD, n = 3, **P < 0.01. (B) Lysates from HEK293 cells expressing PTEN-V5 with empty vector, P-REX2-PH-V5, or P-REX1-PH-V5 were incubated with V5-agarose beads, and phosphatase assays were performed on immunoprecipitated proteins. Levels of free phosphate released are shown. ±SD, n = 3, **P < 0.01.
Previous work has shown that the DHPH domain could activate PI3K signaling in cells via PTEN (19). Therefore, we next examined the ability of the PH domain to rescue PI3K signaling ablated by PTEN expression. Expression of PTEN in U87 cells resulted in decreased phosphorylation of AKT at Thr-308, and expression of either the DHPH or PH domain with PTEN rescued AKT phosphorylation (Fig. S4D). Furthermore, coexpression of P-REX2ΔDHPH with PTEN did rescue phosphorylated AKT levels, but rather resulted in a greater decrease in AKT phosphorylation compared with PTEN alone (Fig. S4E). Next, we determined if inhibition of PTEN activity was specific to the PH domain of P-REX2. Phosphatase assays performed on PTEN-V5 coimmunoprecipitated with P-REX2-PH-V5 or P-REX1-PH-V5, the closest homolog to P-REX2, revealed that the PH domain of P-REX2 inhibited PTEN phosphatase activity, but the PH domain of P-REX1 did not (Fig. 3B and Fig. S5A). Furthermore, P-REX1 did not coimmunoprecipitate with PTEN when coexpressed in HEK293 cells (Fig. S5B).
Insulin- and IGF1-Stimulated PI3K Signaling Is Decreased in Prex2−/− Mouse Embryonic Fibroblasts.
We have shown that inhibition of PTEN activity by both purified P-REX2 and P-REX2 overexpressed in cells is complex and tightly regulated. However, the physiological significance of P-REX2–mediated PTEN inhibition remains to be determined. To examine this, we generated Prex2 knockout mice using the ES cell clone AH0440 purchased from the Sanger Institute Gene Trap Resource. This clone contains a gene trap cassette consisting of a splice acceptor site, a β-galactosidase/neomycin phosphotransferase II fusion ending with a stop codon, and a polyadenylation sequence. This cassette was inserted between exons 4 and 5 of Prex2, which encodes the DH domain of P-rex2 (Fig. S6 A and B). The insertion of this reporter gene also results in a deletion of exons 5 and 6, as shown by PCR of genomic DNA from ES cells (Fig. S6C). The mutant allele was transmitted through the germ line, and heterozygous mutant mice were intercrossed to generate Prex2−/− mice, which were then backcrossed with C57BL/6 mice for eight generations. PCR genotyping is shown (Fig. S6A). We confirmed loss of P-rex2 protein in various tissue samples by Western blot analysis (Fig. S6D). Two different isoforms of P-rex2 are expressed in different tissue types: P-rex2A that runs at 180 kDa, and a shorter isoform that runs at 120 kDa that we will call P-rex2C (Fig. S6D), as predicted by GenBank entry AK138884. This isoform, which is missing most of the IP4P domain, is not the previously described P-rex2B isoform (16), as it does not contain the epitope within the IP4P domain recognized by our antibody. The brain and liver express both P-rex2A and P-rex2C, whereas the lung expresses only P-rex2A, and adipose tissue only expresses P-rex2C. P-rex2 is not detectable in skeletal muscle when using our antibody (Fig. S6D). Alternative splicing may result in a transcript spliced directly from exon 4 to exon 7, bypassing the gene-trap vector. However, the absence of a protein band at the predicted size of 170 kDa suggests that this is likely not occurring (Fig. S6D). Furthermore, due to the loss of exons 5 and 6, which encode a portion of the DH domain, this deleted protein would be expected to be nonfunctional. Prex2−/− mice were fertile and appeared healthy.
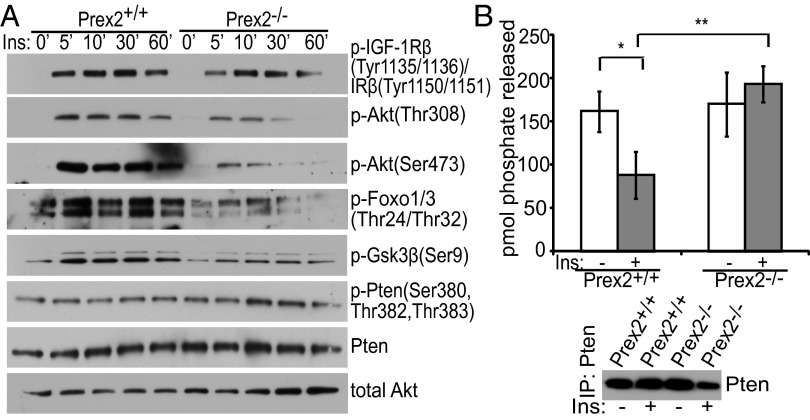
To determine the effect of Prex2 loss on signaling and Pten activity, we first generated Prex2+/+ and Prex2−/− mouse embryonic fibroblasts (MEFs). We confirmed that P-rex2 was expressed in Prex2+/+ cells (Fig. S7A) and found that growth of Prex2−/− MEFs was not impaired under normal growth conditions (Fig. S7B). To examine the effect of Prex2 loss on growth-factor–stimulated PI3K signaling, we assessed levels of phosphorylated Akt after stimulation with 10 µg/mL insulin, 20 ng/mL insulin-like growth factor 1 (IGF1), 20 ng/mL platelet-derived growth factor (PDGF), or 20 ng/mL epidermal growth factor (EGF) at various time points (Fig. 4A and Fig. S7C). In response to insulin, the level and duration of Akt phosphorylation at Thr-308 and Ser-473 was reduced in Prex2−/− MEFs (Fig. 4A). Phosphorylation of downstream Akt targets including forkhead box protein O1 and 3 (Foxo1/3) and glycogen synthase kinase 3β (Gsk3β) were also decreased. No decrease in insulin receptor β (IRβ) activation was observed in Prex2−/− MEFs, suggesting that PI3K signaling is being affected downstream of insulin receptor activation (Fig. 4A). Levels of C-terminally phosphorylated Pten and total Pten levels were not significantly altered in Prex2−/− MEFs (Fig. 4A). Prex2 loss also resulted in reduced IGF1-stimulated phosphorylation of Akt, Foxo1/3a and Gsk3β, but had no effect on PI3K signaling stimulated by EGF or PDGF (Fig. S7C). To further understand this growth factor specificity, we examined the affinity of P-REX2 for activated membrane receptors. Transfected P-REX2-V5 was immunoprecipitated from HEK293 cells stimulated with insulin or EGF. We found that P-REX2 interacted with phosphorylated IR after insulin stimulation, but showed no affinity for phosphorylated EGFR following EGF stimulation (Fig. S7D), suggesting that P-REX2 can associate with activated insulin receptor at the membrane following stimulation.
Fig. 4.
Prex2 loss affects PI3K signaling and Pten activity in fibroblasts. (A) Prex2+/+ and Prex2−/− MEFs were starved for 3 h and then stimulated with insulin (Ins, 10 μg/mL). Lysates were collected at the indicated time points, and levels of phosphorylated Akt and downstream targets were analyzed by Western blot. (B) Lysates from starved or insulin-stimulated Prex2+/+ and Prex2−/− MEFs were incubated with PTEN antibody conjugated to agarose beads overnight. Phosphatase assays were performed on immunoprecipitated protein complexes. Levels of free phosphate released are shown. ±SD, n = 3, *P < 0.05, **P < 0.01. Representative Pten immunoprecipitations are shown.
Pten Regulation by Insulin Stimulation Is Dependent on P-rex2.
To determine if reduced insulin signaling in the absence of P-rex2 is a result of increased Pten activity, we performed phosphatase assays on Pten immunoprecipitated from Prex2+/+ or Prex2−/− MEFs. Pten activity was equal between genotypes when cells were starved. However, following insulin stimulation, Pten activity in Prex2+/+ MEFs was reduced by 45.3%, but remained elevated in Prex2−/− MEFs (Fig. 4B). In concordance with this, we observed an endogenous interaction between P-rex2 and Pten in immortalized Prex2+/+ fibroblasts that appears to increase slightly upon insulin stimulation (Fig. S7E).
Prex2 Deletion Results in Decreased Insulin-Stimulated PI3K Signaling and Increased Pten Activity in Vivo.
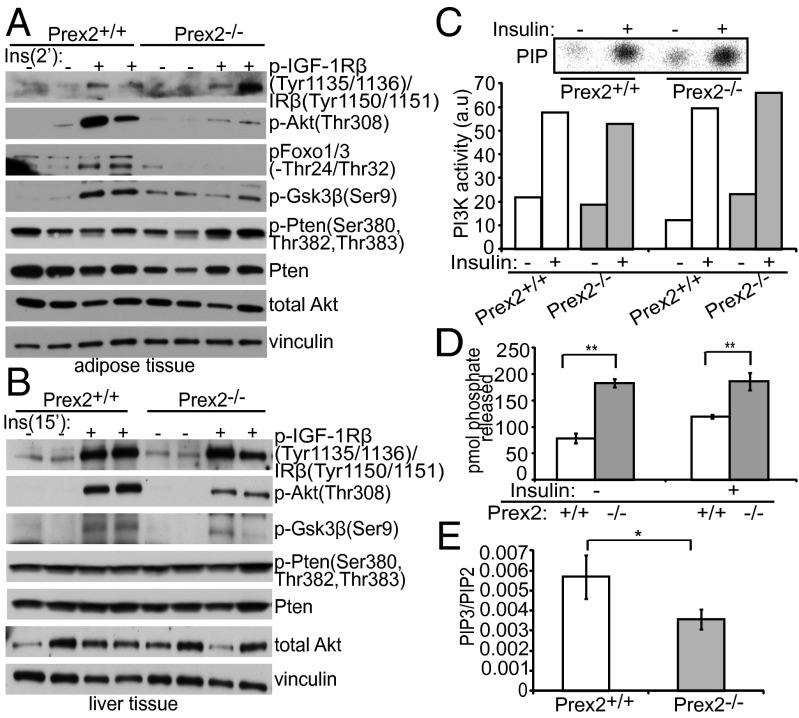
To determine if P-rex2 regulates insulin signaling in vivo, we measured signaling outputs in mouse adipose and liver tissue, which are highly responsive to insulin. We stimulated mice with insulin by i.p. injection and collected tissue samples postinjection. Whereas no differences in IRβ activation were observed, Prex2−/− adipose tissue had decreased Akt phosphorylation at Thr-308, as well as decreased phosphorylation of downstream Akt targets including phosphorylated Foxo1/3a and phosphorylated Gsk3β compared with Prex2+/+ tissue following 2 min of insulin stimulation (Fig. 5A). Prex2 loss in liver tissue also resulted in reduced Akt and Gsk3β phosphorylation after 15 min (Fig. 5B) and 2 min (Fig. S8A) of insulin stimulation, whereas levels of activated IRβ were comparable. Levels of total Pten in both liver and adipose tissue were not affected by P-rex2 loss; however there appeared to be an induction of Pten C-terminal tail phosphorylation in Prex2−/− adipose tissue following insulin stimulation (Fig. 5 A and B). To determine if P-rex2 deletion alters PI3K activity downstream of insulin receptor activation, we performed PI3K assays on immunoprecipitated PI3K from starved and stimulated liver samples using anti–phospho-Tyr antibody. PI3K activity was equivalent in Prex2+/+ and Prex2−/− liver samples (Fig. 5C), suggesting that P-rex2 regulation of insulin signaling occurs downstream of PI3K.
Fig. 5.
Prex2 loss affects PI3K signaling and Pten activity in vivo. (A) Prex2+/+ and Prex2−/− mice were fasted for 16 h and injected intraperitoneally with insulin (Ins) (0.75 mU/g) for 2 min. Levels of phosphorylated Akt, Foxo1/3, Gsk3-β, and IR in collected adipose tissue lysates were analyzed by Western blot. (B) Prex2+/+ and Prex2−/− mice were fasted for 16 h and injected intraperitoneally with insulin (0.75 mU/g) for 15 min. Levels of phosphorylated Akt, Foxo1/3, Gsk3-β, and IRβ in collected liver tissue lysates were analyzed by Western blot. (C) PI3K activity for phosphatidylinositol phosphate substrates was determined for Prex2+/+ and Prex2−/− liver samples during starvation and stimulation with insulin (10 mU/g) for 8 min. Lysates were incubated with anti–phospho-Tyr antibody. A representative autoradiograph is shown, and PI3K activity from two independent experiments are quantified and presented as arbitrary units (a.u. in this figure). (D) Prex2+/+ and Prex2−/− mice were fasted overnight and then injected with insulin (10 mU/g) for 8 min. Lysates from three starved and three stimulated liver samples per genotype were incubated with PTEN antibody conjugated to agarose beads overnight, and phosphatase assays were performed in triplicate on immunoprecipitated protein complexes. Levels of free phosphate released are shown. ±SEM, **P < 0.01. (E) Liver samples collected for the above phosphatase assays were used to compare PIP3 levels from insulin-stimulated mice. Lipids were measured using mass spectrometry and PIP3 values are presented as a ratio of PIP3/PIP2 to account for cell number variability. ±SD, (Prex2+/+, n = 4; Prex2−/−, n = 3), *P < 0.05.
To determine if P-rex2 can regulate Pten activity in vivo, we measured the phosphatase activity of Pten immunoprecipitated from Prex2+/+ and Prex2−/− liver lysates. Pten activity from Prex2−/− livers was 52% and 38% higher than Prex2+/+ livers during fasted and stimulated conditions, respectively (Fig. 5D and Fig. S8B). In addition, an endogenous interaction between Pten and P-rex2 was observed in Prex2+/+ liver lysates (Fig. S8C). We also quantified PIP3 levels from insulin-stimulated liver lysates using high performance liquid chromatography–mass spectrometry. Prex2−/− liver had 37.4% less PIP3 compared with Prex2+/+ liver when normalized to PIP2 (Fig. 5E), supporting our finding that Pten in the liver is less active in the presence of P-rex2.
Deletion of Prex2 Leads to Decreased Glucose Uptake and Insulin Resistance in Vivo.
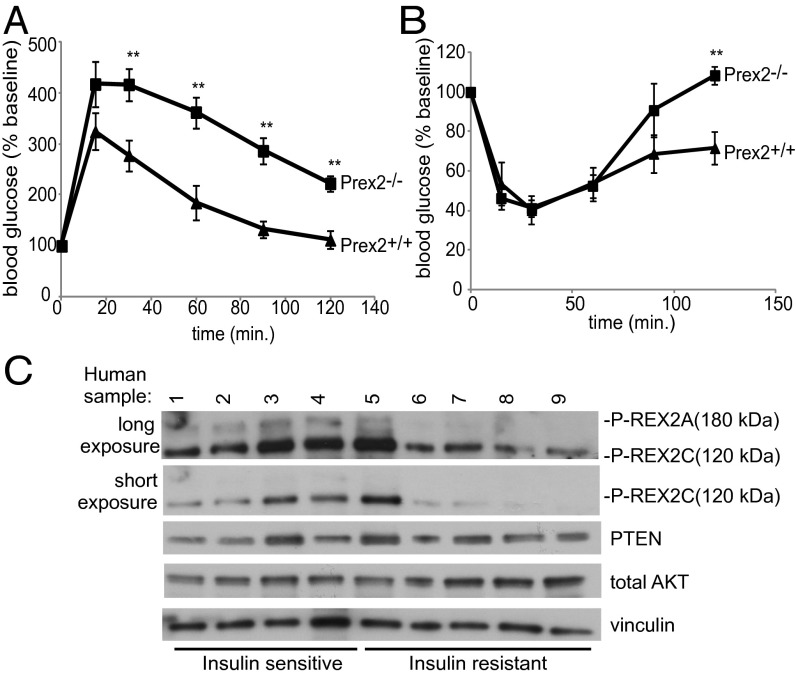
Insulin signaling tightly regulates glucose homeostasis; therefore, we examined whether Prex2−/− mice were defective for glucose uptake by performing glucose challenge tests. Six-week-old Prex2+/+ and Prex2−/− fasted male mice were injected intraperitoneally with a bolus of dextrose, and blood glucose levels were recorded over time and normalized to baseline levels. Whereas there was no difference in baseline glucose, Prex2−/− mice exhibited 22% higher peak in blood glucose levels compared with Prex2+/+ mice 15 min after injection (Fig. 6A). Blood glucose levels of Prex2−/− mice remained elevated throughout the time course, as quantified by area under the curve (P = 0.002, Fig. 6A). Insulin tolerance tests were also performed by injecting 8-wk-old male mice intraperitoneally with insulin and measuring blood glucose levels over time. Blood glucose levels were similar at early time points; however, 120 min postinjection Prex2+/+ blood glucose levels were 29.6% below baseline, whereas blood glucose levels in Prex2−/− mice were 8.4% above baseline (Fig. 6B). These data suggest that reduced insulin signaling in the absence of Prex2 leads to glucose intolerance and decreased insulin response.
Fig. 6.
P-rex2 regulates glucose uptake and insulin sensitivity in vivo. (A) Glucose challenge tests (GCTs) were performed by measuring blood glucose levels following i.p. glucose injection at 2 mg dextrose/g body weight at the indicated time points (n = 6). (B) Insulin tolerance tests (ITTs) were performed by measuring blood glucose levels following i.p. injection of insulin (0.75 mU/g) at the indicated time points (Prex2+/+, n = 4; Prex2−/−, n = 5). For both GCT and ITT tests, blood glucose levels are shown normalized to baseline. ±SEM, **P < 0.01. (C) Lysates from human adipose tissue samples were prepared and levels of P-REX2 and PTEN were analyzed by Western blot.
Having shown that P-rex2 has a role in regulating insulin signaling in mice, we next wanted to examine if P-REX2 protein expression was altered in insulin-resistant human subjects. We obtained adipose tissue from human subjects with clinically determined insulin sensitivities (Table S1) and prepared protein lysates. Western blot analysis showed expression of both the 180-kDa P-REX2A and the 120-kDa P-REX2C isoforms, which is predicted to exist by UniProt Knowledge Base entry B4DFX0_HUMAN. P-REX2A and P-REX2C protein levels in adipose tissue appeared lower in insulin-resistant subjects compared to insulin-sensitive subjects, with P-REX2A expression being significantly decreased (P = 0.019, Fig. 6C). Furthermore, the activity of PTEN immunoprecipitated from adipose tissue was 40% higher in insulin-resistant subjects compared with insulin-sensitive subjects (Fig. S8 D and E). This provides evidence of P-REX2–regulated insulin signaling not only in mice, but also in humans.
Discussion
Here we present a unique mechanism of GEF-mediated phosphatase inhibition. In summary, P-REX2 docks to the PDZ-BD of PTEN through its C-terminal domain, reads the phosphorylation state of the PTEN tail via the DH domain, and inhibits PTEN activity by unleashing the PH domain (Fig. 2F). We hypothesize that negatively charged residues on the DH domain interact with positive charges on the PH domain, blocking the phosphatase-inhibitory region of the PH domain (Fig. 2F, stars) from inhibiting PTEN. However, phosphorylation of the C-terminal tail of PTEN creates a cluster of negative charges that could compete with the DH domain for binding to positive charges on the PH domain, unleashing the PH domain for allosteric inhibition of PTEN catalysis. Interestingly, the formation of intramolecular interactions between the DH and PH domains of other GEFs, including Vav1 and Sos1, has been previously implicated in the regulation of GEF activation of GTPases (20). It is likely that there is a balance in PTEN phosphorylation states, ranging from fully phosphorylated to completely unphosphorylated. Whereas our results are contingent on some measure of tail phosphorylation, it is possible that fully phosphorylated PTEN may be inactive and not regulated in the same fashion. Docking of the IP4P domain of P-REX2 to the PDZ-BD of PTEN likely ensures that the PH domain is positioned in close proximity to PTEN to inhibit phosphatase activity. However, this interaction does not appear to be required for PTEN inhibition, as P-REX2C, which is missing the IP4P domain, is able to regulate PTEN activity and insulin signaling in fat.
The above mechanism of PTEN regulation is provocative because it reveals a unique function for a PH domain. The PH domain is highly represented in the human genome, displays a diverse range of amino acid sequences, and is found in proteins of various functions (21). Therefore, phosphatase inhibition by a PH domain may be broadly relevant to signaling pathway regulation. On the other hand, the sequence specificity of the P-REX2 PH domain may be uniquely adapted to inhibition of PTEN. Our results show that the PH domain of P-REX1, a close homolog of P-REX2 known to bind PIP3, does not inhibit PTEN phosphatase activity, suggesting that a specific PH domain may only be able to regulate the activity of a specific phosphatase, similar to GEF regulation of GTPases. Although P-REX1 did not inhibit PTEN, we cannot eliminate the possibility that there are other PH domains that share the ability to inhibit PTEN. In addition, other PH domains could regulate different phosphatases, tightly controlling various signaling pathways and cellular processes.
Type II noninsulin-dependent diabetes mellitus is a common metabolic disease caused by the inability of liver, muscle, and fat to respond to normal levels of insulin as well as the lack of insulin production by pancreatic β cells to compensate for this insulin resistance (22). Previous studies have shown that PTEN is a critical regulator of insulin signaling, as loss of PTEN in tissues such as liver, fat, and muscle results in increased insulin sensitivity and glucose metabolism and protection from diabetes (23–25). It was therefore not surprising that P-REX2 PH domain inhibition of PTEN is important for the regulation of insulin signaling and the maintenance of glucose homeostasis in vivo. In addition to inhibiting PTEN activity, it is possible that P-REX2 activation of Rac1 may also regulate insulin signaling through several mechanisms including direct activation of p110β (26). Interestingly, decreased P-REX2 protein expression found in adipose tissue from human subjects suggests that deregulated P-REX2 expression could cause insulin resistance in people. Restoration of P-REX2 levels or inhibition of PTEN may prove to be successful therapies for cases of insulin resistance and type II diabetes that show evidence of reduced P-REX2 expression and elevated PTEN phosphatase activity.
Materials and Methods
Mice deleted for Prex2 were generated using the embryonic stem cell line AH0440 purchased from the Wellcome Trust Sanger Institute. This line was created by high throughput gene trapping, resulting in the insertion of a reporter gene containing a β-galactosidase and neomycin phosphotransferase II fusion between exons 4 and 5 of Prex2. The location of the reporter gene was confirmed by RT-PCR. More detailed methods on immunoblotting, coimmunoprecipitation, protein purification, phosphatase assays, and in vivo metabolic studies can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Barry Fine for guidance with experimental protocols and laboratory members for editing, and we acknowledge Agencourt Biosciences for the sequencing of our plasmids. This work was supported by the National Institutes of Health Grants R01 CA155117 (to R.P.), R01 CA082783 (to R.P.), R01 DK71349 (to P.A.K.), UL1 TR000117 (to P.A.K.), T32 CA009503-22 (to C.H.), and GM008224-20 (to C.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213773111/-/DCSupplemental.
References
- 1.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 2.Barr FA, Elliott PR, Gruneberg U. Protein phosphatases and the regulation of mitosis. J Cell Sci. 2011;124(Pt 14):2323–2334. doi: 10.1242/jcs.087106. [DOI] [PubMed] [Google Scholar]
- 3.Ostman A, Frijhoff J, Sandin A, Böhmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem. 2011;150(4):345–356. doi: 10.1093/jb/mvr104. [DOI] [PubMed] [Google Scholar]
- 4.Böhmer F, Szedlacsek S, Tabernero L, Ostman A, den Hertog J. Protein tyrosine phosphatase structure-function relationships in regulation and pathogenesis. FEBS J. 2013;280(2):413–431. doi: 10.1111/j.1742-4658.2012.08655.x. [DOI] [PubMed] [Google Scholar]
- 5.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 7.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 10.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528(1-3):145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 11.Birle D, et al. Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide-induced serine phosphorylation. J Immunol. 2002;169(1):286–291. doi: 10.4049/jimmunol.169.1.286. [DOI] [PubMed] [Google Scholar]
- 12.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20(14):5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahdar M, et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA. 2009;106(2):480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim RH, Mak TW. Tumours and tremors: How PTEN regulation underlies both. Br J Cancer. 2006;94(5):620–624. doi: 10.1038/sj.bjc.6602994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeldt H, Vázquez-Prado J, Gutkind JS. P-REX2, a novel PI-3-kinase sensitive Rac exchange factor. FEBS Lett. 2004;572(1-3):167–171. doi: 10.1016/j.febslet.2004.06.097. [DOI] [PubMed] [Google Scholar]
- 16.Donald S, et al. P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 2004;572(1–3):172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Paik JH, Wang Z, Hla T, Wu D. Role of guanine nucleotide exchange factor P-Rex-2b in sphingosine 1-phosphate-induced Rac1 activation and cell migration in endothelial cells. Prostaglandins Other Lipid Mediat. 2005;76(1-4):95–104. doi: 10.1016/j.prostaglandins.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Donald S, et al. P-Rex2 regulates Purkinje cell dendrite morphology and motor coordination. Proc Natl Acad Sci USA. 2008;105(11):4483–4488. doi: 10.1073/pnas.0712324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325(5945):1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 21.Ingley E, Hemmings BA. Pleckstrin homology (PH) domains in signal transduction. J Cell Biochem. 1994;56(4):436–443. doi: 10.1002/jcb.240560403. [DOI] [PubMed] [Google Scholar]
- 22.Kruszynska YT, Olefsky JM. Cellular and molecular mechanisms of non-insulin dependent diabetes mellitus. J Investig Med. 1996;44(8):413–428. [PubMed] [Google Scholar]
- 23.Stiles B, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected] Proc Natl Acad Sci USA. 2004;101(7):2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurlawalla-Martinez C, et al. Insulin hypersensitivity and resistance to streptozotocin-induced diabetes in mice lacking PTEN in adipose tissue. Mol Cell Biol. 2005;25(6):2498–2510. doi: 10.1128/MCB.25.6.2498-2510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wijesekara N, et al. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol Cell Biol. 2005;25(3):1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fritsch R, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153(5):1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.