Significance
The complement system is a crucial component of the innate immune response in humans. In this study, we report the characterization of an autotransporter protease of Neisseria meningitidis named NalP. We show that NalP is able to cleave the α-chain of the human complement factor C3 in a species-specific manner. As a consequence, the deposition of C3b on the bacterial surface is reduced and, in human serum, the NalP-generated C3b fragment is further degraded by host factors. Our results significantly increase the knowledge of how N. meningitidis can survive and multiply in human serum, evading the innate immune system.
Keywords: serum resistance, immune evasion
Abstract
The complement system is a crucial component of the innate immune response against invading bacterial pathogens. The human pathogen Neisseria meningitidis (Nm) is known to possess several mechanisms to evade the complement system, including binding to complement inhibitors. In this study, we describe an additional mechanism used by Nm to evade the complement system and survive in human blood. Using an isogenic NalP deletion mutant and NalP complementing strains, we show that the autotransporter protease NalP cleaves C3, the central component of the complement cascade. The cleavage occurs 4 aa upstream from the natural C3 cleavage site and produces shorter C3a-like and longer C3b-like fragments. The C3b-like fragment is degraded in the presence of the complement regulators (factors H and I), and this degradation results in lower deposition of C3b on the bacterial surface. We conclude that NalP is an important factor to increase the survival of Nm in human serum.
The Gram-negative capsulated bacterium Neisseria meningitidis (Nm) is one of the major causes of meningitis and septicemia worldwide (1). During the course of infection, the bacterium must adapt to different host niches, a crucial factor for its survival and dissemination (2). Evasion of the complement system is critical for Nm to cause invasive disease. The observation that people deficient in various complement components are highly predisposed to invasive meningococcal disease provides epidemiologic evidence for the role of complement in host defense against this bacterial infection (3, 4). Complement activation can occur through the classical pathway (CP), the lectin pathway, or the alternative pathway (AP). Activation of the CP and lectin pathway results in cleavage of C4; deposited C4b can bind to C2, and cleavage of the latter generates C4b,2a, which functions as a C3 convertase. Cleavage of C3 by convertases is a critical event in complement activation, because it leads to release of the anaphylatoxin C3a and deposition of C3b on the bacterial surface (5). Strict regulation of the complement system is necessary to avoid inappropriate activation and host cell damage. As an example, C3b in the fluid phase is cleaved to iC3b by factor I (fI) in the presence of the cofactor factor H (fH). This cleavage inactivates C3b and prevents C3b from forming AP C3 convertase or AP/CP C5 convertase enzymes (6). The ability to escape to the complement system is a key determinant in the virulence of pathogens. Bacteria can escape recognition by the complement system through the actions of cell surface structures or secreted proteins (7–9). Nm has evolved several redundant mechanisms to evade the host innate responses at sites of colonization and during systemic growth, including expression of fHbp (10, 11) and NspA (12, 13), that facilitate binding of fH, lipooligosaccharide (LOS) sialic acid, which inhibits complement deposition (12), and Opc protein, which binds to vitronectin (2). In a recent functional genomic study, we identified factors involved in Nm survival in human blood (14). One of these factors is NalP, which is an autotransporter with subtilisin-like serine protease activity that is involved in autoproteolytic processing, resulting in secretion of the NalP passenger domain into the bacterial supernatant (15, 16). The expression of NalP is phase-variable because of slipped-strand mispairing of a polycytidine tract in the coding sequence (17). NalP contains a lipobox at the C-terminal end of the signal sequence; the lipid moiety permits anchorage in the outer membrane (16). Lipidated NalP proteins are only temporarily retained at the cell surface; however, the lipid moiety retards the release of NalP passenger domain from the bacterial surface and allows the partial or total cleavage of surface protein targets on bacterial surface (18), including IgA protease Iga, adhesion and penetration protein App (16), autotransporter serine protease AusI (19), Neisserial heparin binding antigen NHBA (20), and lactoferrin binding protein LbpB (21). This activity modulates the expression of meningococcal proteins at bacterial surface and also has recently been implicated in the formation and regulation of Nm biofilm (22). Although several bacterial targets of NalP protease have been well-characterized, host targets of NalP have not been identified thus far. In this work, we elucidate the molecular basis of Nm complement resistance mediated by NalP.
Results
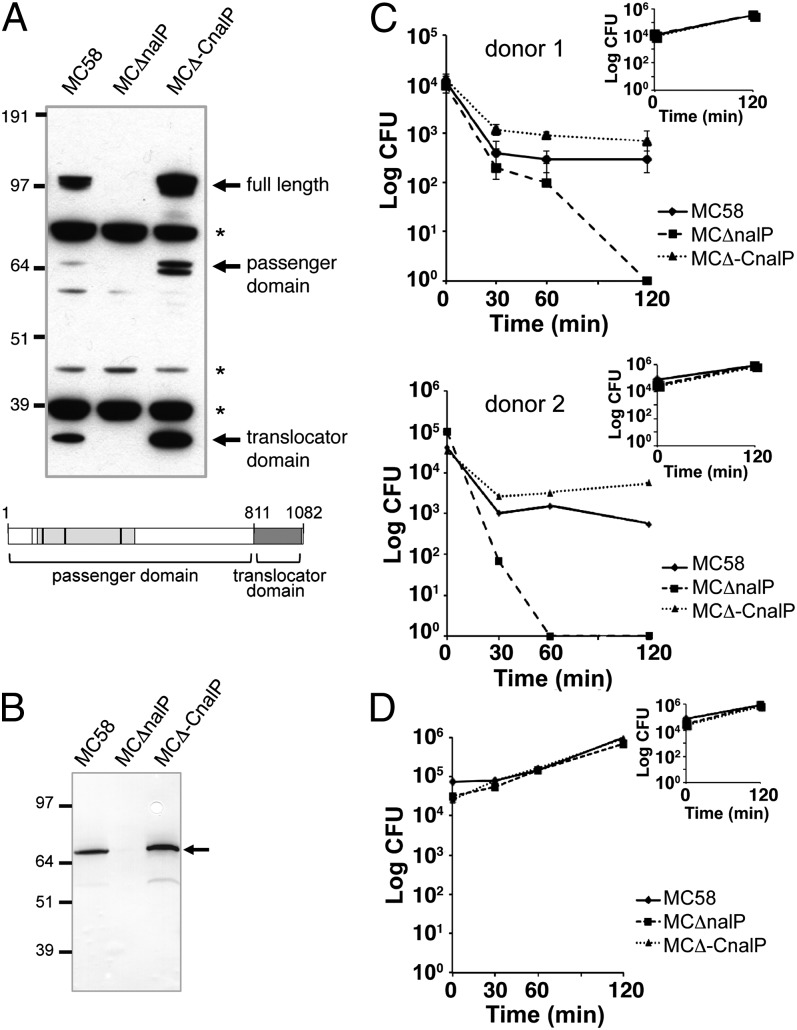
NalP Protease, and Not Its Protein Targets, Is Essential for Survival in Human Serum.
In a previous study of Nm transcriptome analysis in human blood, we showed that the nalP gene was up-regulated during incubation in human blood and that survival of a nalP deletion mutant (named MCΔnalP) in human blood was impaired (14). To further characterize the NalP phenotype, we complemented the strain MCΔnalP with the shuttle vector pFP-12 expressing nalP under the control of a constitutive promoter to generate the complementing strain MCΔ–CnalP. The expression of NalP in the three different isogenic strains was evaluated by Western blot analysis. As shown in Fig. 1A, the complementing strain expressed NalP in its active form. Three different specific fragments were detected in whole-cell protein lysates: the full-length form (∼110 kDa), a ∼70-kDa fragment corresponding to the passenger domain that also contains the catalytic domain, and the ∼30-kDa translocator domain fragment (15, 16). The presence of only the passenger domain but not intact NalP or the translocator domain in the supernatants prepared from the NalP-expressing strains was confirmed by Western blot analysis (Fig. 1B). The complementing strain MCΔ−CnalP expresses higher amounts of NalP compared with the WT strain, likely because of the episomal complementation system used. The WT, mutant, and complementing strains were tested for survival in human serum. As shown in Fig. 1C, the strain lacking NalP was killed in human serum, and survival was restored in the complementing strain, confirming a role for NalP in the survival of Nm in human serum. Heat-inactivated serum (all complement pathways inactivated) was used as control and showed similar survival for all strains, confirming a role for complement in killing of the NalP mutant (Fig. 1D). The survival of the three isogenic Nm strains in human whole blood paralleled with the phenotype observed in human serum (Fig. S1). NalP is a surface-exposed autotransporter with serine protease activity able to cleave different surface-exposed proteins, including Iga, NHBA, AusI, App, and LbpB (15, 16, 20, 21). To determine whether NalP contributed to Nm survival in blood through an indirect effect by cleaving one or more of the known target proteins, deletion mutants of each NalP target were generated individually in strain MC58. Because nalP is prone to phase variation (15, 16), each mutant strain was tested for NalP expression. Western blot confirmed that all of the mutant strains express comparable levels of NalP (Fig. S2A). The WT MC58 and mutant strains were then incubated in human whole blood for 2 h. Samples were obtained at various time points to assess the number of viable colonies (colony forming unit, cfu). As shown in Fig. S2B, bacterial survival for each mutant strain was similar to the MC58 WT strain. This result suggests that the phenotype observed in human blood for the nalP deletion mutant strain is not caused by the cleavage of one of the known NalP targets.
Fig. 1.
Complementation of NalP restores survival in human serum. (A) Western blot analysis showed the expression of NalP in Nm WT and NalP complementing strain but not NalP deletion mutant. Three forms of NalP, corresponding to the full-length protein (100 kDa), the released passenger domain (70 kDa), and the translocator domain (30 kDa), are indicated by arrows. *Not specific bands. A schematic representation of NalP protein with different domains is also shown. The catalytic site of the passenger domain is indicated by a light gray box, and the catalytic triad is indicated by vertical black lines. (B) The presence of only the passenger domain but not intact NalP or the translocator domain in the supernatant was confirmed by Western blot analysis. (C) Nm WT, NalP deletion mutant, and complementing strains were tested for survival in human serum at 40% over a time course of 120 min using serum from two different blood donors. The graphs represent the survival in human serum, whereas Insets represent the growth control in GC-rich medium for the same time course of 120 min. Error bars are indicated by vertical lines. (D) Complement is required for killing of the NalP mutant. All strains survived in heat-inactivated human serum (complement activity abrogated by heat treatment at 56 °C for 30 min).
NalP Specifically Cleaves Human C3.
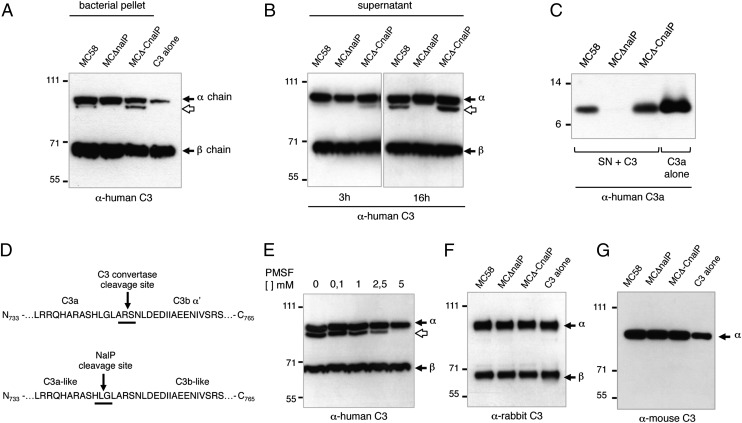
Looking for additional NalP targets, we explored the possibility that NalP could inactivate components of the complement system. The WT MC58 strain and its MCΔnalP deletion mutant and MCΔ−CnalP complementing strain were examined for their ability to cleave complement proteins C1q, C2, C3, and C4, which are involved in activation of the complement cascade. Nm strains were incubated with each of the purified complement molecules for 16 h at 37 °C, and the effect of NalP on the complement factor was determined by Western blot analysis. C1q, C2, and C4 were not cleaved by any of the strains (Fig. S3). In contrast, incubation of WT MC58 with human C3 generated an ∼100-kDa α-chain fragment that was absent in the reaction containing the MCΔnalP deletion mutant strain (Fig. 2A); the cleavage was more evident with the complementing strain MCΔ−CnalP, which expressed the highest amounts of NalP. To evaluate whether the passenger domain of NalP, released in the bacterial supernatant after its autoproteolytic processing, had similar C3 cleaving activity, concentrated supernatants from the WT, deletion mutant, and complementing MC58 strains were assayed for human C3 cleavage. The passenger domain alone that contains the catalytic activity of NalP and is released in the supernatant of WT and MCΔ−CnalP complementing strains was able to cleave human C3 (Fig. 2B). We used rabbit polyclonal α-C3/C3a antibody to immunoprecipitate C3 and its fragments from the reaction mixture, and we observed that a fragment of ∼10 kDa was also generated by bacterial supernatants from WT and NalP complementing strains. This fragment comigrated with the natural C3a (Fig. 2C). To map the cleavage site, the ∼100-kDa C3 fragment generated by NalP was subjected to N-terminal sequence analysis. The sequence obtained (745-GLARSNLDED-754) showed that NalP cleaved the C3 α-chain 4 aa N-terminal to the physiologic cleavage site of C3 convertase, which cleaves C3 between Arg748 and Ser749 (Fig. 2D). Hence, NalP generated a C3a-like molecule that is 4 aa shorter than physiologic C3a and a C3b-like fragment that is 4 aa longer than the natural C3b. Despite several attempts, we were unable to express in Nm a NalP protein that contained a loss of function mutation in the catalytic triad. Hence, to confirm the specificity of the cleavage, the C3 cleavage assay was conducted in the presence of increasing concentrations of the serine protease inhibitor PMSF. We observed a dose-dependent inhibition of C3 cleavage, and cleavage was completely abrogated at 5 mM PMSF (Fig. 2E). To further confirm NalP-specific C3 cleavage, we cloned and expressed the WT and mutated nalP genes in Escherichia coli strain BL21-DE3 using the expression vector pET-21b to generate strain Ec-NalP and Ec-NalPS426A, respectively. Western blot analysis (Fig. S4A) showed that NalP was expressed and localized in the outer membrane protein fraction in strains Ec-NalP and Ec-NalPS426A; mutation of the serine at position 426 to alanine (S426A) abolished autoprocessing and secretion of the passenger domain in the supernatant of strain Ec-NalPS426A. Bacterial pellets and concentrated supernatants from cultures of E. coli strains Ec-NalP, Ec-NalPS426A, and Ec-pET (vector control) were tested for their ability to cleave human C3. The C3 α-chain was cleaved only in the presence of pellet and supernatant fraction prepared from strain Ec-NalP that expressed WT NalP but not the strain that expressed NalP with the S426A mutation, thereby confirming specificity of the cleavage mediated by NalP (Fig. S4B). The interaction of Nm with several human proteins has been shown to be species-specific (13, 23, 24). To investigate whether the cleavage of C3 mediated by NalP was restricted to the human protein, a cleavage assay was performed using C3 purified from rabbit and mouse sera. Culture supernatants from Nm strains MC58, MCΔnalP, and MCΔ−CnalP were incubated with rabbit or mouse C3, and the mixtures were then analyzed by Western blot using anti-rabbit C3 or anti-mouse C3, respectively. NalP failed to cleave both rabbit and mouse C3, suggesting that NalP cleaves C3 in a human-specific manner (Fig. 2 F and G).
Fig. 2.
NalP specifically cleaves human C3. (A) Nm WT, NalP deletion mutant, and complementing strain bacterial pellets were incubated with human C3 at 37 °C for 16 h. Western blot analysis showed that NalP-expressing strains were only able to cleave C3 α-chain and not C3 β-chain (black arrows), releasing a C3b-like α′ of ∼ 100 kDa (open arrow). (B) The supernatants were tested for their ability to cleave human C3 after 3 or 16 h of incubation. Only bacterial supernatants containing NalP passenger domain (i.e., WT and complementing strains) were able to cleave human C3 α-chain. Cleavage was evident even after 3 h of incubation. (C) A molecule with a size similar to C3a is released from C3 after cleavage by NalP. The reaction mixture was immunoprecipitated using rabbit polyclonal α-C3/C3a antibody. The Western blot analysis with anti-human C3a showed a C3a-like molecule of ∼ 10 kDa generated only in reactions that contained supernatants (SNs) from the WT and complementing strains that possessed the passenger domain of NalP. (D) The C3b-like fragment, generated by NalP, was subjected to N-terminal sequencing. The results revealed that NalP cleaved human C3 between Lys744 and Gly745 (4 aa N-terminal to the C3 convertase cleavage site), thereby generating a shorter C3a-like molecule and a longer C3b-like molecule compared with the corresponding physiological counterparts. (E) The C3 cleavage assay using bacterial supernatant obtained from the complementing strain was conducted in the presence of increasing concentrations of the serine protease inhibitor PMSF ranging from 0.1 to 5 mM. Western blot analysis of samples showed a dose-dependent inhibition of human C3 cleavage by NalP, and the cleavage was completely abrogated at 5 mM PMSF. (F and G) Cleavage of C3 by NalP is human-specific. NalP from bacterial supernatants was tested for its ability to cleave C3 isolated from (F) rabbit and (G) mouse serum by Western blot analysis, and no cleavage was observed. In the case of mouse C3, the antibody used does not detect the C3 β-chain.
NalP Decreases C3b Deposition on the Bacterial Surface.
Having shown that NalP is able to cleave human C3, we hypothesized that NalP-mediated C3 cleavage would inhibit complement activation and C3b fragment deposition on the bacterial surface. As shown in Fig. 3A, fewer C3b fragments were deposited on MC58 and MCΔ−CnalP strains compared with the MCΔnalP deletion mutant strain, which was measured by flow cytometry. Thus, NalP expressed on the bacterial surface is able to cleave human C3 and limit deposition of C3b on the bacterial surface.
Fig. 3.
Functional characterization of human C3 cleavage by NalP. (A) Nm WT, NalP deletion mutant, and the complementing strains were incubated with 50% human serum for 60 min. C3b deposition was determined using goat polyclonal α-human C3 antibody and analyzed by FACS. The mean fluorescence intensity for the entire bacterial population is shown (text color corresponds to the color of the histogram). After 16 h of incubation of human C3 with bacterial supernatant from NalP complementing strain, (B) human fH and fI or (C) human C3-depleted serum (that contains both fH and fI) at 2% (vol/vol) was added. Samples were obtained at the different time points indicated, and degradation of the C3b-like fragment generated by NalP (open arrow) was analyzed by Western blotting. (D) Control reactions included samples incubated with fH or fI separately for 1 h.
C3b-Like Molecule Generated by NalP Is Degraded by Serum Host Factors.
One possibility to explain the lower deposition of C3b on the bacterial surface observed above is that the NalP-generated C3b-like fragment is susceptible to degradation by complement regulators. One known mechanism of regulation involves proteolysis of C3b by fI in conjunction with its cofactor fH, which binds C3b and increases the affinity for fI (6). To test this hypothesis, human C3 was first incubated with concentrated supernatant from culture of MCΔ−CnalP to generate the C3b-like cleavage fragment. This mixture was then incubated with human fH and fI either combined or separately (Fig. 3 B and D, respectively) or 2% (vol/vol) human C3-depleted serum that served as a source of fH and fI (Fig. 3C) for periods of time ranging from 0 to 1 h. fH and fI degraded only the C3b-like molecule generated by NalP (Fig. 3C, open arrow) but had no effect on intact C3. These results showed specific degradation of only the C3b-like molecule generated by NalP by fH and fI.
Discussion
NalP is a surface-exposed autotransporter of Nm with serine protease activity that has been shown to cleave several proteins on the outer membrane of Nm. In this work, we have shown that NalP also cleaves a host protein and that this cleavage represents a major mechanism for survival in human serum. We provide evidence that NalP is able to cleave human C3 in a human-specific manner. The cleavage occurs 4 aa upstream from the natural cleavage site and produces a shorter C3a-like and longer C3b-like fragments. The C3b-like fragment generated by NalP is susceptible to degradation by the complement inhibitors, fH and fI. This degradation results in reduced deposition of C3b on the surface of Nm. We have not investigated whether the C3a-like fragment generated by NalP is active; however, it lacks the C-terminal arginine that is known to be essential for its activity. Therefore, we believe that the NalP cleavage of human C3 generates a C3a-like fragment that does not possess the anaphylatoxin activity of C3a generated physiologically by C3 convertases. It is likely that both the decreased C3b deposition and the absence of the anaphylatoxin activity contribute to Nm immune evasion and survival in serum and blood.
Species-Specific Cleavage of Complement Factors.
Proteolytic cleavage of complement components by bacterial proteases is a mechanism used by many human pathogens. Examples are streptococcal cysteine protease SpeB, which degrades C3 to inhibit bacterial clearance (25), the streptococcal cell-associated peptidase ScpA, which cleaves C5a to inhibit neutrophil chemotaxis (26), GelE of Enterococcus faecalis (27) and aureolysin of Staphylococcus aureus, which cleave C3 (28), EspP of enterohemorrhagic E. coli, which cleaves C3 and C5 to impair complement activation (29), and PgtE of Salmonella enterica, which proteolytically cleaves C3b, C4b, and C5 to enhance bacterial resistance to human serum (30).
NalP is the only known Nm protein that cleaves C3 and promotes bacterial survival, because a nalP deletion mutant was not able to cleave C3 or survive in human serum or whole blood. However, the phenotype was restored when nalP was complemented in trans through episomal expression. Although our attempts to generate a complementing strain expressing NalP that was mutated in the catalytic triad were not successful, the specificity of the catalytic domain of NalP in the C3 cleavage was confirmed by expressing WT NalP and a mutant NalP that lacked catalytic activity (S426A) in a nonpathogenic E. coli strain. We also showed that NalP could cleave human C3 but not rabbit or mouse C3, suggesting that the cleavage is species-specific. Although the full amino acid sequence of rabbit C3 is not available, alignment of several available nonhuman C3 amino acid sequences, including mouse C3, shows some diversity in the region surrounding the NalP cleavage site that can explain the species specificity of the cleavage (Fig. S5). The species specificity of NalP is consistent with the other Nm proteins that interact specifically with human factors, which include fHbp, NspA, opacity proteins, and transferring binding protein (13, 24, 31, 32). The specificity of interactions with human factors may explain why Nm is strictly a human pathogen and why antibody-mediated killing of Nm in vitro is more efficient when complement is derived from rabbits, rats, or mice, with fH and C3 that are not recognized by fHbp, NspA, or NalP, respectively.
Molecular Mechanisms for Survival in Serum.
It is likely that C3 that is cleaved by NalP—and particularly, the C3 molecules cleaved in solution by the released passenger domain—does not form covalent bonds with the bacteria, because the thiolate and acylimidazole intermediates that are formed on exposure of internal thioester bond in C3 after release of C3a (or C3a-like) are highly labile with a half-life <100 µs (33) and thus, may undergo hydrolysis by reacting with a water molecule before encountering electron-donating –OH residues on the bacterial surface. Similar to the ability of fH and fI to rapidly cleave C3b that is generated by C3 convertases (6), the C3b-like molecule generated by NalP is also rapidly degraded by fH and fI. Thus, activation of C3 in the fluid phase followed by rapid degradation of the C3b-like molecule by fI and fH (either in solution or bound to the bacterial surface through ligands, such as fHbp and NspA) could constitute a mechanism of complement escape. The C3a molecule produced by C3 convertase is one of the anaphylatoxins of the complement system and has diverse effects ranging from the release of histamine from human mast cells to chemotaxis of basophils and eosinophils to activation of neutrophils (34–37). The activity of C3a in the bloodstream and tissues is tightly controlled by carboxypeptidases, which rapidly remove the C-terminal Arg residue (38, 39). The resulting molecule, called C3a-desArg, is devoid of proinflammatory activity (40). We, therefore, believe that the C3a-like molecule generated by NalP, which lacks the C-terminal 4 aa, would also lack the biological functions observed by intact C3a. Similar to NalP, S. aureus aureolysin also cleaves C3 at a single site. Unlike NalP, aureolysin cleaves C3 2 aa C-terminal to the C3 convertase cleavage site to generate functionally active C3b and C3a (28). In contrast to NalP, the streptococcal cysteine protease SpeB fully degrades C3 as a result of cleavage at multiple sites (25).
NalP Expression in Neisseria Species.
Based on a bioinformatics analysis conducted with the genomic sequences available in the databases, Nm is the only species of the genus Neisseria that possesses a functional nalP (Table S1). nalP is not present in any commensal species but is present in the human pathogen N. gonorrheae. However, none of the N. gonorrheae genome sequences analyzed (17 strains) seem to encode full-length NalP proteins because of the presence of premature stop codons (Table S1). This analysis is in accordance with a previous report that nalP in N. gonorrheae FA1090 is a pseudogene because of the presence of numerous termination codons (15). In the case of Nm, the nalP gene was present in all but one of the strains analyzed (Table S1). About one-half (13 of 27) of the sequences analyzed (listed in Table S1) possess a frameshift in the polycytidine region that results in phase variation. It is conceivable that Nm needs to switch the expression of the nalP gene on or off in different niches of its human host. In the context of adaptation and proliferation in human blood, strains that are phase-off could revert to phase-on phenotype to express nalP and facilitate the survival in human blood. We propose that Nm strains that show high rates of nalP gene phase variation may possess an advantage when causing invasive disease in humans.
In a recent molecular epidemiology study of 647 Nm isolates, nalP was not identified in ∼11% of the strains because of deletion events in the nalP locus (41). The infrequency of the nalP deletion suggests that it is disadvantageous for the bacteria and that, in the nalP-negative strain, the function could be compensated by other factors or other mechanisms. This hypothesis recalls the redundancy in fH binding by Nm, where strains lacking fHbp can use NspA to bind fH (13, 14) to avoid complement activation on bacterial surface.
Role of the NalP-Secreted Passenger Domain and Full-Length Molecule.
The full-length NalP protein is exposed on the meningococcal surface, whereas the passenger domain is released in the supernatant (15, 16). Roussel-Jazédé et al. (18) reported that the lipid moiety of NalP enables it to be retained at the cell surface for a duration sufficient to cleave its targets on bacterial surface. However, none of the NalP targets seem to contribute to serum resistance of Nm. The majority of NalP on the surface is eventually autoprocessed, and the passenger domain with its catalytic domain is released into the supernatant (16, 42). We have shown that the full-length protein on bacterial surface as well as the released passenger domain both cleave human C3. Inhibition of complement activation by NalP occurs by cleavage of C3, which generates a functionally inactive C3a-like molecule, whereas the corresponding C3b-like fragment is inactivated rapidly by fH and fI (Fig. 4). For NalP to function as a complement inhibitor, it is crucial that the passenger domain is secreted in the extracellular environment, and therefore, it can cleave C3 distal to the bacterial surface to generate C3b that is incapable of forming covalent bonds with bacterial targets to generate C3 convertases as described above. We believe that the released passenger domain is mainly responsible for cleavage of C3 in fluid phase. However, C3 cleavage by NalP expressed on bacterial surface may contribute to serum survival by reducing the generation of active C3 convertases on the membrane. It will be of interest to determine if the inhibition of complement activation is the result of increased degradation of the C3b-like molecule, decreased deposition of the C3b-like molecule on bacterial surface, or decreased ability of the C3b-like molecule to interact with factor B to form active C3/C5 convertases. Future studies will be needed to address the importance of NalP in the pathogenesis of Nm infections and how the full-length and secreted forms of NalP contribute to complement inhibition.
Fig. 4.
Schematic representation of the contribution of NalP in Nm complement evasion. For NalP to function as a complement inhibitor, NalP full-length protein on bacterial surface as well as the passenger domain containing the catalytic site and secreted in the extracellular environment must both cleave human C3. Inhibition of complement activation by NalP occurs by cleavage of C3, which generates a functionally inactive C3a-like molecule, whereas the corresponding C3b-like fragment is inactivated rapidly by fH and fI. Thus, the activity of NalP leads to the inactivation of C3 in human blood at surface level and distal to the bacterial surface, resulting in the inhibition of C3b deposition and therefore, serum resistance.
In conclusion, Nm has evolved several redundant mechanisms to evade the host innate responses at sites of colonization and during systemic growth (7, 43). Several molecules, such as the capsule, fHbp, and NspA, cooperate to confer serum resistance. Here, we have shown that NalP is an additional important molecule that contributes to the arsenal of meningococcal defenses to evade the immune system.
Materials and Methods
Bacterial Strains and DNA Manipulation.
The strains, plasmids, and primers used in this study are listed in Tables S2 and S3. Detailed descriptions of bacterial strains, growth conditions, and methods for DNA manipulation and construction of Nm recombinant strains are provided in SI Materials and Methods.
Survival Experiments in Human Whole Blood and Human Serum.
The experiments of survival in human blood and human serum were performed as previously described (14). Bacteria were grown until midexponential phase and then added to human whole blood or human serum at 40% in d-PBS. Cultures were incubated for 2 h; at various time points, an aliquot of the sample was removed, and the number of viable bacteria was determined by plating serial dilutions. Detailed methods and additional details are reported in SI Materials and Methods.
Complement Cleavage Assay.
Nm MC58 WT, NalP deletion mutant, and complementing strains were grown in Gonococcal (GC) Medium Base until midlog phase. Then, bacteria were centrifuged, the pellet was incubated with single complement factors (human factors C1q, C2, C3, and C4 and rabbit and mouse C3) for 16 h at 37 °C while shaking. Complement factor cleavage for each factor was detected by Western blot analysis. The same experiment was performed using 50× concentrated bacterial supernatant. The specificity of human C3 cleavage by NalP expressed on Nm bacterial surface was determined using the serine protease inhibitor PMSF. The activity of purified rabbit and mouse C3 was tested by checking rabbit or mouse C3 deposition on yeast glucan particles in presence of human C3-depleted serum (Fig. S6). Detailed methods are reported in SI Materials and Methods.
Analysis of C3 Fragments.
After incubation of C3 with concentrated bacterial supernatant from Nm NalP complementing strain, C3 fragments (C3a- and C3b-like molecules) were immunoprecipitated and then analyzed by Western blotting. The C3b-like molecule was purified and subjected to N-terminal sequencing (Alphalyse). In addition, the C3b-like molecule was tested for degradation by human host factors by adding fH and fI or 2% (vol/vol) human C3-depleted serum. C3b-like molecule degradation was analyzed by Western blotting. Additional details are reported in SI Materials and Methods.
C3b Deposition on Nm Bacterial Surface.
Nm WT, NalP deletion mutant, and complementing strains were grown in GC medium until midlog phase. Then, bacteria were resuspended in 50% human serum diluted in HBSS++ and incubated at 37 °C for 60 min while shaking. C3b deposition was detected by FACS analysis. Additional details are reported in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Alessandro Muzzi for the bioinformatics analysis of the nalP gene and Dr. Rosane DeOliveira for technical assistance. We thank Mariagrazia Pizza and Vega Masignani for useful discussion and Giorgio Corsi for artwork. I.V. is the recipient of a Novartis Fellowship from the PhD Program in Functional Biology of Molecular and Cellular Systems of the University of Bologna. S.R. is supported by National Institute of Allergy and Infectious Diseases, National Institutes of Health Grants AI054544, AI084048, and AI32725.
Footnotes
Conflict of interest statement: R.R. and D.S. are full-time employees of Novartis Vaccines and Diagnostics.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321556111/-/DCSupplemental.
References
- 1.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369(9580):2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 2.Virji M. Pathogenic neisseriae: Surface modulation, pathogenesis and infection control. Nat Rev Microbiol. 2009;7(4):274–286. doi: 10.1038/nrmicro2097. [DOI] [PubMed] [Google Scholar]
- 3.Figueroa JE, Densen P. Infectious diseases associated with complement deficiencies. Clin Microbiol Rev. 1991;4(3):359–395. doi: 10.1128/cmr.4.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ram S, Lewis LA, Rice PA. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev. 2010;23(4):740–780. doi: 10.1128/CMR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jongerius I, Ram S, Rooijakkers S. Bacterial complement escape. Adv Exp Med Biol. 2009;666:32–48. doi: 10.1007/978-1-4419-1601-3_3. [DOI] [PubMed] [Google Scholar]
- 6.Rambach G, Würzner R, Speth C. Complement: An efficient sword of innate immunity. Contrib Microbiol. 2008;15:78–100. doi: 10.1159/000136316. [DOI] [PubMed] [Google Scholar]
- 7.Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: Learning from staphylococci and meningococci. Nat Rev Microbiol. 2010;8(6):393–399. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- 8.Rooijakkers SH, van Strijp JA. Bacterial complement evasion. Mol Immunol. 2007;44(1-3):23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6(2):132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madico G, et al. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177(1):501–510. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seib KL, et al. Factor H-binding protein is important for meningococcal survival in human whole blood and serum and in the presence of the antimicrobial peptide LL-37. Infect Immun. 2009;77(1):292–299. doi: 10.1128/IAI.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis LA, Carter M, Ram S. The relative roles of factor H binding protein, neisserial surface protein A, and lipooligosaccharide sialylation in regulation of the alternative pathway of complement on meningococci. J Immunol. 2012;188(10):5063–5072. doi: 10.4049/jimmunol.1103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis LA, et al. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 2010;6(7):e1001027. doi: 10.1371/journal.ppat.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echenique-Rivera H, et al. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 2011;7(5):e1002027. doi: 10.1371/journal.ppat.1002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner DP, Wooldridge KG, Ala’Aldeen DA. Autotransported serine protease A of Neisseria meningitidis: An immunogenic, surface-exposed outer membrane, and secreted protein. Infect Immun. 2002;70(8):4447–4461. doi: 10.1128/IAI.70.8.4447-4461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Ulsen P, et al. A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol. 2003;50(3):1017–1030. doi: 10.1046/j.1365-2958.2003.03773.x. [DOI] [PubMed] [Google Scholar]
- 17.Saunders NJ, et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol. 2000;37(1):207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 18.Roussel-Jazédé V, Grijpstra J, van Dam V, Tommassen J, van Ulsen P. Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins. Microbiology. 2013;159(Pt 2):286–295. doi: 10.1099/mic.0.063982-0. [DOI] [PubMed] [Google Scholar]
- 19.van Ulsen P, et al. A novel phase-variable autotransporter serine protease, AusI, of Neisseria meningitidis. Microbes Infect. 2006;8(8):2088–2097. doi: 10.1016/j.micinf.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Serruto D, et al. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci USA. 2010;107(8):3770–3775. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roussel-Jazédé V, Jongerius I, Bos MP, Tommassen J, van Ulsen P. NalP-mediated proteolytic release of lactoferrin-binding protein B from the meningococcal cell surface. Infect Immun. 2010;78(7):3083–3089. doi: 10.1128/IAI.01193-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arenas J, Nijland R, Rodriguez FJ, Bosma TN, Tommassen J. Involvement of three meningococcal surface-exposed proteins, the heparin-binding protein NhbA, the α-peptide of IgA protease and the autotransporter protease NalP, in initiation of biofilm formation. Mol Microbiol. 2013;87(2):254–268. doi: 10.1111/mmi.12097. [DOI] [PubMed] [Google Scholar]
- 23.Boulton IC, et al. Purified meningococcal transferrin-binding protein B interacts with a secondary, strain-specific, binding site in the N-terminal lobe of human transferrin. Biochem J. 1999;339(Pt 1):143–149. [PMC free article] [PubMed] [Google Scholar]
- 24.Granoff DM, Welsch JA, Ram S. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect Immun. 2009;77(2):764–769. doi: 10.1128/IAI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terao Y, et al. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J Biol Chem. 2008;283(10):6253–6260. doi: 10.1074/jbc.M704821200. [DOI] [PubMed] [Google Scholar]
- 26.Wexler DE, Chenoweth DE, Cleary PP. Mechanism of action of the group A streptococcal C5a inactivator. Proc Natl Acad Sci USA. 1985;82(23):8144–8148. doi: 10.1073/pnas.82.23.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, et al. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J Immunol. 2008;181(9):6328–6336. doi: 10.4049/jimmunol.181.9.6328. [DOI] [PubMed] [Google Scholar]
- 28.Laarman AJ, et al. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186(11):6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 29.Orth D, et al. EspP, a serine protease of enterohemorrhagic Escherichia coli, impairs complement activation by cleaving complement factors C3/C3b and C5. Infect Immun. 2010;78(10):4294–4301. doi: 10.1128/IAI.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramu P, et al. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Lett. 2007;581(9):1716–1720. doi: 10.1016/j.febslet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 31.Gray-Owen SD, Schryvers AB. The interaction of primate transferrins with receptors on bacteria pathogenic to humans. Microb Pathog. 1993;14(5):389–398. doi: 10.1006/mpat.1993.1038. [DOI] [PubMed] [Google Scholar]
- 32.Sadarangani M, Pollard AJ, Gray-Owen SD. Opa proteins and CEACAMs: Pathways of immune engagement for pathogenic Neisseria. FEMS Microbiol Rev. 2011;35(3):498–514. doi: 10.1111/j.1574-6976.2010.00260.x. [DOI] [PubMed] [Google Scholar]
- 33.Sim RB, Twose TM, Paterson DS, Sim E. The covalent-binding reaction of complement component C3. Biochem J. 1981;193(1):115–127. doi: 10.1042/bj1930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daffern PJ, Pfeifer PH, Ember JA, Hugli TE. C3a is a chemotaxin for human eosinophils but not for neutrophils. I. C3a stimulation of neutrophils is secondary to eosinophil activation. J Exp Med. 1995;181(6):2119–2127. doi: 10.1084/jem.181.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsner J, et al. C3a activates reactive oxygen radical species production and intracellular calcium transients in human eosinophils. Eur J Immunol. 1994;24(3):518–522. doi: 10.1002/eji.1830240304. [DOI] [PubMed] [Google Scholar]
- 36.Elsner J, Oppermann M, Czech W, Kapp A. C3a activates the respiratory burst in human polymorphonuclear neutrophilic leukocytes via pertussis toxin-sensitive G-proteins. Blood. 1994;83(11):3324–3331. [PubMed] [Google Scholar]
- 37.Klos A, et al. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009;46(14):2753–2766. doi: 10.1016/j.molimm.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bokisch VA, Müller-Eberhard HJ. Anaphylatoxin inactivator of human plasma: Its isolation and characterization as a carboxypeptidase. J Clin Invest. 1970;49(12):2427–2436. doi: 10.1172/JCI106462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews KW, et al. Expression of the third complement component (C3) and carboxypeptidase N small subunit (CPN1) during mouse embryonic development. Dev Comp Immunol. 2004;28(6):647–655. doi: 10.1016/j.dci.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Sayah S, et al. Two different transduction pathways are activated by C3a and C5a anaphylatoxins on astrocytes. Brain Res Mol Brain Res. 2003;112(1–2):53–60. doi: 10.1016/s0169-328x(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 41.Oldfield NJ, et al. Prevalence and phase variable expression status of two autotransporters, NalP and MspA, in carriage and disease isolates of Neisseria meningitidis. PLoS One. 2013;8(7):e69746. doi: 10.1371/journal.pone.0069746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain S, et al. Polar localization of the autotransporter family of large bacterial virulence proteins. J Bacteriol. 2006;188(13):4841–4850. doi: 10.1128/JB.00326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo H, Tang CM, Exley RM. Mechanisms of avoidance of host immunity by Neisseria meningitidis and its effect on vaccine development. Lancet Infect Dis. 2009;9(7):418–427. doi: 10.1016/S1473-3099(09)70132-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.