Significance
The P-element–induced wimpy testis (PIWI) proteins and their bound small RNAs (PIWI-interacting RNAs, piRNAs) are known to repress transposon expression in the germline, yet they likely have broader regulatory functions. We show that the PIWI–piRNA pathway functions in the stem cells of an early diverging animal. We demonstrate that Hydra has two PIWI proteins that are localized in the cytoplasm of all adult stem/progenitor cell types. We identified putative targets of the pathway, both transposon and nontransposon, by sequencing piRNAs and mapping them to a newly assembled Hydra transcriptome. Finally we demonstrate that Hydra PIWI is essential in the somatic lineages. This study supports the existence of a common regulatory pathway ancestral to both stem and germ cells.
Abstract
PIWI proteins and their bound PIWI-interacting RNAs (piRNAs) are found in animal germlines and are essential for fertility, but their functions outside of the gonad are not well understood. The cnidarian Hydra is a simple metazoan with well-characterized stem/progenitor cells that provides a unique model for analysis of PIWI function. Here we report that Hydra has two PIWI proteins, Hydra PIWI (Hywi) and Hydra PIWI-like (Hyli), both of which are expressed in all Hydra stem/progenitor cells, but not in terminally differentiated cells. We identified ∼15 million piRNAs associated with Hywi and/or Hyli and found that they exhibit the ping-pong signature of piRNA biogenesis. Hydra PIWI proteins are strictly cytoplasmic and thus likely act as posttranscriptional regulators. To explore this function, we generated a Hydra transcriptome for piRNA mapping. piRNAs map to transposons with a 25- to 35-fold enrichment compared with the abundance of transposon transcripts. By sequencing the small RNAs specific to the interstitial, ectodermal, and endodermal lineages, we found that the targeting of transposons appears to be largely restricted to the interstitial lineage. We also identified putative nontransposon targets of the pathway unique to each lineage. Finally we demonstrate that hywi function is essential in the somatic epithelial lineages. This comprehensive analysis of the PIWI–piRNA pathway in the somatic stem/progenitor cells of a nonbilaterian animal suggests that this pathway originated with broader stem cell functionality.
P-element–induced wimpy testis (PIWI) proteins and their bound small PIWI-interacting RNAs (piRNAs) are central players in a regulatory pathway that is essential for germline establishment and maintenance. Loss of PIWI proteins in Drosophila, mice, and zebrafish leads to a loss of fertility, due to a disruption in germline stem cell (GSC) formation or maintenance, arrest in meiosis, and other gametogenic defects (1). Piwi is also expressed outside the germline, largely in various kinds of stem and progenitor cells. For example, piwi genes are expressed in the pluripotent stem cells of planarians, sponges, and tunicates, and are required for regeneration (2). Piwi expression is also found in hematopoietic stem cells in humans, mesenchymal stem cells in mice, and somatic stem cells in cnidarians and ctenophores (3–6). However, detailed analyses have been largely confined to the function of the PIWI–piRNA pathway in the germline and the gonadal somatic cells in a few model bilaterians, with a focus on transposon silencing (7). The potential significance of the pathway in stem cells outside the gonad and on nontransposon sequences is largely unexplored.
Hydra is a morphologically simple multicellular organism belonging to the phylum Cnidaria, which is the sister group to bilaterians (SI Appendix, Fig. S1A). The adult Hydra polyp is composed of three distinct cell lineages: the two epithelial lineages (ectoderm and endoderm) and the interstitial lineage (Fig. 1A). The multipotent interstitial stem cells that support the interstitial lineage give rise to three somatic cell types (nerves, nematocytes, and gland cells) and to germ cells (Fig. 1A) (8). The epithelial lineages do not have a true stem cell population, but they are mitotic along the entire length of the body column and these progenitor/stem cells are responsible for maintaining the lineage (9). These cells indefinitely self-renew and retain the capability of differentiating into the nonmitotic cells that function in the tentacles and foot (Fig. 1A). In this study we provide a comprehensive analysis of both PIWI protein expression and piRNA sequences in Hydra, which demonstrates that the PIWI–piRNA pathway has ancient and broadly conserved stem cell functions, including somatic functions.
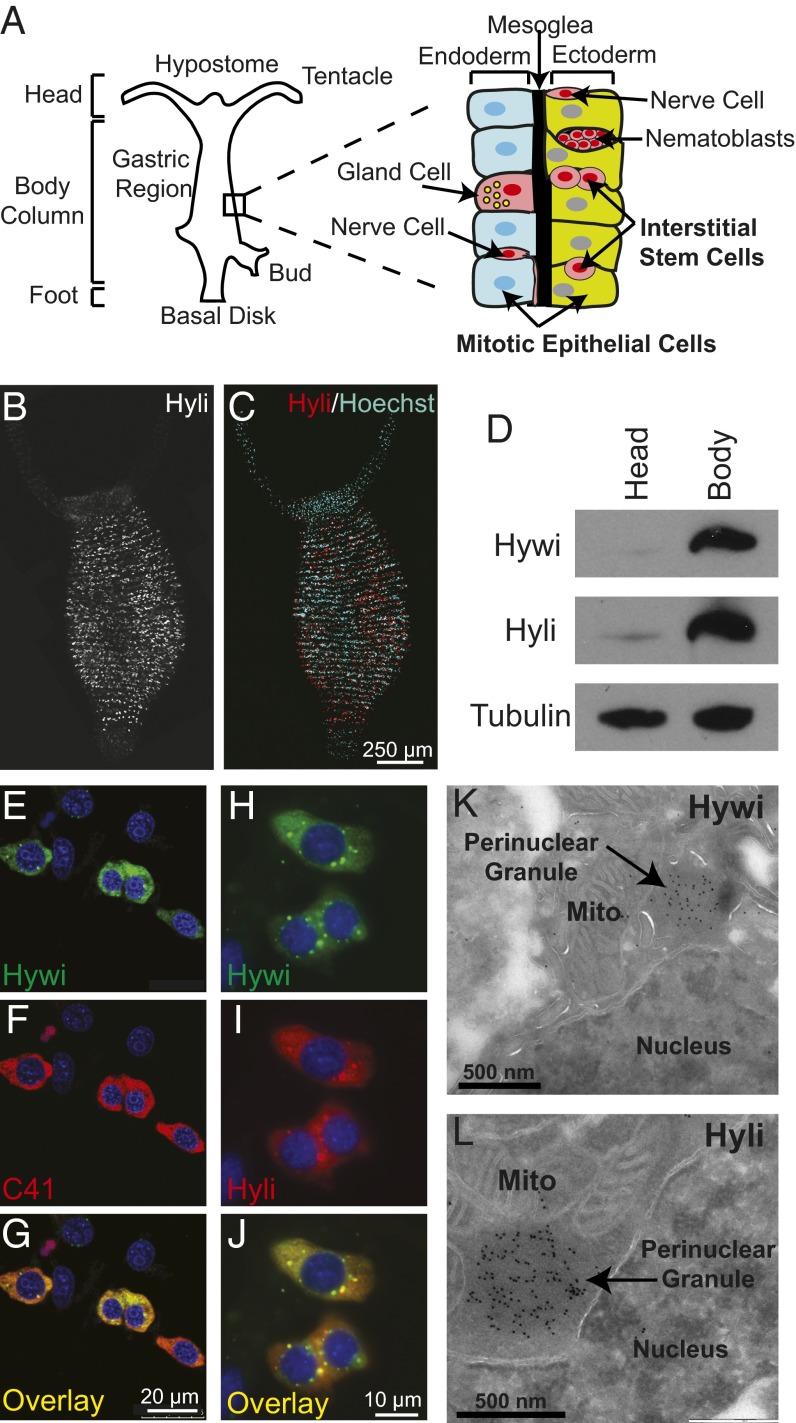
Fig. 1.
PIWI proteins are expressed in the interstitial stem cells and are enriched in perinuclear granules. (A) A schematic of Hydra showing that it is composed of three cell lineages. The ectodermal (green) and endodermal (blue) epithelial cell lineages form the inside and outside of the body column. All of the epithelial cells in the body column are mitotic and maintain the lineage (9). As they divide they are displaced toward the extremities where they become postmitotic, differentiate, and eventually are sloughed from the tips of the tentacles and the foot. The interstitial cell lineage (pink) consists of the interstitial stem cells, which give rise to the differentiated nerve cells, gland cells, nematocytes (from precursor nematoblast nests), and germ cells (8). The expression of Hydra PIWI protein, Hyli and Hywi, is restricted to the body column as shown by Hyli whole-mount immunofluorescence (B and C) and Hywi and Hyli immunoblot analysis (D). (E–G) Hywi (green) and Hyli (SI Appendix, Fig. S2 D–F) are expressed in the interstitial stem cells as shown by colabeling with the C41 antibody, which labels interstitial stem cells (red) (11). Hywi (green) and Hyli (red) proteins are distributed diffusely in the cytoplasm of interstitial stem cells, and are enriched in perinuclear granules, as demonstrated by immunofluorescence (H–J) and immunoelectron microscopy (K and L). DNA is labeled with Hoechst 33342.
Results
Hydra PIWI Proteins, Hywi and Hyli, Are Expressed in Multipotent Stem Cells.
Computational searches of the Hydra genome (10) revealed four Argonaute proteins: two Argonuate family proteins (Hy-ago1 and Hy-ago2) and two PIWI family proteins (SI Appendix, Fig. S1B). The Hydra PIWI family proteins were named Hydra PIWI (Hywi) and Hydra PIWI-like (Hyli) for their PIWI and PIWI-like orthologs (SI Appendix, Fig. S1B). We generated polyclonal antibodies against the N-terminal and mid domains of both Hywi and Hyli and demonstrated their specificity with immunoprecipitation experiments (SI Appendix, Fig. S1 C–G). The antibodies stained numerous cells throughout the body column, but not in the extremities (Fig. 1 B and C and SI Appendix, Fig S2 A–C). The restriction of Hywi and Hyli expression to the body column, where the stem/progenitor cells reside, was also seen by immunoblot analysis of body columns and heads (Fig. 1D). Colabeling with C41 antibody, an interstitial stem cell marker (11), demonstrated that Hywi and Hyli are expressed in interstitial stem cells (Fig. 1 E–G and SI Appendix, Fig. S2 D–F). In addition, both Hywi and Hyli are expressed in nematoblast nests, which are nematocyte progenitor cells of the interstitial lineage (SI Appendix, Fig. S3) (12, 13). Hywi and Hyli proteins are diffusely distributed in the cytoplasm of interstitial stem cells and are enriched in punctate foci around the nucleus (Fig. 1 H–J). Immuno-electron microscopy demonstrated that both Hywi and Hyli are associated with electron-dense perinuclear structures similar to what is seen in the germlines of several animals, including Drosophila, mice, and zebrafish (Fig. 1 K and L) (14–20).
Hywi and Hyli Accumulate in Perinuclear Granules of Epithelial Stem/Progenitor Cells.
Hywi and Hyli staining is prominent in the interstitial stem cells and nematoblasts (Fig. 1 E–G and SI Appendix, Fig S2 D—F and Fig. S3), but immunoblotting of Hydra that are depleted of the interstitial lineage revealed Hywi and Hyli accumulation outside this lineage (Fig. 2A). For more detailed cell type analysis, we used transgenic animals with lineage-specific GFP or Discosoma species Red2 (DsRed2) expression and dissociated whole animals into single cells for both immunoblotting and immunostaining (Fig. 2B) (21, 22). Both Hywi and Hyli were detected by immunoblotting in both ectodermal and endodermal cell populations isolated by FACS (Fig. 2C and SI Appendix, Fig. S4). Furthermore, both Hyli and Hywi proteins accumulate in puncta around the nuclei of ectodermal and endodermal epithelial cells, but do not accumulate significantly elsewhere in the cytoplasm (Fig. 2 E–J and SI Appendix, Fig. S2 G–J). Immunostaining experiments revealed that both Hywi and Hyli are absent from the nucleus (Fig. 1 H–J and Fig. 2 E–J). This is in contrast to PIWI proteins in Drosophila and the mouse, some of which are nuclear and likely act as epigenetic regulators (23–25). To test if the cytoplasmic localization of Hywi and Hyli in situ is due to antigen masking or low abundance in the nucleus, we analyzed nuclear and cytoplasmic fractions by immunoblotting and found that Hywi and Hyli are apparently exclusively cytoplasmic (Fig. 2D).
Fig. 2.
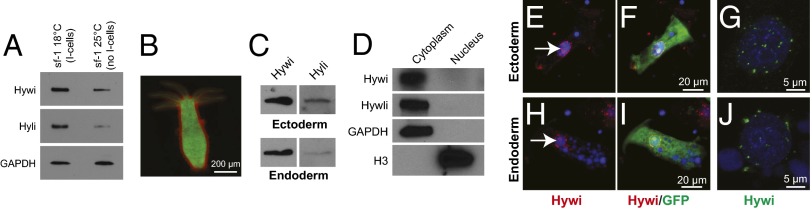
PIWI proteins are cytoplasmic and expressed in the mitotically active somatic epithelial cells. (A) Expression of Hywi and Hyli protein is detected by immunoblot in epithelialized mutant strain (sf-1) Hydra, which lose the interstitial lineage when cultured at 25 °C (46, 47). (B) To test for epithelial expression of Hywi and Hyli, ectodermal cells expressing DsRed2 and endodermal cells expressing GFP were isolated by FACS (SI Appendix, Fig. S4) and subjected to immunoblot analysis with Hywi and Hyli antibodies. (C) Hywi and Hyli were detected in both the ectodermal and endodermal epithelial cells. (D) To test the subcellular localization of Hywi and Hyli, nuclear (histone H3) and cytoplasmic (GAPDH) fractions were probed with the Hywi and Hyli antibodies; both proteins were detected selectively in the cytoplasmic fractions. To determine the subcellular localization of Hywi in epithelial cells, staining was performed on transgenic Hydra that express GFP in either the ectodermal (E–G) or endodermal (H–J) epithelial cells. (E–J) Hywi accumulates in perinuclear granules (arrows in E and H) in ectodermal (E–G) and endodermal (H–J) epithelial cells. (G and J) Hywi-positive granules are detected around the nucleus of epithelial cells in confocal Z-stack projections. (E–J) DNA is labeled with Hoechst 33342; vacuoles in endodermal cells (H and I) are also Hoechst-positive.
Isolation and Characterization of Hydra piRNAs Reveals Conserved Mechanisms of piRNA Biogenesis.
To investigate the function of the PIWI–piRNA pathway in Hydra, piRNAs bound to Hywi and Hyli were isolated by immunoprecipitation and sequenced (SI Appendix, Fig. S5A). Analysis of the size distribution revealed that Hywi and Hyli bind piRNAs of different sizes, which is consistent with PIWI proteins in Drosophila, mice, and zebrafish (Fig. 3A) (14, 16, 23). Over 90% of piRNAs bound to Hywi have a uridine at their 5′ end (SI Appendix, Fig. S5D), and over 80% of piRNAs bound to Hyli have an adenine at their 10th position (SI Appendix, Fig. S5D). Furthermore, we found a complementary 10-base pair overlap between the 5′ ends of Hywi-bound and Hyli-bound piRNAs (Fig. 3B). These features are identical to the ping-pong signature of biogenesis that was first described in Drosophila (14, 26) and also observed in mice and zebrafish (16, 23, 27). Previous sequencing of total Nematostella vectensis and Hydra RNAs identified putative piRNAs (28, 29). Here we have identified bona fide cnidarian piRNAs bound to specific PIWI proteins, thus allowing for comparisons between piRNAs bound to different PIWI proteins. Finally, we show that Hydra piRNAs are 2′-O-methylated at their 3′ ends similar to bilaterian piRNAs (SI Appendix, Fig S5E) (30, 31). Our data definitively demonstrate that Hywi and Hyli participate in ping-pong biogenesis and prove that this mechanism has a deep evolutionary origin in metazoans.
Fig. 3.
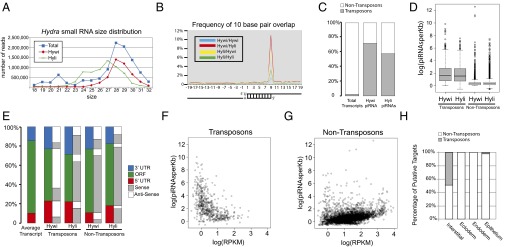
Sequencing and mapping of Hywi- and Hyli-bound piRNAs reveals conserved mechanisms of piRNA biogenesis and candidate posttranscriptional targets. Small RNAs isolated by size and piRNAs bound to Hywi or Hyli were sequenced. (A) Analysis of the size distribution of total small RNAs (blue squares) in Hydra reveals a peak at 21 nucleotides in length and a peak at 28 nucleotides in length. piRNAs bound to Hywi (red diamonds) have a peak at 28 nucleotides, and piRNAs bound to Hyli (green rectangles) have a peak at 27 nucleotides. (B) Hywi- and Hyli-bound piRNAs have a high frequency of complementary overlap 10 bases from their 5′ end. (C) A Hydra transcriptome was assembled and piRNAs were mapped to it. Transposon sequences represent 2.2% of the sequences in the transcriptome. By contrast, 72% of mapping Hywi-bound piRNAs and 58% of mapping Hyli-bound piRNAs map to transposons. (D) A box and whisker plot analyzing the number of piRNAs mapping per kilobase of sequence demonstrated that, on average, 46 Hywi-bound piRNAs and/or 36 Hyli-bound piRNAs mapped to each kilobase of transposon transcript. By contrast, nontransposon transcripts have on average only two piRNAs mapping per kilobase, but 371 and 536 transcripts have more than 10 Hywi- or Hyli-bound piRNAs mapped per kilobase, respectively (with 188 transcripts common to both populations). (E) piRNAs mapping to the UTRs are slightly overrepresented compared with the coding region per kilobase of transcript; the architecture of the average transcript in the assembly is represented by the first bar. For transposon transcripts, the majority of Hywi-bound piRNAs map in the antisense orientation (white), and the majority of Hyli-bound piRNAs map in the sense orientation (gray). The majority of both Hywi- and Hyli-bound piRNAs that map to nontransposon transcripts map in the sense orientation (see SI Appendix, Table S1 for percentages). (F and G) Number of piRNAs bound per transcript (normalized by transcript size) as a function of transcript abundance as measured by RPKM (reads per kilobase per million reads) value. (F) The majority of transposon transcripts are in low abundance and have a high number of piRNAs mapping to them. (G) By contrast, the majority of nontransposon transcripts show a correlation between transcript abundance and the number of piRNAs mapping to them, suggesting that some transcripts may be processed due to their abundance. (H) To identify lineage-specific targets of the PIWI–piRNA pathway, small RNAs were isolated from FACS separated interstitial, ectodermal, and endodermal lineages. Small RNAs greater than 23 nucleotides long were mapped to the transcriptome. Approximately 50% of the putative targets in the interstitial lineage are transposons, whereas no transposon targets were identified as specific to the ectoderm or endoderm. One putative transposon target was identified in the epithelium (combination of ectoderm and endoderm) that is enriched over the interstitial lineage.
The Hydra PIWI–piRNA Targets Transposon Transcripts.
The prevailing model posits that ping-pong piRNA biogenesis results in decreased transposon expression due to posttranscriptional processing of transposon RNAs into piRNAs (14). To test if Hywi and Hyli function in posttranscriptional transposon repression, we first mapped the piRNAs to the Hydra genome (10). Approximately 50% of the sequenced piRNAs were mapped to unique sites in the Hydra genome. From 55% to 65% of Hydra piRNAs map to repeat sequences that were previously identified by RepeatMasker (SI Appendix, Fig. S5 B and C). Because the total repeat content in the Hydra genome is 57%, this mapped population of piRNAs is not significantly enriched for repeat sequences (10).
To better characterize the piRNA targets in Hydra, we focused our attention on transcripts that are expressed in the adult. To this end, we sequenced and assembled a Hydra transcriptome containing ∼27,000 sequences, which we curated to obtain a set of 9,986 transcripts with a significant BLAST (1×e−5) match to the Swiss–Prot database. This allowed for definition of ORFs and transcript orientation. Of the curated transcriptome dataset, 622 transcripts were identified as arising from transposons by BLAST (1×e−5) analysis against the Hydra transposons in Repbase. Of our sequenced piRNAs, 1.7 million mapped to the transcriptome when allowing up to a three-base pair mismatch. Among these, 72% of Hywi-bound piRNAs and 58% of Hyli-bound piRNAs map to transposon transcripts, which is a significant enrichment over the abundance of transposons in the transcriptome (Fig. 3C). Furthermore, significantly more piRNAs map per transposon transcript than per nontransposon transcript (Fig. 3D). The majority of Hywi-bound piRNAs map to transposons in the antisense orientation, whereas the Hyli-bound piRNAs map largely in the sense orientation (Fig. 3 E and F and SI Appendix, Table S1); this sense/antisense bias is consistent with the ping-pong model for piRNA biogenesis and posttranscriptional repression of transposons (14). Although the majority of transposons are lowly expressed, they have a high number of piRNAs mapping to their transcripts (Fig. 3 D and F). This is also consistent with the ping-pong model, which posits that transposon mRNAs are repressed by processing them into piRNAs (14). Taken together, these data strongly suggest that one role of the Hydra Piwi–piRNA pathway is to regulate transposon expression.
Identification of Candidate Nontransposon PIWI–piRNA Pathway Targets.
The processing of mRNAs into piRNAs is also a possible mechanism of posttranscriptional repression for nontransposon genes. We found that both Hywi- and Hyli-bound piRNAs predominantly map to the nontransposon genes of the transcriptome in the sense orientation, which suggests that piRNAs are being made from these transcripts, similar to observations in Drosophila and mice (Fig. 3E and SI Appendix, Table S1) (32). A group of nontransposon transcripts with more than 10 piRNAs mapping per kilobase were selected as putative targets and subjected to gene ontology (GO) analysis (Fig. 3D and SI Appendix, Table S2 and Table S3). We find significant differences in the enriched GO categories between transcripts with high numbers of Hywi piRNAs mapping to them compared with those with high numbers of Hyli piRNAs mapping to them. This suggests selectivity in the mRNAs that are processed into piRNAs. However, we also found a correlation between the expression level of nontransposon transcripts and the number of piRNAs mapped to them (Fig. 3G). Therefore, some piRNA production may occur from highly expressed transcripts simply due to their high abundance.
To test if the PIWI–piRNA pathway in Hydra has targets that are specific to each developmental lineage, we isolated each lineage by FACS for small RNA sequencing. Transgenic Hydra were used that express GFP in the endoderm and DsRed2 in the ectoderm (Fig. 2B and SI Appendix, Fig. S4A). The interstitial lineage was collected as the population of cells without fluorescence (SI Appendix, Fig. S4A). We found that the most abundant small RNAs in the interstitial lineage are between 26 and 32 nucleotides in length, with a peak at 28. By contrast, in both the ectodermal and endodermal lineages, the most abundant small RNAs are between 26 and 34 nucleotides, with a peak at 32 (SI Appendix, Fig. S6A). For all three lineages, there is a bias for uridine at the 5′ end of small RNAs between 26 and 34 nucleotides long (SI Appendix, Fig. S6B). To test for potential lineage-specific targets of the PIWI–piRNA pathway, we mapped small RNAs greater than 23 nucleotides from each lineage to the transcriptome. Transcripts that had at least 10 times more mapped piRNAs from one lineage compared with the other two lineages were considered putative lineage-specific targets. Approximately 50% of the targets specific to the interstitial lineage are transposons, whereas only one putative transposon target was enriched in epithelial cells (Fig. 3H). Generally, more piRNAs from the interstitial lineage map to transposons in the transcriptome compared with piRNAs from the epithelial lineages, and this trend was not observed for nontransposon transcripts (SI Appendix, Fig. S6 D and E). These data suggest that transposon regulation is largely specific to the interstitial lineage, which is further supported by the observation that the ping-pong biogenesis signature is significantly stronger in the interstitial lineage (SI Appendix, Fig. S6C). In addition, we identified putative nontransposon targets and subjected these to gene ontology analysis; the results strongly suggest that the pathway has specific functions in each lineage (SI Appendix, Table S4).
Hywi Has an Essential Function in Hydra Epithelial Cells.
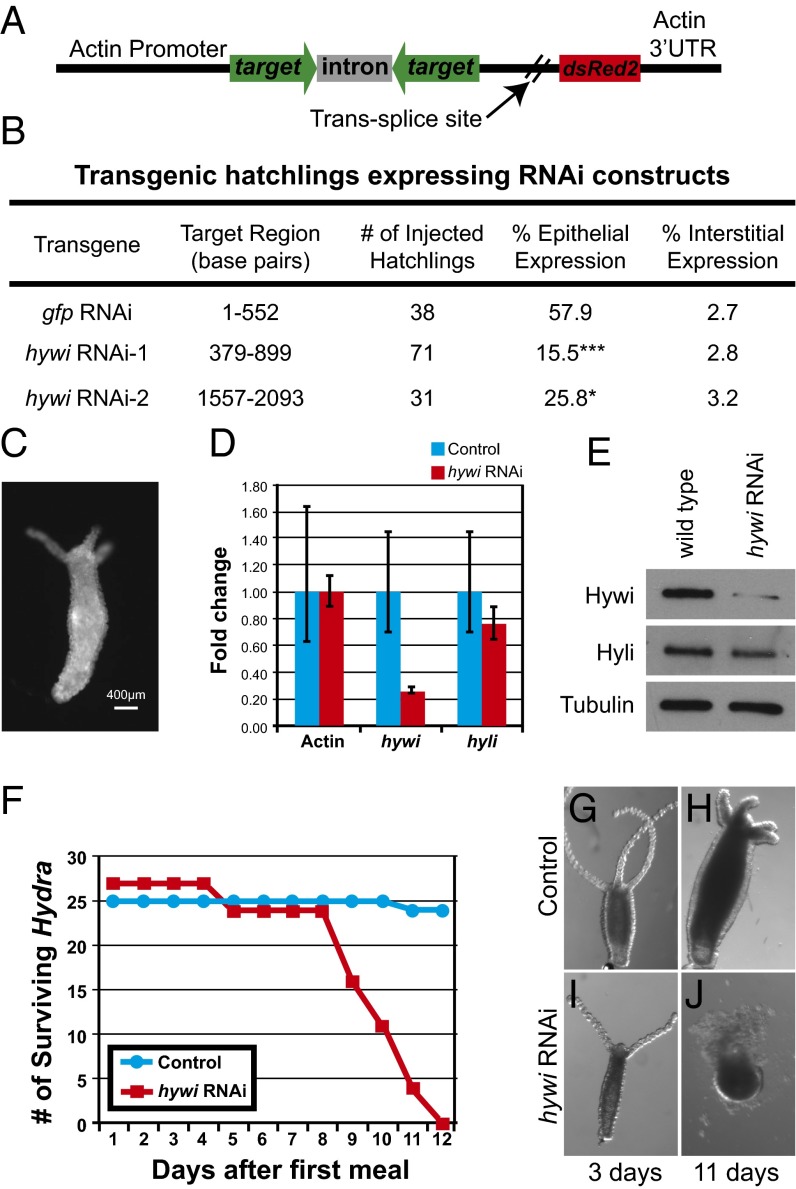
To gain insight into the function of the PIWI–piRNA pathway in Hydra somatic cells, we sought to knock down hywi expression in the epithelial lineages. We modified our previously described operon vector by placing an RNA hairpin in the upstream position and the DsRed2 gene in the downstream position to mark transgenic cells (Fig. 4A) (21). Expression of the two genes is driven by an actin promoter that is not active in the interstitial stem cells, but is active in the differentiated cells of the interstitial lineage and throughout the ectodermal and endodermal lineages (SI Appendix, Fig. S7 A–C). Therefore, the RNAi transgene is predicted to affect hywi expression in the epithelial cell lineages, but not the interstitial lineage. Injection of plasmid DNA into early Hydra embryos results in random integration and the generation of mosaic patches of stably transgenic tissue (33). We tested two different constructs targeting hywi and one control construct with a hairpin from the GFP gene. Hatchlings from these injections were scored for transgene (DsRed2) expression in the epithelial cells (Fig. 4B). Fifty-eight percent of control hatchlings showed DsRed2 expression in the epithelial cells, whereas significantly fewer hatchlings from the hywi RNAi injections showed DsRed2 expression in the epithelial cells (15.5% and 25.8%; Fig. 4B). By contrast, the hywi RNAi and control transgenes were integrated into the interstitial lineage at the same rate (Fig. 4B). Fully transgenic ectodermal or endodermal lines are established by asexual propagation and continual selection of buds with the most transgenic tissue (33). From the initial hatchlings expressing the GFP control transgene, we established lines that are fully transgenic in the ectoderm or endoderm; thus, the control transgene does not negatively affect either tissue. However, we were unable to establish lines with hywi knocked down in the ectoderm or endoderm. These data suggest that hywi is essential in the epithelial lineages.
Fig. 4.
Hywi has an essential function in Hydra epithelial cells. (A) Schematic of the RNAi construct used to knock down hywi in the epithelial cells (ectoderm and endoderm). Expression of the transgene is driven by an actin promoter, which is active in all cells except the interstitial stem cells (SI Appendix, Fig. S7 A–C). The RNA hairpin (inverted repeats separated by an actin intron spacer) and the DsRed2 transcript are arranged in an operon configuration that is spliced apart after transcription. (B) Hydra embryos were injected with control (gfp RNAi) and hywi RNAi knockdown plasmids, and the percentage of injected hatchlings that have transgene expression was quantified. Significantly fewer hatchlings expressed the hywi knockdown transgene in the epithelium compared with the control; the P values for epithelial expression of the hywi RNAi constructs are 0.0001 (hywi RNAi-1) and 0.04 (hywi RNAi-2). (C) The hywi RNAi-1 transgene was stably incorporated into the interstitial lineage and underwent germline transmission (SI Appendix, Fig S8 D–F). This resulted in F1 Hydra expressing the transgene in both the ectodermal and endodermal epithelial layers, but not the interstitial stem cells (SI Appendix, Fig S8 G–M). (D) By qRT-PCR, the hywi mRNA levels (normalized to GAPDH) are reduced by ∼80% in transgenic F1 hatchlings (samples taken 7 d after hatchling), and (E) the protein levels are reduced. (D and E) By contrast, hyli RNA and protein levels are similar between control and hywi knockdown F1 animals. (F) hywi knockdown F1 animals all die between 8 and 12 d after eating their first meal, whereas control hatchlings are normal. (G and H) Control F1 animals (nontransgenic siblings) look normal after 11 d. (I and J) hywi knockdown F1 animals are initially normal, but lose epithelial integrity between 8 and 12 d after eating.
We established three lines expressing either the hywi RNAi-1 or the hywi RNAi-2 construct in the interstitial lineage (as observed by fluorescence in the differentiated cells) (Fig. 4B and SI Appendix, Fig. S7D). In one of these lines, the hywi RNAi-1 transgene is regularly transmitted through the germline, which results in F1 hatchlings that are uniformly transgenic in both the endodermal and ectodermal epithelial layers (Fig. 4C and SI Appendix, Fig. S7 E and F). Both quantitative RT-PCR (qRT-PCR) and Western blot analysis of transgenic F1 hatchlings demonstrated significant down-regulation of hywi compared with nontransgenic F1 siblings (Fig. 4 D and E and SI Appendix, Fig. S7M). By contrast, the RNA and protein levels of hyli are not significantly affected (Fig. 4 D and E). Hywi is not detected in the epithelial cells by immunostaining, but is still detected in interstitial stem cells as expected (SI Appendix, Fig. S7 G–L). Hywi knockdown F1 hatchlings initially appear normal and eat shortly after hatching similar to nontransgenic F1 control siblings (Fig. 4 G and I). However, the hywi knockdown F1 hatchlings begin to lose epithelial integrity as early as 5 d, and die between 8 and 12 d after their first meal (Fig. 4 F, H, and J). A small number of both control and knockdown hatchlings never eat; all of these animals die of starvation rather than loss of epithelial integrity. These observations provide further evidence that hywi is an essential gene in the somatic epithelium of this organism.
Discussion
The PIWI–piRNA pathway is best known for repressing transposon expression in the germline to maintain genomic integrity (7). In this study we report that Hydra PIWI proteins accumulate in the cytoplasm of all stem/progenitor cells of the adult and are essential for the animal. These data reveal crucial functions of the PIWI proteins beyond transposon silencing and strongly suggest that the primary function of the PIWI–piRNA pathway in Hydra stem cells is in posttranscriptional regulation. These data also imply that cytoplasmic function of the pathway is primitive and that nuclear function is derived, although sampling from more nonbilaterian taxa is required before definitive conclusions can be made. Beyond a handful of well-studied bilaterian models, very little is known about the localization of PIWI proteins (reviewed in ref. 34). Interestingly, Drosophila Piwi protein and the zebrafish PIWI protein Zili can be nuclear or cytoplasmic depending on the developmental stage (16, 35). Thus, it is possible that during Hydra embryonic development either Hywi or Hyli has a nuclear function. Nonetheless, our data point to a conserved broader functional importance for this pathway in the cytoplasm of adult stem cells.
We found that the subpopulation of piRNAs that map to the transcriptome are highly enriched for transposon transcripts, which is in contrast to no enrichment for transposon/repeat sequences when we map the total population of piRNAs to the genome. Aside from Drosophila, genomic mapping of putative piRNAs in several other organisms also revealed very little enrichment for repeat sequences over total genomic content (reviewed in ref. 34). Therefore, when considering piRNAs associated with cytoplasmic PIWI proteins, our transcriptome-mapping approach may be preferable to genomic mapping for drawing conclusions about the function of the pathway. Our results support the conclusion that the PIWI–piRNA pathway targets transposon mRNAs via a cytoplasmic pathway in Hydra. This function appears to be largely specific to the interstitial lineage, which is of particular interest because this lineage is capable of giving rise to the germline. Thus, the control of transposon expression is likely an ancient function of the PIWI–piRNA pathway in germ cells.
In addition to transposon expression, putative nontransposon targets were identified in the interstitial lineage including several involved in cell cycle regulation (SI Appendix, Table S4). This is consistent with studies in Drosophila GSCs and in mouse mesenchymal stem cells that demonstrate a potential role for the pathway in controlling cell division (6, 36, 37). Our data also suggest that the PIWI–piRNA pathway has an essential function in the two strictly somatic epithelial lineages, which is likely due to a function in regulating nontransposon genes. The putative targets in the ectoderm are enriched for genes encoding cell adhesion proteins and extracellular matrix components (ECMs) (SI Appendix, Table S4). In the endoderm there is an enrichment of both ECM and proteolysis genes among putative targets. The misregulation of the genes in these categories may lead to the defects observed in the hywi knockdown hatchlings, perhaps due to loss of epithelial integrity.
The presence of genes with shared expression in the germline and in stem cells has led to speculation that these cells have a common evolutionary origin, with germ cells arising as a lineage-restricted stem cell population (38, 39). In addition, several lines of evidence suggest that germline genes are also more broadly expressed in metazoan stem cells. For example, piwi, vasa, and nanos are expressed and often required in many multipotent and pluripotent stem cells, both with and without germline potential (2, 40). A handful of expression studies in ctenophores and cnidarians reveal piwi expression in somatic stem and/or progenitor cells, which suggests an ancient role for PIWI in stem cell regulation (3, 4, 40). Our study provides a comprehensive analysis of PIWI proteins and piRNAs in a cnidarian. These data strongly suggest that the PIWI–piRNA pathway has ancient and conserved stem cell functions beyond the germline and sets the stage for a mechanistic understanding of the pathway in adult somatic stem cells.
Materials and Methods
Animals and Culturing Conditions.
Hydra magnipapillata strain 105 and Hydra vulgaris strain AEP were cultured by standard procedures (41). See SI Appendix, SI Materials and Methods for details.
Hywi and Hyli Antibody Generation.
His-tagged recombinant proteins were made to raise antisera in rabbits (Hywi) or guinea pigs (Hyli). See SI Appendix, SI Materials and Methods for details on protein purification, antibody purification, immunoblotting procedures, immunofluorescence procedures, and immunoelectron microscopy.
Nuclear-Cytoplasmic Fractionation.
Fractionation was done using the ProteoExtract Subcellular Proteome Extraction Kit 539790. See SI Appendix, SI Materials and Methods for details.
FACS.
For small RNA sequencing, animals were prepared as previously described (42). For immunoblot analysis, transgenic Hydra were dissociated with 0.25% Trypsin–EDTA solution. See SI Appendix, SI Materials and Methods for details.
Immunoprecipitation and piRNA Sequencing.
TRIzol-LS was added directly to the Protein A bead/antibody complexes to isolate total RNA. Small RNA libraries were prepared using Illumina Small RNA Preparation Kit v1.5 following the manufacturer’s protocol. Libraries were gel-purified and sequenced using the Genome Analyzer II. See SI Appendix, SI Materials and Methods for further details about procedures, bioinformatics analysis, and genomic mapping of piRNAs.
Sequencing of Lineage-Specific Small RNAs.
Each lineage was collected by FACS, and RNA was isolated using TRIzol and used to generate small RNA libraries using the TruSeq Small RNA Sample Prep Kit according to the manufacturer’s protocol. The libraries were sequenced using the HiSeq 2000. See SI Appendix, SI Materials and Methods for details.
Assembly of the Hydra Transcriptome and Small RNA Mapping.
The transcriptome was assembled using a previously described pipeline (43). piRNA and lineage-specific small RNA mapping was done using Bowtie 0.1.0 (44). Gene ontology analysis of putative PIWI–piRNA pathway targets was done using The Database for Annotation, Visualization and Integrated Discovery (DAVID) (45). See SI Appendix, SI Materials and Methods for details.
Generation of Transgenic Hydra.
The generation of transgenic Hydra was performed as previously described (21, 33). See SI Appendix, SI Materials and Methods for details on plasmid construction and injection methods.
Note Added in Proof.
While this paper was in production, a report on Hydra Piwi proteins and piRNAs by Lim et al. was also accepted for publication by Developmental Biology (48).
Supplementary Material
Acknowledgments
We thank Catherine Dana for excellent assistance with Hydra methods, Ee-Chun Cheng for assistance with FACS, and Grahm Morven for performing immuno-EM. We thank Sneha Mani, Stefan Materna, S. Zachary Swartz, and James Gagnon for critical feedback on the manuscript and members of the H.L., R.E.S., and G.M.W. labs for helpful research discussions. This work was supported by a G. Harold & Leila Y. Mathers Award (to H.L.), National Institutes of Health (NIH) Grant R24 GM080527 (to R.E.S.), NIH Grant 2R01HD028152 and a National Science Foundation Grant IOS-1120972 (to G.M.W.), and an Ellison Senior Scholar Award (to R.A.R.). C.E.J. was a National Research Service Award Postdoctoral Fellow (NIH F32GM9037222).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (NCBI BioProject no. PRJNA213706).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320965111/-/DCSupplemental.
References
- 1.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juliano C, Wang J, Lin H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alié A, et al. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: Ancient association of “germline genes” with stemness. Dev Biol. 2011;350(1):183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Seipel K, Yanze N, Schmid V. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol. 2004;48(1):1–7. doi: 10.1387/ijdb.15005568. [DOI] [PubMed] [Google Scholar]
- 5.Sharma AK, et al. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97(2):426–434. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- 6.Wu Q, et al. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem Biophys Res Commun. 2010;396(4):915–920. doi: 10.1016/j.bbrc.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 8.David CN. Interstitial stem cells in Hydra: Multipotency and decision-making. Int J Dev Biol. 2012;56(6-8):489–497. doi: 10.1387/ijdb.113476cd. [DOI] [PubMed] [Google Scholar]
- 9.Holstein TW, Hobmayer E, David CN. Pattern of epithelial cell cycling in hydra. Dev Biol. 1991;148(2):602–611. doi: 10.1016/0012-1606(91)90277-a. [DOI] [PubMed] [Google Scholar]
- 10.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464(7288):592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David CN, Fujisawa T, Bosch TC. Interstitial stem cell proliferation in hydra: Evidence for strain-specific regulatory signals. Dev Biol. 1991;148(2):501–507. doi: 10.1016/0012-1606(91)90268-8. [DOI] [PubMed] [Google Scholar]
- 12.Bosch T, David C. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol. 1987;121(1):182–191. [Google Scholar]
- 13.David CN, Murphy S. Characterization of interstitial stem cells in hydra by cloning. Dev Biol. 1977;58(2):372–383. doi: 10.1016/0012-1606(77)90098-7. [DOI] [PubMed] [Google Scholar]
- 14.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 15.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27(20):2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129(1):69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 19.Unhavaithaya Y, et al. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284(10):6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19(8):640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dana CE, Glauber KM, Chan TA, Bridge DM, Steele RE. Incorporation of a horizontally transferred gene into an operon during cnidarian evolution. PLoS ONE. 2012;7(2):e31643. doi: 10.1371/journal.pone.0031643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glauber KM, et al. A small molecule screen identifies a novel compound that induces a homeotic transformation in Hydra. Development. 2013;140(23):4788–4796. doi: 10.1242/dev.094490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang XA, et al. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24(5):502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 27.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 28.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishna S, et al. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Res. 2013;41(1):599–616. doi: 10.1093/nar/gks1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohara T, et al. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14(4):349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 31.Saito K, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21(13):1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robine N, et al. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol. 2009;19(24):2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103(16):6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani SR, Juliano CE. Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev. 2013;80(8):632–664. doi: 10.1002/mrd.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 36.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 38.Agata K, et al. Two different evolutionary origins of stem cell systems and their molecular basis. Semin Cell Dev Biol. 2006;17(4):503–509. doi: 10.1016/j.semcdb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Extavour CG. Evolution of the bilaterian germ line: Lineage origin and modulation of specification mechanisms. Integr Comp Biol. 2007;47(5):770–785. doi: 10.1093/icb/icm027. [DOI] [PubMed] [Google Scholar]
- 40.Juliano C, Wessel G. Developmental biology. Versatile germline genes. Science. 2010;329(5992):640–641. doi: 10.1126/science.1194037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenhoff HM, Brown RD. Mass culture of hydra: An improved method and its application to other aquatic invertebrates. Lab Anim. 1970;4(1):139–154. doi: 10.1258/002367770781036463. [DOI] [PubMed] [Google Scholar]
- 42.Hemmrich G, et al. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012;29(11):3267–3280. doi: 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 43.Dunn CW, Howison M, Zapata F. Agalma: an automated phylogenomics workflow. BMC Bioinformatics. 2013;14(1):330. doi: 10.1186/1471-2105-14-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 46.Marcum BA, Fuijsawa T, Sugiyama T. A mutant strain (sf-1) containing temperature-sensitive interstitial cells. In: Tardent P, Tardent R, editors. Developmental and Cellular Biology of Coelenterates. Amsterdam: Elsevier; 1980. pp. 429–434. [Google Scholar]
- 47.Terada H, Sugiyama T, Shigenaka Y. Genetic analysis of developmental mechanisms in hydra. XVIII. Mechanism for elimination of the interstitial cell lineage in the mutant strain Sf-1. Dev Biol. 1988;126(2):263–269. doi: 10.1016/0012-1606(88)90137-6. [DOI] [PubMed] [Google Scholar]
- 48.Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2013 doi: 10.1016/j.ydbio.2013.12.007. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.