Significance
The pineal gland is dedicated to the production of melatonin. Submammalian pineal glands can also detect light, and the retinas of many species can make melatonin. From this finding and others, it is seems that both tissues evolved from a common ancestral photodetector. A key factor driving their independent evolution may have been the evolution of melatonin synthesis and more specifically, the timezyme, a form of arylalkylamine N-acetyltransferase (AANAT) that plays a key role in controlling rhythmic production of melatonin. The current report indicates that the timezyme evolved from a primitive form of AANAT over 500 Mya in chordate evolution through a process of gene duplication followed by rapid neofunctionalization and that it was not a posthoc acquisition.
Abstract
Melatonin (N-acetyl-5-methoxytrypamine) is the vertebrate hormone of the night: circulating levels at night are markedly higher than day levels. This increase is driven by precisely regulated increases in acetylation of serotonin in the pineal gland by arylalkylamine N-acetyltransferase (AANAT), the penultimate enzyme in the synthesis of melatonin. This unique essential role of AANAT in vertebrate timekeeping is recognized by the moniker the timezyme. AANAT is also found in the retina, where melatonin is thought to play a paracrine role. Here, we focused on the evolution of AANAT in early vertebrates. AANATs from Agnathans (lamprey) and Chondrichthyes (catshark and elephant shark) were cloned, and it was found that pineal glands and retinas from these groups express a form of AANAT that is compositionally, biochemically, and kinetically similar to AANATs found in bony vertebrates (VT-AANAT). Examination of the available genomes indicates that VT-AANAT is absent from other forms of life, including the Cephalochordate amphioxus. Phylogenetic analysis and evolutionary rate estimation indicate that VT-AANAT evolved from the nonvertebrate form of AANAT after the Cephalochordate–Vertebrate split over one-half billion years ago. The emergence of VT-AANAT apparently involved a dramatic acceleration of evolution that accompanied neofunctionalization after a duplication of the nonvertebrate AANAT gene. This scenario is consistent with the hypotheses that the advent of VT-AANAT contributed to the evolution of the pineal gland and lateral eyes from a common ancestral photodetector and that it was not a posthoc recruitment.
Eyes have evolved in all animals to facilitate interactions with the photic environment (1, 2). However, among animals, vertebrates are unique in that they also possess a photoneuroendocrine structure, the pineal gland (3). It converts the 24-h rhythm in environmental lighting into a 24-h rhythm in circulating melatonin, thereby providing a unique and valuable signal of the photic environment. The details of pineal evolution are not clear (4, 5). However, it has been posited that an essential element was arylalkylamine N-acetyltransferase (AANAT; E.C. 2.3.1.87), the penultimate enzyme in the melatonin biosynthesis pathway (6–8); this scenario is referred to as the AANAT hypothesis of pineal evolution (7, 8).
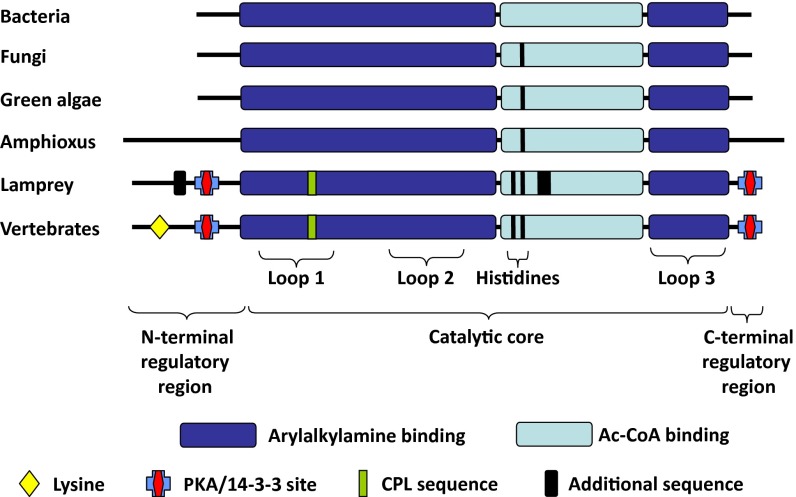
AANAT catalyzes the N-acetylation of arylalkylamines using acetyl CoA (AcCoA) as the acetyl group donor. The AANAT family, which belongs to the GCN5 superfamily (9, 10), is composed of two subfamilies termed vertebrate (VT) AANAT and nonvertebrate (NV) AANAT.† This nomenclature reflects the phylogenetic distribution of the family members (13–17). The most striking differences between VT- and NV-AANAT are found in regulatory and catalytic regions of the encoded proteins (Fig. 1), consistent with different metabolic roles (7, 8).
Fig. 1.
Schematic organization of VT- and NV-AANATs. Major structural differences between NV- and VT-AANAT include the addition of PKA/14-3-3 binding sites flanking the core of the enzyme, which mediate regulation, the addition of histidines to facilitate catalysis by promoting removal of protons generated during the transfer of the acetyl group, and the addition of a short peptide sequence (Cys-Pro-Leu), which creates a floppy loop that facilitates arylalkylamine binding and product release in the active site. The highly conserved lysine found in the N-terminal flanking regions of VT-AANAT is thought to facilitate regulation of proteasomal proteolysis by serving as a ubiquitination site (10, 13, 14, 25, 26, 28, 29). Modified from ref. 51.
The NV-AANAT is thought to perform a detoxification function through acetylation of a broad range of endogenous and exogenous arylalkylamines and polyamines (13–16). It has been found in Gram-positive bacteria, protists, fungi, some lower plants, algae, and cephalochordates. The broad detoxification role of NV-AANAT contrasts with the dedicated role that VT-AANAT plays in melatonin synthesis, which was seen in teleosts and tetrapods (6, 18).
Melatonin is recognized as the vertebrate hormone of the night, because circulating levels are markedly elevated at night. In bony fish and tetrapods, VT-AANAT is highly expressed in the pineal gland, the primary source of circulating melatonin (6, 18). The unique and essential role that VT-AANAT plays in vertebrate timekeeping is recognized by the moniker the timezyme (10). VT-AANAT is also found in the retina, where N-acetylserotonin and/or melatonin may play paracrine roles (19).
The AANAT hypothesis of pineal evolution holds that AANAT originally served a detoxification function in an ancestral photodetector and that its function evolved from broad acetylation of potentially toxic amines into selective acetylation of serotonin and the production of melatonin (7, 8). According to this hypothesis, as melatonin became recognized as a hormonal signal of darkness, the conflict of selective pressures to optimize both photodetection and melatonin signaling was resolved by the cellular separation of these functions and resulted in the independent evolution of the pinealocyte and retina.
There is, however, no evidence available to determine whether VT-AANAT coevolved with the eyes and pineal gland or whether it was recruited after these structures evolved. The VT-AANAT is absent from the genomes of the urochordate Ciona intestinalis (sea squirt) and the cephalochordate Branchiostoma floridae (amphioxus), suggesting that VT-AANAT appeared after the split of these lineages from vertebrates. In the current report, we sought to determine whether VT-AANAT is present in representatives of early divergent vertebrate classes of Agnathans (jawless vertebrates: lampreys) and Chondrichthyes (cartilaginous fish: chimaeras and sharks), which share a common ancestor dating from nearly 500 Mya in the early Cambrian period (20, 21). The results provide clues to how and when VT-AANAT evolved and address the validity of the AANAT hypothesis of pineal evolution.
Results
VT-AANAT Is Present in Agnathans and Chondrichthyes.
Sequence analysis.
VT-AANAT cDNA was cloned from retinal cDNA libraries of the catshark Scyliorhynus canicula (1,708 bp; GenBank accession no. EU378921) and the elephant shark Callorhinchus milii (624 bp; GenBank accession no. KF246475) as described in Materials and Methods. The encoded proteins (206 and 207 aa) exhibited the two defining features of the GCN5 N-acetyltransferase superfamily, motifs A and B (SI Appendix, Fig. S1) (9, 22–29). In addition, the sequences exhibited defining characteristics of the VT-AANATs of bony vertebrates, including motifs C and D, PKA sites nested in 14-3-3 binding domains, a Cys-Pro-Leu peptide sequence in a floppy loop within the binding region, and closely positioned histidine residues in the active site. An additional feature of VT-AANATs (a lysine located close to the N terminus) was also found in AANATs cloned from Chondrichthyes. Altogether, the VT-AANAT sequences of Chondrichthyes were similar in length to most AANATs of bony vertebrates, including teleost AANAT1a and AANAT2, and other tetrapod AANATs, with 60–73% sequence identity to other VT-AANATs.
The sequence of the cloned sea lamprey Petromyzon marinus VT-AANAT (GenBank accession no. KF246473) is ∼50% identical to Chondrichthyes, teleost, and mammalian AANAT sequences (SI Appendix, Fig. S1), and also, it has two nonhomologous sequences: one (63 aa) at the N terminus and a second (62 aa) in the catalytic core. Similar nonhomologous sequences were also found in the river lamprey Lampetra fluviatilis VT-AANAT (GenBank accession no. KF290562). A truncated sequence, lacking the C-terminal end of the skate VT-AANAT, was also identified from the skate genome and transcriptome databases; this 163-aa sequence displayed 87% identity with the corresponding catshark VT-AANAT sequence (SI Appendix, Fig. S1). The organization and the defining sequence features of selected AANATs are summarized in Fig. 1. In addition to sequence conservation, the lamprey and other vertebrate VT-AANAT genes exhibit identical intron/exon structure.
Structural similarity of C. milii VT-AANAT to VT-AANATs was also supported by modeling, in which the ovine AANAT crystal structure was used as the template. Of 166 residues modeled in C. milii VT-AANAT, 107 residues (64%) are nearest to an identical residue in the ovine template (SI Appendix, Fig. S2).
Biochemical analysis.
The cloned VT-AANAT transcripts were expressed, and substrate specificity was determined using phenylethylamines, indolethylamines, and alkylamines. The expressed VT-AANATs efficiently acetylated phenylethylamines and indolethylamines, but they did not acetylate alkylamines, including butylamine and two diamines (spermidine and putrescine) (Table 1 and SI Appendix, Fig. S3). This substrate preference profile is similar to the profile found in bony vertebrates. In addition, the pH dependencies of C. milii VT-AANAT (peak at 6.8) and P. marinus and S. canicula VT-AANAT (peak between 6 and 8.5) resembled the pH dependencies of bony vertebrate VT-AANATs (SI Appendix, Fig. S4).
Table 1.
Kinetics of recombinant VT- and NV-AANAT
| Substrates | KM (mM)* | kcat (s−1)* | Ki (mM)* | kcat/KM (×103⋅s−1⋅mol−1) |
||
| Mean | 95% CI |
|||||
| + | − | |||||
| VT-AANAT P. marinus | ||||||
| Phenylethylamines | ||||||
| PEA | 2.35 ± 2.05 | 4.77 ± 2.22 | 30.5 ± 43.2 | 2.03 | 11.34 | 1.17 |
| Tyramine | 1.44 ± 0.67 | 5.64 ± 1.29 | 25.4 ± 17.3 | 3.92 | 3.46 | 1.43 |
| Indoleethylamines | ||||||
| Tryptamine | 0.11 ± 0.07 | 3.75 ± 0.58 | 42.8 ± 41.5 | 33.4 | 54.4 | 13.4 |
| Serotonin | 0.54 ± 0.44† | 4.93 ± 2.25† | 3.03 ± 1.48† | 9.05† | 28.48† | 4.98† |
| OMS | 0.05 ± 0.02 | 3.36 ± 0.33 | 9.63 ± 3.54 | 67.7 | 59.5 | 22.1 |
| Alkylamines | n.a. (Putrescine, spermidine, butylamine) | |||||
| VT-AANAT C. milii | ||||||
| Phenylethylamines | ||||||
| PEA | 3.62 ± 0.97 | 33.73 ± 2.67 | — | 9.31 | 3.37 | 2.02 |
| Dopamine | 10.7 ± 1.41 | 16.52 ± 0.86 | — | 1.55 | 0.24 | 0.19 |
| Tyramine | 1.32 ± 0.11 | 39.09 ± 0.95 | — | 29.57 | 0.51 | 0.5 |
| Indoleethylamines | ||||||
| Tryptamine | 0.71 ± 0.09 | 17.64 ± 0.66 | 201 ± 72 | 25.0 | 3.5 | 2.8 |
| Serotonin | 0.59 ± 0.14 | 23.98 ± 0.9 | — | 40.66 | 2.04 | 1.86 |
| OMS | 0.58 ± 0.08 | 21.52 ± 0.97 | 92.5 ± 33.7 | 37.12 | 5.8 | 4.53 |
| Alkylamines | n.a. (Putrescine, spermidine, butylamine) | |||||
| NV-AANAT C. milii | ||||||
| Phenylethylamines | ||||||
| PEA | 1.61 ± 0.89 | 0.026 ± 0.004 | — | 0.016 | 0.019 | 0.006 |
| Dopamine | n.a. | — | — | — | ||
| Tyramine | 44.87 ± 13.7 | 0.20 ± 0.03 | — | 0.004 | 0.0003 | 0.0003 |
| Indoleethylamines | ||||||
| Tryptamine | 2.6 ± 0.73 | 0.71 ± 0.005 | — | 0.027 | 0.011 | 0.006 |
| Serotonin | 59.46 ± 91.4 | 0.63 ± 0.89 | 4.62 ± 8.03 | 0.011 | 0.007 | 0.004 |
| OMS | 7.44 ± 1.66 | 0.2 ± 0.02 | — | 0.027 | 0.008 | 0.005 |
| Alkylamines | ||||||
| Putrescine | 5.19 ± 1.43 | 0.5 ± 0.08 | — | 0.1 | 0.04 | 0.02 |
| Spermidine | 6.38 ± 2.63 | 0.39 ± 0.05 | — | 0.061 | 0.019 | 0.04 |
| Butylamine | 2.09 ± 1.25 | 0.026 ± 0.004 | — | 0.012 | 0.017 | 0.005 |
The catalytic efficiency (kcat) for the expressed VT-AANAT enzyme from Agnathans and Chondrichthyes was higher for indolethylamines than phenylethylamines; neither acetylated alkylamines. The kcat values were generally lower for NV-AANAT than the corresponding values for VT-AANAT; kcat values for alkylamines were higher than for arylalkylamines. KM, kcat, and Ki values were inferred from figures as shown in SI Appendix, Fig. S1 using Prism6.01 (GraphPad software). CI was calculated using GraphPad Prism6 (KM and kcat) and Quickcalcs (http://www.graphpad.com/quickcalcs/) software. CI, confidence interval; n.a., no activity; OMS, O-methylserotonin; PEA, phenylethylamine.
Mean ± CI.
Strong autoinhibition was observed with serotonin; accordingly, the KM, kcat, and kcat/KM values are based on product formation below 1 mM and the Ki value is based on product formation above this value.
Distribution in the retina and pineal organ.
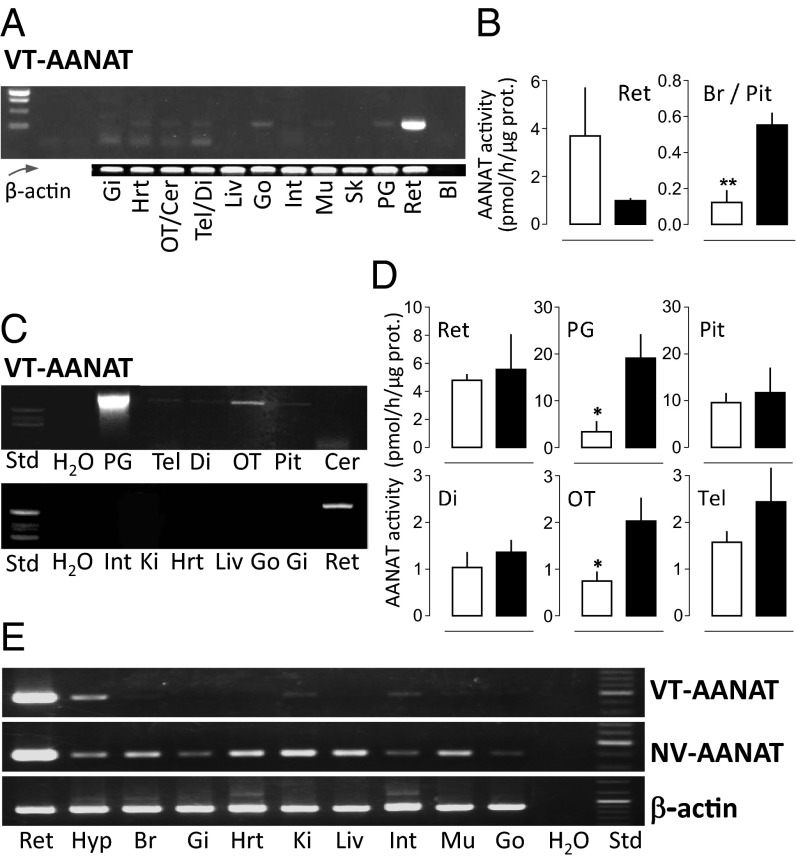
VT-AANAT transcripts were detected by PCR in extracts of lamprey, catshark, and elephant shark tissues (Fig. 2 A, C, and E). In the lamprey and elephant shark, a strong amplification signal was obtained from retinal extracts. In contrast, a much weaker signal was obtained from other brain or peripheral tissue extracts relative to the β-actin signal (the diencephalon of the elephant shark being a notable exception). In the catshark, the signal obtained from pineal extracts was by far the strongest. Amplification was also obtained from retinal extracts and to a lesser degree, brain (except cerebellum) and pituitary extracts; no evidence of the presence of VT-AANAT mRNA was obtained from extracts of the peripheral tissues investigated (Fig. 2 A, C, and E).
Fig. 2.
Tissue distribution of VT-AANAT in lamprey, catshark, and elephant shark. (A, C, and E) Presence of VT-AANAT mRNA in (A) lamprey, (C) catshark, and (E) elephant shark tissues as indicated by RT-PCR. (B and D) VT-AANAT activity in (B) lamprey and (D) catshark neural organs and tissues. Sampling was performed at midday (white bars) and midnight (black bars). Mean ± SEM (n = 5). Student t test: *P < 0.05; **P < 0.01. Additional details in SI Appendix. Bl, blank; Cer, cerebellum; Di, diencephalon; Gi, gills; Go, gonads; Hrt, heart; Hyp, hypothalmus; Int, intestine; Ki, kidney; Liv, liver; Mu, muscle; OT, optic tectum; Pit, pituitary; PG, pineal gland; Ret, retina; Sk, skin; Tel, telencephalon.
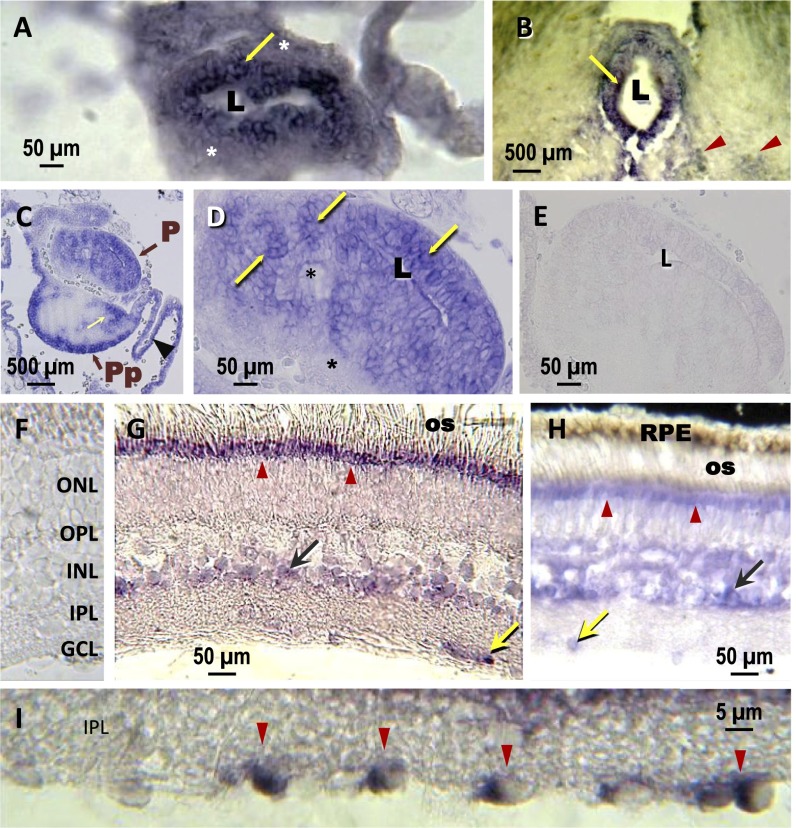
In situ hybridization confirmed a high expression of VT-AANAT in the retina and pineal gland of the lamprey and catshark. The catshark pineal gland (Fig. 3 A and B) exhibited distinct labeling in cells located at the center of the pineal epithelium, which is in close contact with the lumen of the pineal vesicle (i.e., where the photoreceptor cells are located) (30). A similar situation occurred in the pineal complex (i.e., pineal and parapineal organs) (31) of the lamprey (Fig. 3 C and D). In the retina of both lamprey and catshark, unambiguous expression was detected in the most apical part of the outer nuclear layer; this region corresponds to the ellipsoid of the photoreceptor cells (Fig. 3 G–I). A weaker labeling was observed in the inner nuclear layer, which could be amacrine and/or bipolar and/or Müller cells (Fig. 3 G and H). A subpopulation of cells in the ganglion cell layer was also labeled (Fig. 3I). The labeling in the lamprey and catshark retina resembles the pattern seen in the teleost retina (32).
Fig. 3.
Localization of VT-AANAT mRNA in the pineal complex and retina of lamprey and catshark by in situ hybridization. (A) Catshark pineal end vesicle and (B) stalk. Strongly labeled cells (yellow arrows) are seen in the inner epithelium around the pineal lumen (L). The cells at the periphery of the organ (asterisk) are unlabeled. Note the labeled nuclei (red arrowheads in B) in the epithalamus. C shows a frontal section through the lamprey pineal (P) and parapineal (Pp) organs. In C, positive reactions are seen in cells at the base of the Pp and surrounding dorsal sac (arrowhead), possibly corresponding to ependymal cells (31); the yellow arrow points to unidentified cells of the Pp rostral bulb. D is a higher magnification of the P end vesicle, where densely labeled cells appear in clusters (arrows); other unlabeled cells (asterisks) are seen in the central and basal epithelium. (E and F) Controls. There is no labeling in sections hybridized with a sense probe in the (E) lamprey pineal organ and (F) the catshark retina. (G and H) The labeling displays a similar pattern in the (G) catshark and (H) lamprey retinas. It is seen in the most apical part of the photoreceptor cells (red arrowheads) of the ONL at the level of the ellipsoid and just below the os layer; this labeling is particularly strong in the catshark. It is also seen in cells at the basal part of the INL and GCL (green and yellow arrows, respectively). (I) High magnification of the GCL in the catshark retina. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer; os, outer segments; RPE, retinal pigment epithelium.
Native enzyme activity.
In agreement with the RT-PCR identification of AANAT transcripts, AANAT activity was measured in the retina and brain of lamprey and catshark (Fig. 2 B and D). The levels of activity were similar in the corresponding organs of both species. Activity was 5- to 10-fold higher in the retina than the brain; it was three- to fourfold higher in the pineal organ compared with the retina as measured in the catshark (Fig. 2D). AANAT activity was not detected in the lamprey pineal end vesicles (pool of three organs); this apparent absence of activity may reflect limited tissue, low expression levels, or a combination (Fig. 2A) (see above). Midday/midnight differences are consistent with previous observations made in bony fish and tetrapods (6, 8, 32).
NV-AANAT Is Present in Chondrichthyes.
Sequence analysis.
The NV-AANAT sequences cloned from C. milii (572 bp; GenBank accession no. KF246476) and C. monstrosa (650 bp; GenBank accession no. KF246474) retinal cDNAs encoded 166-aa sequences (SI Appendix, Fig. S1), and the nucleotide sequences were 90% identical. The encoded proteins were 87% identical to each other and 30–50% identical to other known NV-AANAT proteins. The alignment of the VT- and NV-AANATs from C. milii showed low identity (∼15%) across the complete aligned sequences (SI Appendix, Fig. S1). In particular, although the generic AcCoA binding motifs A and B were clearly conserved, the VT-AANAT–specific substrate binding motifs C and D were not recognizable in NV-AANAT (Fig. 1). The deduced C. milii and C. monstrosa NV proteins lacked the defining features of the VT-AANAT identified above (Fig. 1).
Modeling revealed that only 37 of 163 (23%) C. milii NV-AANAT residues modeled are identical to the corresponding ovine residue (SI Appendix, Fig. S2). Consequently, the NV-AANAT model must be interpreted with caution, although it shows two columns of conserved residues that span the β-sheet and the conservation of the inner surface of a helix, consistent with a conserved fold.
It was not possible to identify an NV-AANAT in the lamprey and catshark using PCR amplification of genomic DNA or cDNA from different tissues with primers selected on conserved regions of NV-AANATs. Moreover, the search of the sequence databases from lamprey (genome: ftp://ftp.ensembl.org/pub/release-68/fasta/petromyzon_marinus/dna) and catshark (transcriptome: http://skatebase.org/) also gave negative results. Thus, it seems that NV-AANAT was lost during the evolution of these lineages. In contrast, a BLAST search of the skate genome and transcriptome databases using either chimaera NV-AANAT sequence identified a skate NV-AANAT that was ∼60% identical to the Holocephali sequence and 35% identical to amphioxus NV-AANATβ (SI Appendix, Fig. S1). The NV-AANAT transcript from the elephant shark displayed a widespread and intense distribution in extracts from all of the tissues examined, which was evidenced by RT-PCR (Fig. 2E).
Biochemical analysis.
C. milii NV-AANAT acetylated arylalkylamines; in contrast to VT-AANAT, this NV-AANAT also efficiently acetylated alkylamines. The catalytic efficiency for all substrates was 100- to 1,000-fold lower than observed for the VT-AANATs (Table 1 and SI Appendix, Fig. S3), which was previously shown for NV-AANATs from amphioxus (14) and yeast (13). Although the KM of C. milii NV-AANAT for AcCoA was similar to the KM found for VT-AANATs (0.5 mM), its optimal pH was ∼9, which is identical to the optimal pH of amphioxus and yeast NV-AANATs and in marked contrast with the pH maximum values of C. milii and tetrapod VT-AANATs (SI Appendix, Fig. S4).
Phylogenetic Analysis and Evolutionary Rate Estimation of VT- and NV-AANATs.
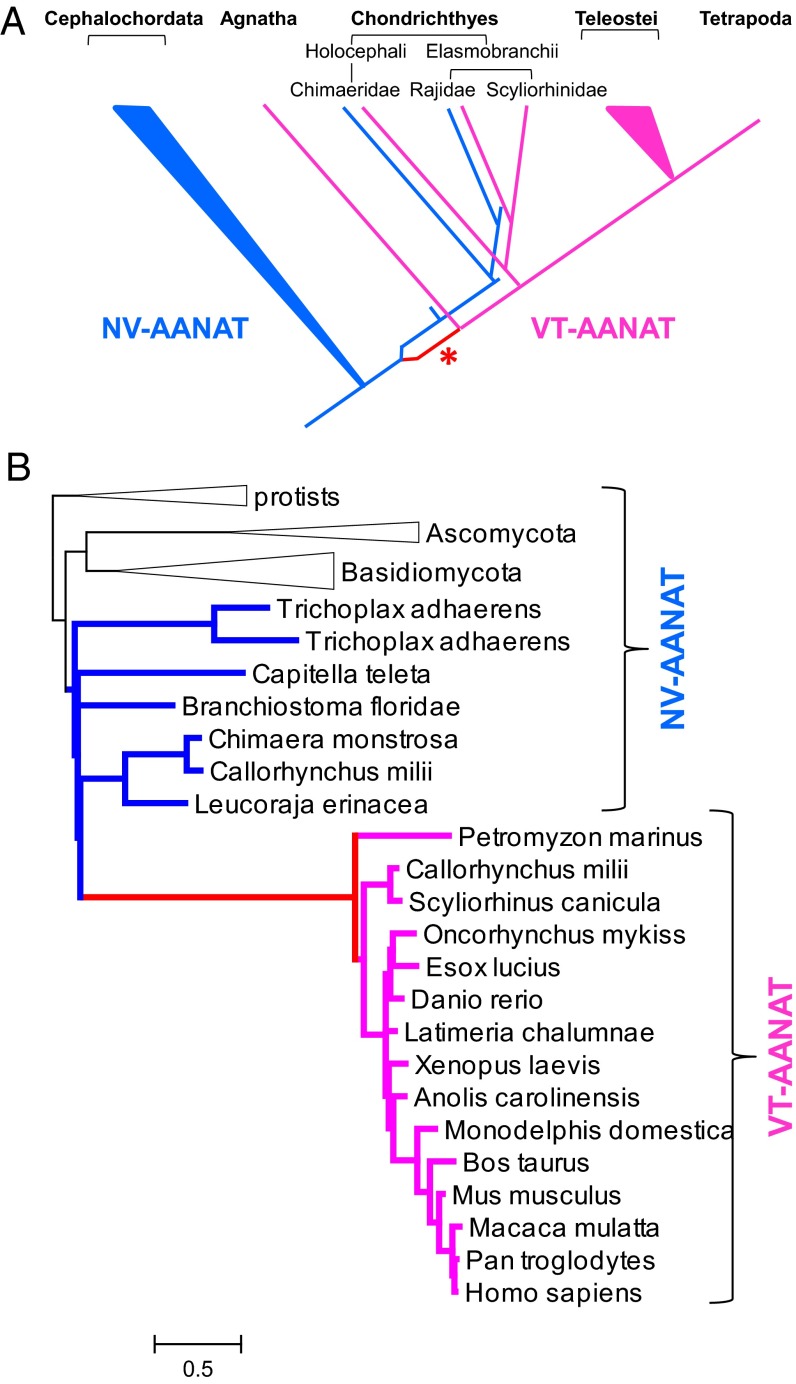
Although VT-AANAT shows relatively weak sequence similarity to NV-AANAT, these two subfamilies of the AANAT family seem to include orthologous genes, because the respective sequences show the greatest similarity to one another by a significant margin in genome-wide comparisons of protein sequences. Accordingly, we turned to phylogenetic analysis to reconstruct the evolution of the AANAT family. Sequences of putative orthologs of VT- and NV-AANATs from diverse eukaryotes were extracted from the National Center for Biotechnology Information nonredundant protein sequence database. An approximate maximum likelihood phylogenetic tree of 362 AANATs was constructed (SI Appendix, Fig. S5). Analysis of this preliminary tree indicates partitioning of the family into the NV- and VT-AANAT subfamilies, with the latter characterized by a very long basal branch. To reconcile the preliminary tree with the consensus phylogeny of eukaryotes (33, 34), we proposed an evolutionary scenario that involved a duplication of the ancestral NV-AANAT early in the vertebrate lineage, the loss of NV-AANAT in Agnathans and bony vertebrates, and a dramatic acceleration of evolution of one of the duplicates (VT-AANAT) (Fig. 4A, asterisk). A maximum likelihood phylogenetic tree that was reconstructed for a representative set of 41 sequences using the RAxML program under the constraints of this scenario (Fig. 4B) could not be rejected at a 2% significance level according to the Shimodaira–Hasegawa test implemented in RAxML.
Fig. 4.
Phylogenetic analysis. (A) Scenario of VT- and NV-AANAT evolution. Blue, metazoan NV-AANAT; magenta, VT-AANAT; red branch marked with an asterisk, VT-AANAT base branch (accelerated evolution). (B) Constrained maximum likelihood tree of representative AANAT sequences. Black, nonmetazoan NV-AANAT; blue, metazoan NV-AANAT; magenta, VT-AANAT; red, VT-AANAT base branch. The scale bar indicates the number of substitutions per site.
Calculation of the evolutionary rates revealed an acceleration of evolution by approximately an order of magnitude in the stem of the VT-AANAT subfamily (i.e., at the base of the vertebrates) (SI Appendix, Fig. S6 and Table S3). Two other branches within the VT-AANAT clade were also associated with elevated evolution rates, albeit not nearly as dramatically as the VT-AANAT stem [namely the branch leading from Tetrapoda to Amniota and the branch leading from Eutheria to Euarchontoglires (the base of the common ancestor of primates and rodents)].
Discussion
The current study fills an essential gap in our understanding of VT- and NV-AANAT evolution, because the AANAT family members in Agnathans and Chondrichthyes have not been previously characterized. The sequence analysis and biochemical results presented here establish that VT-AANAT is present in Agnathans and Chondrichthyes and that both NV- and VT-AANAT are present in some taxa within Chondrichthyes; in addition, it is shown that VT-AANAT is expressed in the retina and pineal glands of members of these groups. These findings indicate that the rhythmic production and release of melatonin by the lamprey pineal gland (35, 36) are likely to reflect the presence of VT-AANAT and not the presence of other acetyltransferases capable of acetylating serotonin (37).
Taken together with the presence of NV-AANAT in cephalochordates (14), these findings indicate that both enzymes were present in the common ancestor of vertebrates before the split of Agnatha and Gnathostomata. The parsimonious evolutionary scenario involves duplication at the base of the vertebrate lineage followed by at least two independent losses of NV-AANAT (in Agnatha and Teleostomi) and a dramatic acceleration of the evolution of VT-AANAT (Fig. 4) that implies a major functional shift. The only conceivable alternative could be recruitment of VT-AANAT from a different acetyltransferase family that is distantly related to NV-AANAT. However, given that NV-AANAT is the closest nonvertebrate homolog of VT-AANAT, there seems to be no support for such a scenario.
These findings pinpoint the origin of VT-AANAT as a distinct functional entity and imply that VT-AANAT evolved from the ancestral NV-AANAT before or concomitant with the emergence of lateral eyes and pineal gland in the Cambrian period, rather than at a later stage of evolution. The large distribution of NV-AANAT in the elephant shark supports the idea of a detoxification role for this enzyme. The more restricted distribution of VT-AANAT in primitive chordates (Agnatha and Chondrichthyes) is consistent with the hypothesis that the functional shift of the encoded protein from a role in detoxification to a role in melatonin synthesis was closely associated with the evolution of the pinealocyte and retinal photoreceptors from a common ancestral photodetector (7, 8).
Detoxification of arylalkylamines in photodetectors is thought to be important, because arylalkylamines can complex with and inactivate retinaldehyde, an essential component of light capture (38). Other retinaldehyde-related genes are also thought to have appeared as a result of duplication and neofunctionalization including an isomerase (Rpe65), retinoid metabolizing enzymes (Rdh5, Rhd8, Rdh12, and Lrat), and retinoid binding proteins (Rlbp1 and Rbp3) (39). Several of these genes are expressed in both the pineal gland and retina, which was indicated by microarray (40) and RNA sequencing (National Center for Biotechnology Information/Gene Expression Omnibus accession no. GSE46069). Notably, the genes associated with the retinoid cycle are thought to have appeared in the primitive vertebrate eye as a result of duplication of ancestral genes that were unrelated to vision (39). This innovation of vertebrate photoreception is critical for the visual cycle. Accordingly, it would seem that the evolutions of retinoid cycle-associated genes and of arylalkylamine N-acetyltransferase share a common selective pressure, which is to enhance visual sensitivity.
Acetylation of arylalkylamines might also provide selective advantage by preventing interactions with polyunsaturated fatty acids, including arachidonic acid and docosahexaenoic acid (41, 42). Docosahexaenoic acid is essential to the structure and function of photoreceptor cells in both the retina and the pineal organ (43–46). Accordingly, VT-AANAT might have functioned to enhance the efficiency of the primitive vertebrate’s photodetector by preventing arylalkylamine adduct formation with several essential photodetection molecules.
The absence of the NV-AANAT in Agnathans, catshark, and Teleostomi most likely reflects gene loss (47). In these vertebrates, the detoxification function of NV-AANAT might have been relegated to other enzymes (e.g., spermine/spermidine acetyltransferases) (48, 49). From an evolutionary theoretical viewpoint, the history of the AANAT family is an interesting case of neofunctionalization (50); after a gene duplication, one of the copies evolves to assume a new function, and uncharacteristically, the other copy is lost rather than retaining the ancestral function. These gene losses might have been instigated by a diminishing importance of amine detoxification in vertebrates. The nature of this putative functional shift remains to be elucidated. In contrast, the retention of NV-AANAT in several taxa within Chondrichthyes might reflect continued pressure for amine acetylation linked to environmental or metabolic factors.
Gene loss is likely to also explain the apparent absence of AANAT from Ciona. This absence, along with the low sequence similarity between VT-AANAT and NV-AANAT, contributed to the hypothesis that AANAT was acquired by vertebrates by horizontal gene transfer (17) and might have been recruited after evolution of the eye and retina. The results of the present analysis do not seem to be compatible with this scenario but rather, indicate that the unusual aspect of the AANAT family history involved an explosive acceleration of evolution in the VT-AANAT branch that was apparently associated with the functional shift from amine detoxification to melatonin synthesis.
Materials and Methods
Animals.
Adult sea lampreys (P. marinus), catsharks (S. canicula), ratfish (Chimaera monstrosa), and elephant sharks (C. milii) were used; details regarding source and handling are in SI Appendix.
Methods and Chemicals.
See SI Appendix.
Supplementary Material
Acknowledgments
We want to thank J. A. Donald, T. Toop, and J. D. Bell for assistance in providing specimens of C. milii and also M. Gabriel Diaz (fisherman in Port-Vendres, France) for providing the S. canicula individuals. This study was supported by French Research National Agency Grant 07-BLAN-0097-01, the Centre National de la Recherche Scientifique, Institut français de recherche pour l'exploitation de la mer and Université Pierre et Marie Curie-Paris 6 Grants GDR2821 and UMR7232, the Japan Society for the Promotion of Science, and the intramural programs of the National Institute of Child Health and Human Development, the National Library of Medicine, and the Center for Information Technology (all at the National Institutes of Health).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database, www.ncbi.nlm.nih.gov/genbank/ (accession nos. EU378921, KF246473–KF246476, KF290562, BK008791, and BK008792).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312634110/-/DCSupplemental.
†The AANAT family that is the subject of the current report is distinct from another family also named AANAT, which includes enzymes involved in acetylation of dopamine and other arylalkylamines in insects (11, 12). These two protein families are only distantly related to each other, although both belong to the GCN5 superfamily of acetyltransferases.
References
- 1.Gehring WJ. The genetic control of eye development and its implications for the evolution of the various eye-types. Int J Dev Biol. 2002;46(1):65–73. [PubMed] [Google Scholar]
- 2.Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15(9):371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien PJ, Klein DC. Pineal and Retinal Relationships. Orlando, FL: Academic; 1986. [Google Scholar]
- 4.Ekström P, Meissl H. Evolution of photosensory pineal organs in new light: The fate of neuroendocrine photoreceptors. Philos Trans R Soc Lond B Biol Sci. 2003;358(1438):1679–1700. doi: 10.1098/rstb.2003.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collin JP, et al. Pineal transducers in the course of evolution: Molecular organization, rhythmic metabolic activity and role. Arch Histol Cytol. 1989;52(Suppl):441–449. doi: 10.1679/aohc.52.suppl_441. [DOI] [PubMed] [Google Scholar]
- 6.Klein DC, et al. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–357. [PubMed] [Google Scholar]
- 7.Klein DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland—a tale of conflict and resolution. J Biol Rhythms. 2004;19(4):264–279. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- 8.Klein DC. Evolution of the vertebrate pineal gland: The AANAT hypothesis. Chronobiol Int. 2006;23(1–2):5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- 9.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: A structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein DC. Arylalkylamine N-acetyltransferase: “The timezyme.”. J Biol Chem. 2007;282(7):4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- 11.Dai FY, et al. Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J Biol Chem. 2010;285(25):19553–19560. doi: 10.1074/jbc.M109.096743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amherd R, Hintermann E, Walz D, Affolter M, Meyer UA. Purification, cloning, and characterization of a second arylalkylamine N-acetyltransferase from Drosophila melanogaster. DNA Cell Biol. 2000;19(11):697–705. doi: 10.1089/10445490050199081. [DOI] [PubMed] [Google Scholar]
- 13.Ganguly S, Mummaneni P, Steinbach PJ, Klein DC, Coon SL. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3.1.87) J Biol Chem. 2001;276(50):47239–47247. doi: 10.1074/jbc.M107222200. [DOI] [PubMed] [Google Scholar]
- 14.Pavlicek J, et al. Evolution of AANAT: Expansion of the gene family in the cephalochordate amphioxus. BMC Evol Biol. 2010;10:154. doi: 10.1186/1471-2148-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Sutton A, Sternglanz R. A yeast polyamine acetyltransferase. J Biol Chem. 2005;280(17):16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- 16.Smith TJ. Phylogenetic distribution and function of arylalkylamine N-acetyltransferase. Bioessays. 1990;12(1):30–33. doi: 10.1002/bies.950120107. [DOI] [PubMed] [Google Scholar]
- 17.Iyer LM, Aravind L, Coon SL, Klein DC, Koonin EV. Evolution of cell-cell signaling in animals: Did late horizontal gene transfer from bacteria have a role? Trends Genet. 2004;20(7):292–299. doi: 10.1016/j.tig.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Falcón J. Cellular circadian clocks in the pineal. Prog Neurobiol. 1999;58(2):121–162. doi: 10.1016/s0301-0082(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 19.Shen J, et al. N-acetyl serotonin derivatives as potent neuroprotectants for retinas. Proc Natl Acad Sci USA. 2012;109(9):3540–3545. doi: 10.1073/pnas.1119201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallström BM, Janke A. Gnathostome phylogenomics utilizing lungfish EST sequences. Mol Biol Evol. 2009;26(2):463–471. doi: 10.1093/molbev/msn271. [DOI] [PubMed] [Google Scholar]
- 21.Inoue JG, et al. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): A mitogenomic perspective. Mol Biol Evol. 2010;27(11):2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 22.Cazaméa-Catalan D, et al. Functional diversity of Teleost arylalkylamine N-acetyltransferase-2: Is the timezyme evolution driven by habitat temperature? Mol Ecol. 2012;21(20):5027–5041. doi: 10.1111/j.1365-294X.2012.05725.x. [DOI] [PubMed] [Google Scholar]
- 23.Cazaméa-Catalan D, et al. Unique arylalkylamine N-acetyltransferase-2 polymorphism in salmonids and profound variations in thermal stability and catalytic efficiency conferred by two residues. J Exp Biol. 2013;216(Pt 10):1938–1948. doi: 10.1242/jeb.080960. [DOI] [PubMed] [Google Scholar]
- 24.Hickman AB, Klein DC, Dyda F. Melatonin biosynthesis: The structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism. Mol Cell. 1999;3(1):23–32. doi: 10.1016/s1097-2765(00)80171-9. [DOI] [PubMed] [Google Scholar]
- 25.Hickman AB, Namboodiri MAA, Klein DC, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 1999;97(3):361–369. doi: 10.1016/s0092-8674(00)80745-x. [DOI] [PubMed] [Google Scholar]
- 26.Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell. 2001;105(2):257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- 27.Zilberman-Peled B, Bransburg-Zabary S, Klein DC, Gothilf Y. Molecular evolution of multiple arylalkylamine N-acetyltransferase (AANAT) in fish. Mar Drugs. 2011;9(5):906–921. doi: 10.3390/md9050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf E, De Angelis J, Khalil EM, Cole PA, Burley SK. X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition. J Mol Biol. 2002;317(2):215–224. doi: 10.1006/jmbi.2001.5371. [DOI] [PubMed] [Google Scholar]
- 29.Pavlicek J, et al. Evidence that proline focuses movement of the floppy loop of arylalkylamine N-acetyltransferase (EC 2.3.1.87) J Biol Chem. 2008;283(21):14552–14558. doi: 10.1074/jbc.M800593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falcón J. L'organe pinéal du Brochet (Esox lucius, L.) II. Etude en microscopie électronique de la différenciation et de la rudimentation des photorécepteurs; conséquences possibles sur l'élaboration des messages sensoriels. Reprod Nutr Dev. 1979;19(3A):661–688. [Google Scholar]
- 31.Cole WC, Youson JH. Morphology of the pineal complex of the anadromous sea lamprey, Petromyzon marinus L. Am J Anat. 1982;165(2):131–163. doi: 10.1002/aja.1001650205. [DOI] [PubMed] [Google Scholar]
- 32.Besseau L, et al. Melatonin pathway: Breaking the ‘high-at-night’ rule in trout retina. Exp Eye Res. 2006;82(4):620–627. doi: 10.1016/j.exer.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3(11):838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 34.Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 35.Bolliet V, Ali MA, Anctil M, Zachmann A. Melatonin secretion in vitro from the pineal complex of the lamprey Petromyzon marinus. Gen Comp Endocrinol. 1993;89(1):101–106. doi: 10.1006/gcen.1993.1013. [DOI] [PubMed] [Google Scholar]
- 36.Samejima M, Tamotsu S, Uchida K, Moriguchi Y, Morita Y. Melatonin excretion rhythms in the cultured pineal organ of the lamprey, Lampetra japonica. Biol Signals. 1997;6(4–6):241–246. [PubMed] [Google Scholar]
- 37.Cassone VM, Natesan AK. Time and time again: The phylogeny of melatonin as a transducer of biological time. J Biol Rhythms. 1997;12(6):489–497. doi: 10.1177/074873049701200602. [DOI] [PubMed] [Google Scholar]
- 38.Saari JC. Vitamin A metabolism in rod and cone visual cycles. Annu Rev Nutr. 2012;32:125–145. doi: 10.1146/annurev-nutr-071811-150748. [DOI] [PubMed] [Google Scholar]
- 39.Albalat R. Evolution of the genetic machinery of the visual cycle: A novelty of the vertebrate eye? Mol Biol Evol. 2012;29(5):1461–1469. doi: 10.1093/molbev/msr313. [DOI] [PubMed] [Google Scholar]
- 40.Bailey MJ, et al. Night/day changes in pineal expression of >600 genes: Central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284(12):7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters GH, et al. Binding of serotonin to lipid membranes. J Am Chem Soc. 2013;135(6):2164–2171. doi: 10.1021/ja306681d. [DOI] [PubMed] [Google Scholar]
- 42.Verhoeckx KC, et al. Presence, formation and putative biological activities of N-acyl serotonins, a novel class of fatty-acid derived mediators, in the intestinal tract. Biochim Biophys Acta. 2011;1811(10):578–586. doi: 10.1016/j.bbalip.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Jeffrey BG, Weisinger HS, Neuringer M, Mitchell DC. The role of docosahexaenoic acid in retinal function. Lipids. 2001;36(9):859–871. doi: 10.1007/s11745-001-0796-3. [DOI] [PubMed] [Google Scholar]
- 44.Falcón J, Henderson RJ. Incorporation, distribution, and metabolism of polyunsaturated fatty acids in the pineal gland of rainbow trout (Oncorhynchus mykiss) in vitro. J Pineal Res. 2001;31(2):127–137. doi: 10.1034/j.1600-079x.2001.310206.x. [DOI] [PubMed] [Google Scholar]
- 45.Bennett MP, Mitchell DC. Regulation of membrane proteins by dietary lipids: Effects of cholesterol and docosahexaenoic acid acyl chain-containing phospholipids on rhodopsin stability and function. Biophys J. 2008;95(3):1206–1216. doi: 10.1529/biophysj.107.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soubias O, Teague WE, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281(44):33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 47.Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100(5):605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- 48.Lien YC, Ou TY, Lin YT, Kuo PC, Lin HJ. Duplication and diversification of the spermidine/spermine N1-acetyltransferase 1 genes in zebrafish. PLoS One. 2013;8(1):e54017. doi: 10.1371/journal.pone.0054017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medina MA, Urdiales JL, Rodríguez-Caso C, Ramírez FJ, Sánchez-Jiménez F. Biogenic amines and polyamines: Similar biochemistry for different physiological missions and biomedical applications. Crit Rev Biochem Mol Biol. 2003;38(1):23–59. doi: 10.1080/713609209. [DOI] [PubMed] [Google Scholar]
- 50.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 51.Coon SL, Klein DC. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol Cell Endocrinol. 2006;252(1–2):2–10. doi: 10.1016/S0303-7207(06)00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.