Significance
Mammalian skeletal muscle is a dynamic and plastic tissue, capable of responding to physiological demands and pathophysiological stresses. This response relies on the muscle’s ability to activate myogenic progenitor cells (MPCs) resulting in myogenesis. In this study, we demonstrate that cytoglobin, a stress-responsive hemoprotein abundantly expressed in MPCs, is capable of modulating MPCs’ viability and proliferative/differentiative capacity. Collectively, our data demonstrate that cytoglobin serves an important role in muscle regeneration. Thus, an enhanced understanding of cytoglobin’s role in myogenesis may enable the development of therapeutic approaches for treating patients with muscle injuries and other neuromuscular disorders.
Keywords: myogenesis, myocyte differentiation/proliferation, nuclear protein, redox signaling
Abstract
Mammalian skeletal muscle can remodel, repair, and regenerate itself by mobilizing satellite cells, a resident population of myogenic progenitor cells. Muscle injury and subsequent activation of myogenic progenitor cells is associated with oxidative stress. Cytoglobin is a hemoprotein expressed in response to oxidative stress in a variety of tissues, including striated muscle. In this study, we demonstrate that cytoglobin is up-regulated in activated myogenic progenitor cells, where it localizes to the nucleus and contributes to cell viability. siRNA-mediated depletion of cytoglobin from C2C12 myoblasts increased levels of reactive oxygen species and apoptotic cell death both at baseline and in response to stress stimuli. Conversely, overexpression of cytoglobin reduced reactive oxygen species levels, caspase activity, and cell death. Mice in which cytoglobin was knocked out specifically in skeletal muscle were generated to examine the role of cytoglobin in vivo. Myogenic progenitor cells isolated from these mice were severely deficient in their ability to form myotubes as compared with myogenic progenitor cells from wild-type littermates. Consistent with this finding, the capacity for muscle regeneration was severely impaired in mice deficient for skeletal-muscle cytoglobin. Collectively, these data demonstrate that cytoglobin serves an important role in muscle repair and regeneration.
The regenerative capacity of adult skeletal muscle is not unlimited. Exhaustion of muscle progenitor cells (MPCs) is thought to be an important factor contributing to the progressive weakness and atrophy of peripheral skeletal muscle that occurs with various skeletal myopathies and during normal aging (1–9). A better understanding of the processes and proteins involved in myogenesis and muscle regeneration may enable the design of innovative therapies to improve the care of victims of severe muscle trauma, patients with skeletal myopathies, and the elderly.
Mammalian skeletal muscle is composed of a heterogeneous set of multinucleate muscle fiber types, each with a distinct set of contractile and metabolic properties. Over the course of a mammal’s lifetime, the maintenance, adaptation, and repair of skeletal muscle depend on the continual ability to initiate myogenesis (4, 5, 10–12). Skeletal muscle injury can occur as a consequence of direct myotrauma (e.g., after a burn or intense physical activity) or genetic mutation of key cytoskeleton proteins as in the muscular dystrophies (1, 5, 10, 12–16). In response to damage or to a change in demand, quiescent MPCs residing between the sarcolemma and the basal lamina are activated and begin to proliferate (17–19). Once activated, MPCs, also known as “satellite cells,” can either fuse to existing myofibers or form myofibers de novo.
Muscle regeneration is a highly organized process that can be divided broadly into two phases—degenerative and regenerative—regardless of the initiating event (e.g., acute trauma or chronic, genetically induced myopathy) (5, 17, 18, 20–22). The degenerative phase, which generally occurs within 48 h of an acute injury, is characterized by cell death (both necrotic and apoptotic) and inflammation. During the regenerative phase (2–14 d postinjury) activated MPCs contribute to the repair of injured muscle fibers, generate new myofibers, and self-renew a population of quiescent MPCs (4, 5, 22). In adult skeletal muscle, quiescent, mitotically inactive MPCs express a set of myogenic-specific transcription factors including forkhead box protein K1 (Foxk1) and paired box transcription factor 3 (Pax3) and 7 (Pax7) (4, 5, 22). Upon activation, MPCs start to express transcription factors from the myogenic regulatory factor (MRF) family [myogenic differentiation antigen (MyoD), myogenic factor 5 (Myf5), myogenin, and MRF4]. This expression initiates a cascade of molecular events resulting in the activation, proliferation, and eventual differentiation of MPCs into mature, terminally differentiated myotubes (4, 5, 13–15, 22, 23).
Recently, we demonstrated that hypoxia induces the expression of the gene encoding cytoglobin (Cygb) in C2C12 myoblasts, an immortalized cell line derived from MPCs (24, 25). Cygb is a heme-containing protein of the globin family that is structurally similar to myoglobin (Mb) and is conserved across species (mammals, fish, amphibians, reptiles, and birds) (26, 27). In mammals it is expressed in a variety of tissues, including skeletal muscle, although the Cygb transcript level in the adult skeletal muscle is low compared with the levels in heart and brain (24, 26–29). Mechanistic studies defining the function of Cygb are limited. In vitro studies indicate it is able to scavenge free radicals, and overexpression of Cygb in various cell lines preserves cell viability under conditions of oxidative stress (30–34). The objective of the present study was to determine the role of Cygb in skeletal muscle. We present in vitro and in vivo evidence that Cygb protects the viability of activated MPCs and is essential for effective muscle regeneration.
Results
Cygb Is Expressed Within the Nucleus of MPCs and Proliferating Myoblasts.
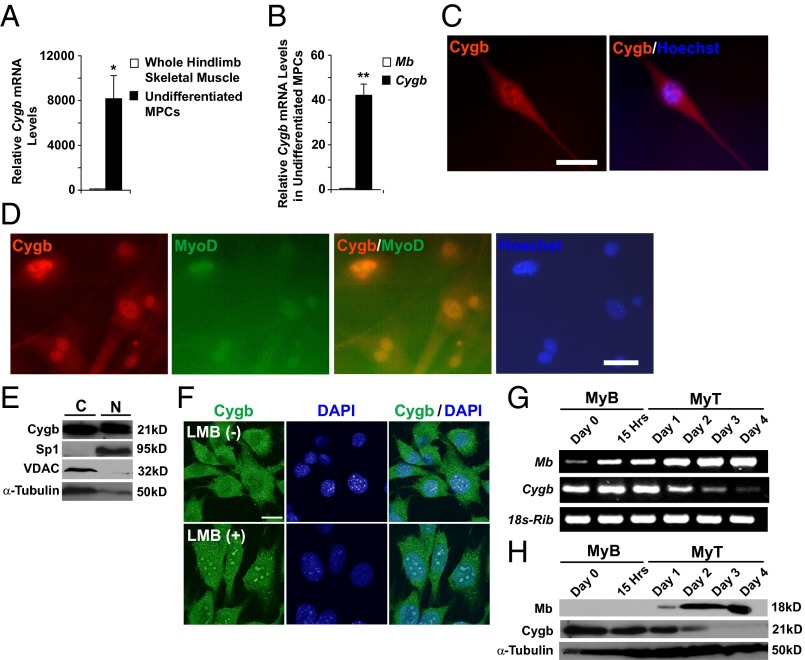
Primary MPCs were isolated from the hindlimb muscles of WT mice. Transcript levels for Cygb were assessed by real-time quantitative RT-PCR (qRT-PCR). Undifferentiated MPCs were markedly enriched in Cygb transcript (8,000-fold) compared with whole hindlimb skeletal muscle (Fig. 1A). In addition, Cygb was more abundant than Mb within undifferentiated MPCs (Fig. 1B). Immunocytochemistry (ICC) demonstrated that Cygb protein is expressed in both the cytosol and the nucleus of primary, undifferentiated MPCs (Fig. 1C). In addition, ICC and Hoechst staining of isolated MPCs showed that Cygb colocalized with MyoD, an MPC-specific transcription factor belonging to the MRF family, suggesting that under these conditions Cygb is a nuclear protein (Fig. 1D). Although Cygb has been considered primarily cytoplasmic, our data are consistent with previous reports demonstrating Cygb protein in the nuclei of both hepatocytes and neurons (35, 36).
Fig. 1.
Cygb is found in both the cytoplasm and nucleus of primary MPCs, and its expression decreases upon differentiation of myoblasts. (A) qRT-PCR was used to compare the Cygb transcript level in undifferentiated MPCs with that in hindlimb skeletal muscle of adult WT mice. (B) Relative transcript levels of Mb and Cygb were compared in undifferentiated WT MPCs. (C) Representative ICC image showing Cygb localized to both the cytosol and nuclei of isolated WT MPCs. (Scale bar: 40 μm.) (D) Cygb and MyoD colocalize to the nuclei of MPCs. The Cygb signal is tagged red, and the MyoD signal is green. Nuclear Hoechst staining is labeled blue. (Scale bar: 40 μm.) (E) Western blot analysis indicates Cygb protein in both the cytosolic (C) and nuclear (N) enriched fractions from C2C12 myoblasts. Specialty protein 1 (Sp1) and VDAC were used to assess the purity of fractionation; α-tubulin was used as the loading control. (F) Nuclear abundance of Cygb increased after C2C12 myoblasts were treated with Leptomycin B (LMB) to inhibit nuclear export. The Cygb signal is green, and the nuclear DAPI signal is blue. (Scale bar: 20 μm.) (G and H) RT-PCR (G) and Western blot analysis (H) indicated a decrease in Cygb transcript and protein levels during differentiation of C2C12 myoblasts (MyB) to myotubes (MyT). Mb expression is shown to verify myoblast differentiation. 18s-ribosome (18s-Rib) and α-tubulin were used as loading controls. *P < 0.005 undifferentiated MPCs vs. whole skeletal muscle; **P < 0.005 Cygb expression vs. Mb expression; n = 3 in each group.
Western blot analysis supported the observation that Cygb protein is expressed in the both the cytosol and nuclei of C2C12 myoblasts, an immortalized cell line derived from MPCs (Fig. 1E) (25). Exposure of C2C12 myoblasts to Leptomyocin B, a nonspecific nuclear-export inhibitor, resulted in increased accumulation of Cygb within the nuclei of these cells, suggesting that the Cygb flux between the cytosol and nucleus is dynamic in nature (Fig. 1F). Finally, to determine whether Cygb levels change during differentiation of myoblasts, we assessed Cygb transcript and protein levels in differentiating C2C12 myoblasts (Fig. 1 G and H). Both Cygb transcript and protein levels were abundant in C2C12 myoblasts; however, over the course of differentiation to myotubes, both transcript and protein levels declined and were reciprocal to the well-described temporal pattern of Mb expression, which increases during myotube differentiation (37, 38). Taken together, these data indicate that Cygb is present in MPCs and C2C12 myoblasts during their proliferative state but declines as myotubes are established.
Cygb Expression Is Induced in a Murine Model of Muscle Regeneration.
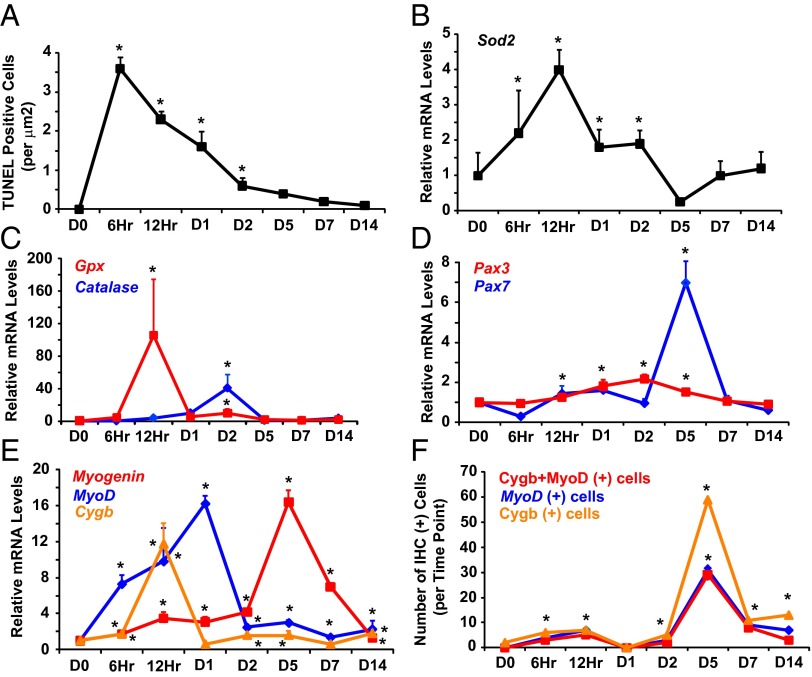
Cardiotoxin (CTX) is a myotoxin derived from the venom of the Taiwanese Cobra (Naja atra) and has been used widely by scientific investigators to induce muscle injury and regeneration in a reproducible manner (39–44). Intramuscular injection of CTX into the hindlimb of WT adult males caused apoptosis, necrosis, and extensive inflammation within the first 48 h postinjury (Fig. 2A and Fig. S1 A and B; degenerative phase). Transcript levels of superoxide dismutase-2 (Sod2), catalase, and glutathione peroxidase (Gpx) also peaked during this phase, indicating mobilization of defense mechanisms against oxidative stress (Fig. 2 B and C). Over the course of the following 2 wk (days 2–14), there was clearance of necrotic tissue, formation of new myotubes (myocytes with centrally located nuclei) on day 5, and complete restoration of the muscle architecture by day 14 (Fig. S1A; regenerative phase).
Fig. 2.
Cygb expression increases during muscle regeneration and colocalizes with MyoD-positive cells. (A–C) TUNEL assays (A) and qRT-PCR data (B and C) indicate the induction of apoptosis and oxidative stress following CTX-induced muscle injury in the hindlimb muscles of WT mice. (D and E) qRT-PCR revealed increased Pax3, Pax7, myogenin, MyoD, and Cygb levels after muscle injury. The transcript levels were normalized to 18s-ribosomal RNA. *P < 0.05 postinjury vs. baseline level; n = 3–5 in each group. (F) IHC was undertaken at various time points after CTX injury to assess the number cells coexpressing both Cygb and MyoD. The number of dual-staining cells was quantified. *P < 0.05 postinjury vs. baseline level; n = 3–5 sections at each time period.
Within the first 5 d after CTX injury, transcript levels of the MPC markers Pax 3 and Pax 7 increased (Fig. 2D). Likewise, there were marked increases in mRNA levels of MyoD and myogenin, which are hallmarks of MPC activation (Fig. 2E). There was a 12-fold increase in Cygb mRNA levels with a temporal expression profile that preceded the expression of most of the myogenic markers (Fig. 2E). Importantly, immunohistochemistry (IHC) demonstrated colocalization of Cygb protein and MyoD protein, which was greatest at day 5, coinciding with the appearance of centrally nucleated myocytes (Fig. 2F and Fig. S1C). At this time point, the majority of MyoD-positive cells also were Cygb positive, providing further evidence that Cygb is expressed in activated MPCs.
Muscle Regeneration Is Impaired in Mice with Knockout of Skeletal Muscle-Specific Cygb.
To investigate the in vivo role of Cygb during skeletal muscle regeneration, a conditional Cygb-knockout mouse line was engineered using standard Cre/loxP technology (Fig. S2). Homozygous floxed mice (Cygbflox/flox) were crossed with Myo-Cre transgenic mice to generate mice with skeletal muscle-specific Cygb knockout [Cygbflox/flox: Myo-Cre (+) or Cygb−/−]. The muscle-specific Myo-Cre driver line expresses Cre-recombinase under the control of the myogenin promoter and a Mef2 enhancer, and it is expressed in somites as early as embryonic day 9.5 (45). The presence and correct insertion of the targeting vector were confirmed by both semiquantitative PCR and Southern blot analysis (Fig. S2 B–D). Cygb−/− mice were viable and were normal under baseline conditions. Depletion of Cygb from whole adult skeletal muscle was confirmed by qPCR and Western blot analysis (Fig. S2 F and G). It was not possible to tell whether the Cre/loxP system already had been activated in MPCs in vivo; however, once isolated and placed in culture, depletion of the Cygb transcript in the MPCs was confirmed by qPCR and Western blot analysis (Fig. 3 A and B).
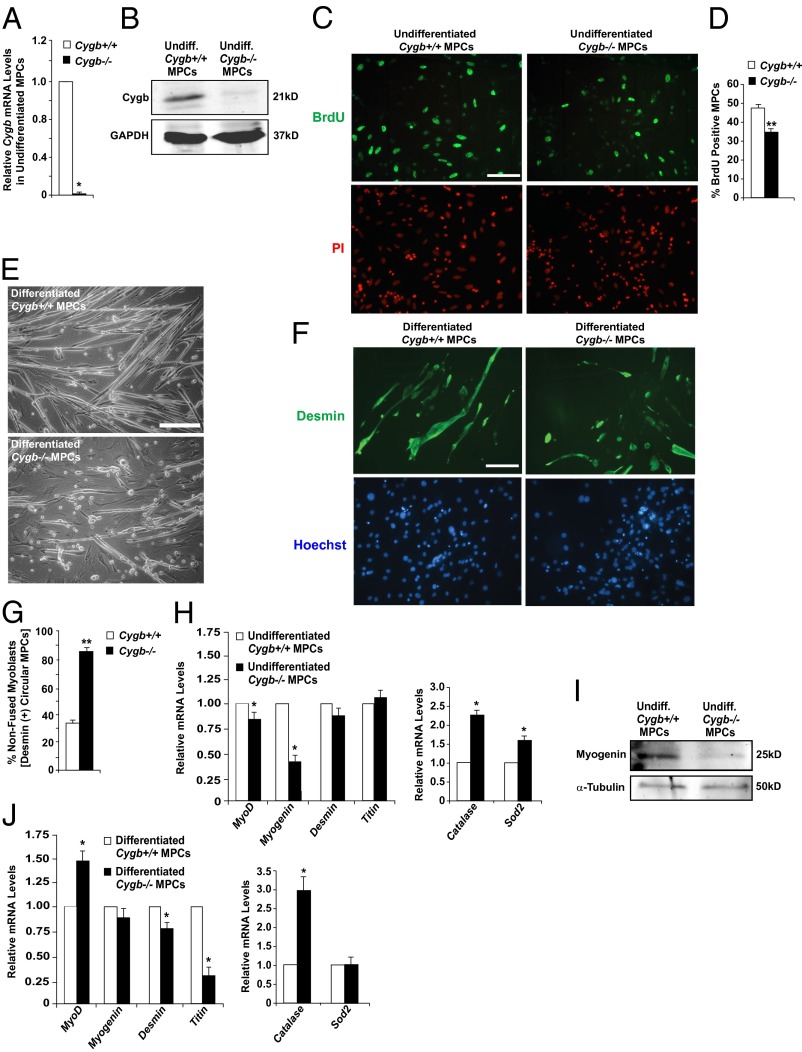
Fig. 3.
Differentiation of Cygb-deficient myogenic progenitor cells is impaired. (A and B) qRT-PCR and Western blot data confirmed knockdown of Cygb in undifferentiated (undiff) MPCs isolated from the hindlimb muscles of Cygb−/− mice. (C and D) BrdU incorporation in undifferentiated MPCs isolated from Cygb+/+ and Cygb−/− muscle revealed a decreased proliferative capacity in Cygb−/− MPCs. (E) Phase-contrast images comparing differentiation of MPCs isolated from the hindlimb muscles of Cygb+/+ and Cygb−/− mice. MPC cultures were incubated in differentiation medium for 5 d. (F and G) Desmin staining of differentiated MPCs revealed a greater number of nonfused myoblasts in differentiated Cygb−/− MPCs. (H and I) qRT-PCR (H) and Western blot analysis (I) comparing expression of myogenin and other MRF family genes, myocyte structural genes, and oxidative stress marker genes in undifferentiated Cygb+/+ and Cygb−/− MPCs. (J) qRT-PCR analysis comparing expression of myogenin and other MRF family genes, myocyte structural genes, and oxidative stress marker genes in Cygb+/+ and Cygb−/− MPCs after 5 d of differentiation. *P < 0.05 Cygb−/− MPCs vs. Cygb+/+ MPCs; n = 3 in each group. **P < 0.05 Cygb−/− MPCs vs. Cygb+/+ MPCs; n = 20–30 in each group. (Scale bar: 200 μm.)
To assess the effect of Cygb deficiency on the ability of MPCs to proliferate, BrdU staining was undertaken on undifferentiated MPCs isolated from the hindlimb muscles of Cygb−/− and control [Cygbflox/flox: Myo-Cre (−) or Cygb+/+] mice. The results revealed a 30% reduction in the number of BrdU-positive Cygb−/− MPCs compared with Cygb+/+ MPCs, suggesting that the proliferative capacity of Cygb-deficient MPCs is impaired (Fig. 3 C and D). In addition, Cygb−/− MPCs were not able to differentiate fully into myotubes (Fig. 3 E–G). As MPCs differentiate into mature, elongated myotubes, the cells express desmin; however, the appearance of desmin-expressing nonfused cells indicates incomplete differentiation. ICC showed twice as many desmin-staining nonfused circular cells in undifferentiated Cygb−/− MPCs incubated for 5 d in differentiating medium than in control MPCs (Fig. 3 F and G), suggesting a defect in the ability of Cygb−/− MPCs to differentiate fully. Supporting the observation made by ICC, qRT-PCR data demonstrated dysregulated transcript levels of various MRF family members in undifferentiated and differentiated Cygb−/− MPCs (Fig. 3 H and J). In particular, the transcript and protein levels of myogenin were decreased markedly in undifferentiated Cygb−/− MPCs as compared with control MPCs (Fig. 3 H and I). In addition, Cygb−/− MPCs incubated for 5 d in differentiating medium showed a significant decrease in the transcript levels of myocyte structural genes (i.e., desmin and titin) as compared with differentiated Cygb+/+ MPCs, supporting the observation that Cygb−/− MPCs fail to differentiate fully into myotubes (Fig. 3J). Finally, the transcript levels of the oxidative stress markers catalase and Sod2 were increased in Cygb−/− MPCs, suggesting that redox states are altered in these cells (Fig. 3 H and J). Collectively, these data suggest that Cygb-deficient MPCs have impaired proliferative and differentiative capacities.
To confirm further the findings from Cygb-deficient MPCs, we used a second model of C2C12 myoblast differentiation. Cygb was knocked down efficiently in C2C12 myoblasts using small, interfering RNA (siRNA) directed against Cygb (siCygb) (Fig. S3A), and mRNA expression of several differentiation-specific genes (Pax3, Pax7, MyoD, and myogenin) was followed although the course of C2C12 differentiation. The data indicated severely dysregulated expression of all analyzed genes in siCygb cells, supporting our previous observation that Cygb-deficient MPCs show impaired differentiation (Fig. S4 A–D). Similarly, mRNA expression of the oxidative stress markers catalase and Sod2 also showed marked up-regulation in siCygb cells throughout the course of differentiation, indicating elevated oxidative stress in these cells as compared with control cells (Fig. S4 E and F). Overexpression of Mb, a globin structurally similar to Cygb, could not reverse the impairment in differentiation in Cygb-deficient MPCs (Fig. S5). This observation suggests that the impaired differentiative capacity in these cells is specific to the loss of Cygb and that Mb is unable to rescue the phenotype.
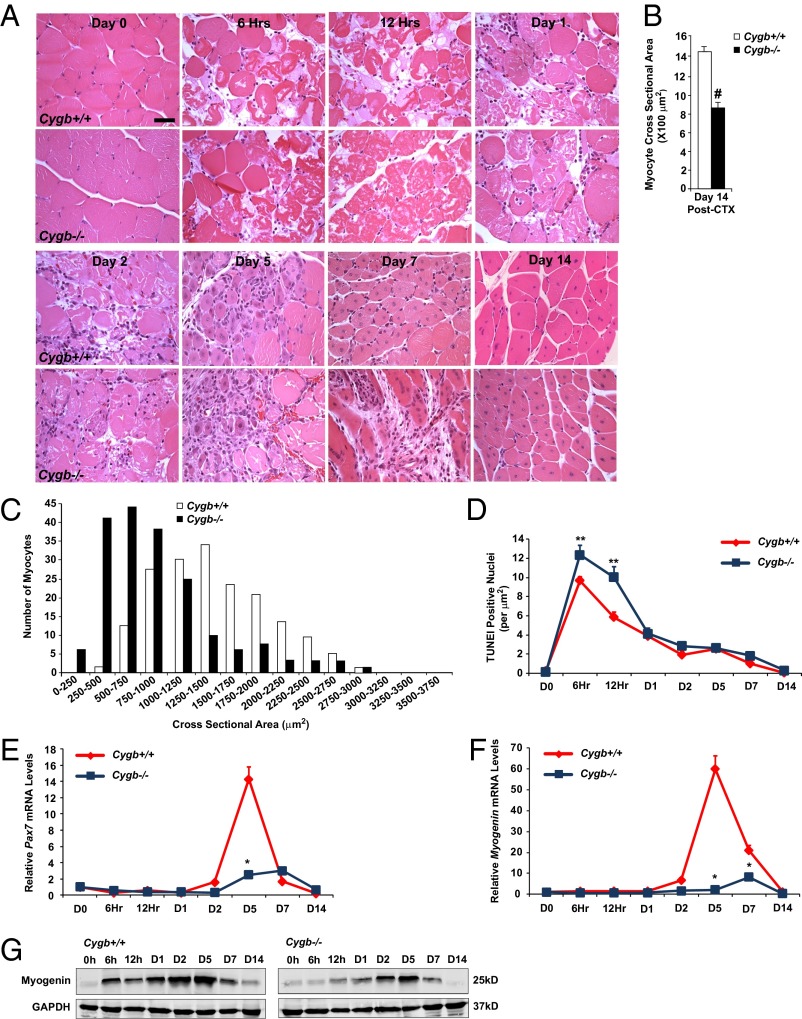
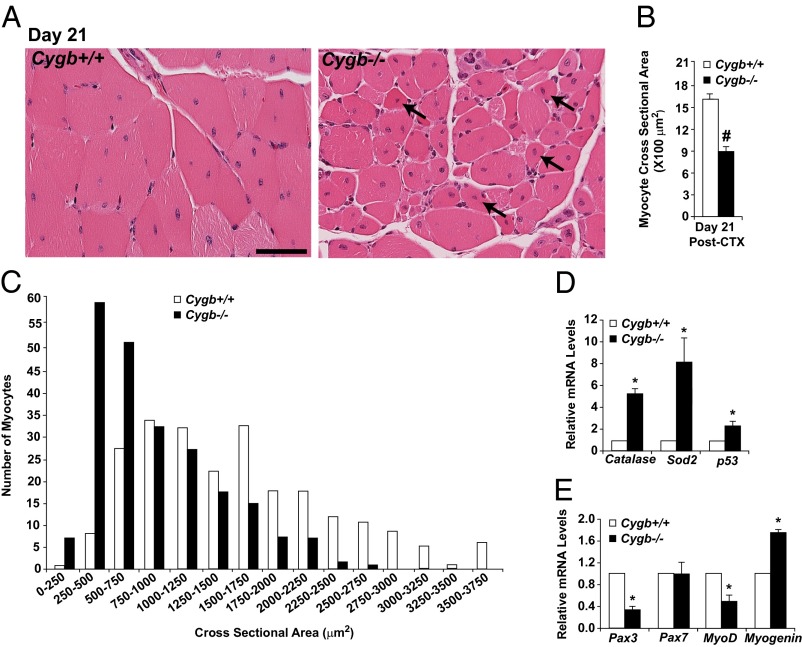
The Cygb−/− mice then were assessed for their ability to repair and regenerate skeletal muscle in vivo using the CTX injury model. The hindlimb muscles from Cygb+/+ and Cygb−/− mice were injected with CTX as previously described. Histological and molecular markers of oxidative stress and muscle regeneration were assessed as a function of time postinjury (Fig. 4A and Fig. S2 H–K). H&E staining of injured skeletal muscle from both Cygb+/+ and Cygb−/− mice revealed extensive necrosis and inflammation during the first 2 d postinjury (Fig. 4A). However, morphometric analysis of the myocyte cross-sectional area (CSA) within the area of injury 14 d postinjury indicated an ∼40% reduction in myocyte size among Cygb−/− mice as compared with injured Cygb+/+ mice (Fig. 4B). Analysis of myocyte CSA clustering demonstrated that the majority of Cygb+/+ myocytes were 1,000–2,000 μm2 in size, whereas the CSA for Cygb−/− myocytes was distributed between 500 and 1,250 μm2 (Fig. 4C). Thus, after CTX injury, Cygb−/− mice demonstrate impaired muscle regeneration that is marked by considerably smaller myocytes (Fig. 4 B and C). Finally, the CTX-injured Cygb−/− mice continued to demonstrate evidence of ongoing myonecrosis and apoptosis up to day 21 postinjury. A time course of TUNEL-positive cells suggested an increase in the magnitude of cell death and a broadening of the window of time during which cell death was elevated (Fig. 4D).
Fig. 4.
Cell death is enhanced, and regeneration is blunted in CTX-injured mice with knockout of skeletal muscle-specific Cygb. (A) H&E staining demonstrated delayed regeneration in Cygb−/− muscle 14 d after CTX injury. Note the persistence of centrally nucleated myocytes (evidence of ongoing muscle regeneration) in the Cygb−/− muscle. (Scale bar: 40 μm.) (B and C) The average myocyte CSA (B) and the distribution of the CSA (C) revealed significantly smaller myocytes within the hindlimbs of Cygb−/− mice than in Cygb+/+ mice at 14 d postinjury. (D) Time course of TUNEL activity following CTX injury in Cygb+/+ and Cygb−/− mice. (E and F) qRT-PCR data analysis was used to quantify Pax7 and myogenin transcript levels after CTX injury in Cygb+/+ and Cygb−/− mice. (G) Western blot analysis revealed a decrease in myogenin protein levels in Cygb−/− mice after CTX injury. GAPDH was used as a loading control. #P < 0.05 Cygb−/− myocytes vs. Cygb+/+ myocytes, n = 200 in each group; *P < 0.05 Cygb−/− vs. Cygb+/+, n = 6 in each group at each time point; **P < 0.05 Cygb−/− vs. Cygb+/+, n = 8–10 in each group at each time point.
A time course of changes in gene expression indicated a peak in Pax7 and myogenin mRNA levels at day 5 in Cygb+/+ hindlimb muscle (Fig. 4 E and F). Activation of these two markers of muscle regeneration was delayed in the Cygb−/− mice and was profoundly blunted, suggesting that Cygb expression is required for the initiation of an appropriate regenerative program. The blunted mRNA-expression profile of myogenin was confirmed by Western blot analysis (Fig. 4G).
The architecture of the Cygb+/+ hindlimb muscle was almost completely restored to normal 14 d after CTX-induced injury. In contrast, the Cygb−/− muscle still had evidence of continued muscle degeneration/regeneration and failure to restore the normal muscle architecture even 21 d postinjury, as characterized by a greater number of centrally nucleated myocytes and smaller myocyte size than seen in the Cygb+/+ animals (Fig. 5 A–C). The Cygb−/− skeletal muscle at this time point still had elevated expression of oxidative stress markers (i.e., catalase, Sod2, and p53) indicative of sustained oxidative stress (Fig. 5D). The expression of Pax3, MyoD, and myogenin also was altered in the Cygb−/− muscle as compared with Cygb+/+ muscle, suggesting sustained differences in the myogenic program 21 d after CTX injury (Fig. 5E). Collectively, the in vivo studies demonstrated that both the timing and quality of muscle repair and regeneration are defective in Cygb-deficient skeletal muscle.
Fig. 5.
Impaired muscle regeneration in mice with skeletal muscle-specific deletion of Cygb. (A) H&E staining revealed persistent muscle injury 21 d after CTX injury in Cygb−/− mice. Arrows indicate centrally nucleated myocytes. (Scale bar: 40 μm.) (B and C) The average myocyte CSA (B) and distribution of the CSA (C) revealed significantly smaller myocytes within the hindlimbs of Cygb−/− mice than in Cygb+/+ mice at 21 d postinjury. (D and E) qRT-PCR data supported increased oxidative stress and altered expression of transcription factors responsible for muscle regeneration in Cygb−/− mice 21 d after CTX injury. #P < 0.05 Cygb−/− myocytes vs. Cygb+/+ myocytes, n = 225 in each group; *P < 0.05 Cygb−/− vs. Cygb+/+, n = 6 in each group.
Depletion of Cygb from C2C12 Myoblasts Increases Cellular Reactive Oxygen Species and Promotes Cell Death.
In vitro studies were initiated to elucidate the function of Cygb within activated MPCs. siCygb was used to deplete 70% of the Cygb transcript from C2C12 myoblasts (Fig. S3A). Although two different Cygb-targeting siRNA oligonucleotides (siCygb3 and siCygb4) were identified as being able to knock down Cygb expression within these cells, the majority of the in vitro studies were undertaken using siCygb4. This particular siRNA oligonucleotide was more effective in knocking down Cygb expression and hereafter is referred to simply as “siCygb.” Finally, we demonstrate that the scrambled, control siRNA had no significant effects on cell death over a wide range of concentrations (0–80 nM) (Fig. S3B).
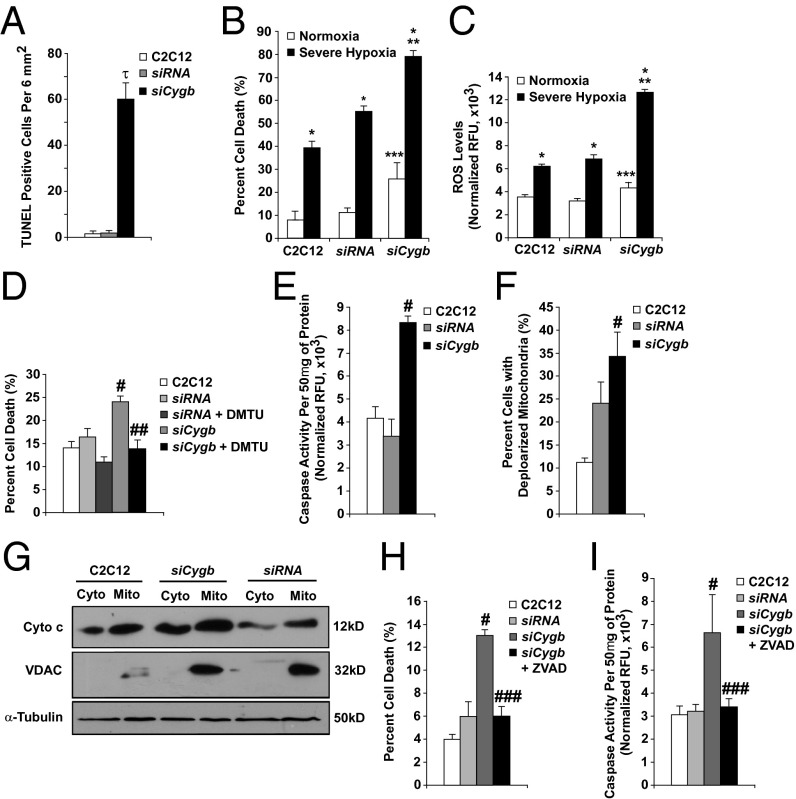
Knockdown of Cygb resulted in a significant increase in cell death as assessed by both TUNEL staining and FACS analysis of fragmented DNA (Fig. 6 A and B and Fig. S3 B and C). Cygb depletion also increased cell death under normoxic conditions in HeLa and COS cells (Fig. S3 D and E). Upon exposure to severe hypoxia (0.1% O2, 5% CO2), cell death was greater in siCygb myoblasts than in control myoblasts (nontransfected C2C12 myoblasts and siRNA myoblasts; Fig. 6B). Similarly, the siCygb-transfected C2C12 myoblasts were more sensitive to menadione, a synthetic vitamin K derivative capable of promoting reactive oxygen species (ROS) generation (Fig. S3F).
Fig. 6.
siRNA depletion of Cygb increases apoptotic activity in C2C12 myoblasts. C2C12 cultures were treated with a control siRNA (siRNA) or with siRNA targeted against Cygb (siCygb). (A) TUNEL activity was used as an indicator of apoptotic cell death under normoxic growth conditions. (B) FACS analysis was used to assess cell death under normoxic conditions as well as upon exposure to severe hypoxia (<0.1% O2 for 12 h). (C) ROS levels were quantified under normoxic and hypoxic conditions using H2DCFDA and were normalized to cell number. (D) DMTU treatment improved the viability of siCygb-depleted myoblasts. (E and F) Caspase activity (E) and mitochondrial membrane potential (F) in normoxic siCygb myoblasts were compared with control myoblasts. (G) Cytochrome c release into the cytosol of siCygb-depleted cells was compared with controls by Western blot. (H and I) FACS analysis (H) and caspase activity assays (I) were used to assess the ability of ZVAD treatment to improve viability of siCygb-treated cells. τP < 0.005 siCygb vs. siRNA or C2C12 cells; n = 5–7 sections in each group. *P < 0.005 hypoxic vs. normoxic cells within each group, **P < 0.05 hypoxic siCygb vs. hypoxic control cells, and ***P < 0.05 normoxic siCygb vs. normoxic control cells; n = 6 in each group. #P < 0.05 siCygb vs. control cells, ##P < 0.05 siCygb+DMTU vs. siCygb cells, ###P < 0.05 siCygb+ZVAD vs. siCygb; n = 6 in each group. C2C12, nontransfected C2C12 myoblasts; siRNA, siRNA-transfected myoblasts, the control for siCygb cells.
The transcript levels of various oxidative stress markers were assessed in siCygb myoblasts under basal conditions Using qRT-PCR. Several genes known to promote stress, including Alox12 and Nox5, were up-regulated (Fig. S3G). Conversely, the expression of several genes involved in reducing oxidative stress, including Gpr156, Ptgs1, and Prdx6, was decreased (Fig. S3H). These data suggested that Cygb-deficient myoblasts are under more oxidative stress than control cells and thus presumably have altered redox states. We pursued this line of investigation and used H2DCFDA, a fluorescent cell-permeable indicator of ROS, to compare ROS generation in control and siCygb C2C12 myoblasts. Under normoxic conditions there was a small but statistically significant increase in total intracellular ROS levels in siCygb myoblasts as compared with control myoblasts (Fig. 6C). This difference was greatly enhanced under hypoxic conditions. To test whether this increase in cellular ROS influenced cell viability, control and siCygb C2C12 myoblasts were treated with dimethyl thiourea (DMTU), a ROS scavenger. DMTU attenuated the increase in cell death in siCygb myoblasts (Fig. 6D).
TUNEL assays and FACS analysis of DNA fragmentation are reliable indicators of cell death but do not distinguish among mechanisms of cell death. Studies evaluating caspase activation, mitochondrial depolarization, and cytochrome c release into the cytosol were used to assess apoptosis. There was an approximately threefold increase in caspase activity in siCygb C2C12 myoblasts as compared with control myoblasts (Fig. 6E). The population of cells with low mitochondrial membrane potential also was increased (Fig. 6F). Western blot analysis of mitochondrial and cytoplasmic cellular fractions showed that more cytochrome c was present in the cytoplasm of siCygb myoblasts than in control myoblasts, suggesting activation of the intrinsic apoptotic death pathway (Fig. 6G). We also found evidence of increased cleavage of both caspase-3 and poly-ADP ribose polymerase, a protein involved in DNA repair, in siCygb myoblasts (Fig. S6 A and B). Treating siCygb myoblasts with zVAD, a caspase inhibitor, improved cell survival, further implicating the apoptotic death pathway as the mode of death in siCygb cells (Fig. 6 H and I). These data are supported by the observation that there was an induction of proapoptotic genes (i.e., Nod1, Pak7, Pycard7, and Trp73) and a down-regulation of antiapoptotic genes (Bcl-2, Birc2, and Nol3) in siCygb myoblasts as compared with control siRNA myoblasts (Fig. S6C). Collectively, these data indicate that depletion of Cygb from C2C12 myoblasts increases their susceptibility to oxidative stress and to ROS-mediated activation of the intrinsic apoptotic death pathway.
Increased Expression of Cygb Protects C2C12 Myoblasts.
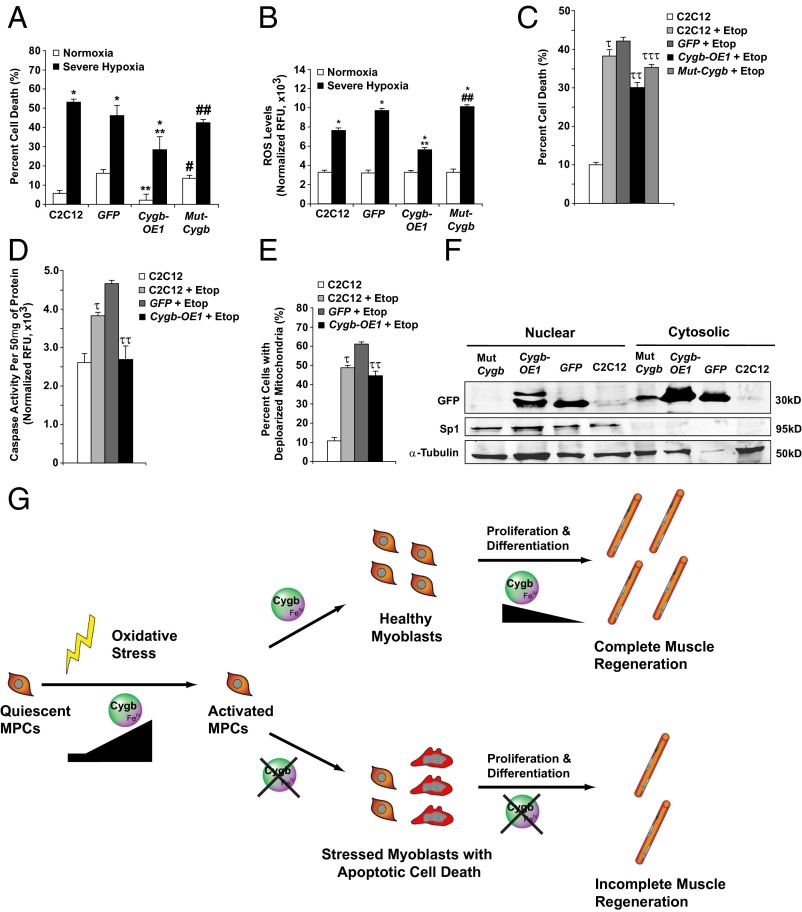
Transient transfection with a Cygb-GFP expression plasmid (Cygb-OE1) was used to overexpress Cygb in C2C12 myoblasts (Fig. S6D). Transfection with GFP alone was used as a control. Transfection with Cygb-OE1 reduced cell death and ROS generation in C2C12 myoblasts exposed to severe hypoxia (Fig. 7 A and B). Cygb overexpression using Cygb-OE1 also protected against menadione-induced cell death (Fig. S6E). The expression of a mutant Cygb-GFP fusion protein [Mut-Cygb; mutation of two evolutionary conserved histidine residues (His81 and His113) and a conserved cysteine (Cys83) within the heme-binding domain of Cygb] was no longer protective, demonstrating the importance of an intact heme-binding domain to the cellular function of Cygb (Fig. 7 A–C). Of note, the heme-binding domain of Cygb serves as the binding site for various ligands; including oxygen, nitric oxide, and ROS (27, 46).
Fig. 7.
Overexpression of Cygb is protective in stressed C2C12 myoblasts. (A) FACS analysis was used to assess cell death in C2C12 myoblasts transfected with no vector or with vectors expressing control GFP, Cygb fused to GFP (Cygb-OE1), or Cygb with a mutant heme-binding domain (mutant Cygb fused to GFP; Mut-Cygb) under normoxia and severe hypoxia (<0.1% O2 for 12 h). (B) ROS levels also were assessed in these four experimental groups under both normoxic and severe hypoxic conditions. (C–E) Transfected C2C12 myoblasts were treated with etoposide (Etop) and assessed for cell death (C), caspase activity (D), and mitochondrial membrane potential (E). (F) Western blot analysis revealed the nuclear localization of Cygb-OE1 and the inability of the mutated Mut-Cygb to localize to the nuclei in C2C12 myoblasts. Sp1 was used to assess the purity of fractionation, and α-tubulin was used as the loading control. (G) Schematic model depicting the role of Cygb during MPC activation and muscle regeneration. *P < 0.005 hypoxic vs. normoxic cells within each group; **P < 0.05 normoxic or hypoxic Cygb-OE1 vs. normoxic or hypoxic control GFP cells, respectively; #P < 0.05 normoxic Mut-Cygb vs. normoxic Cygb-OE1 cells; ##P < 0.05 hypoxic Mut-Cygb vs. hypoxic Cygb-OE1 cells; τP < 0.05 C2C12+Etop vs. C2C12 cells; ττP < 0.05 Cygb-OE1+Etop vs. GFP+Etop; τττP < 0.05 Mut-Cygb+Etop vs. Cygb-OE1+Etop; n = 6 in each group.
Cygb-OE1 transfection into C2C12 myoblasts also improved survival after treatment with etoposide, a cancer drug that promotes apoptosis (Fig. 7C). Consistent with this protective effect, Cygb-OE1 reduced caspase activation and loss of mitochondrial membrane potential caused by etoposide (Fig. 7 D and E). Similarly, transfection of C2C12 myoblasts with an myc-tagged Cygb expression construct (Cygb-OE2) reduced cell death upon exposure to severe hypoxia and menadione (Fig. S4 F and G).
Having established the importance of an intact heme-binding domain for Cygb’s cytoprotective properties, we sought to determine the importance of the heme-binding domain in Cygb’s ability to translocate into the nucleus. Western blot analysis was undertaken on cytosolic and nuclear fractions of C2C12 myoblasts transfected with either the wild-type Cygb-GFP construct (Cygb-OE1) or the mutated Cygb-GFP construct (Mut Cygb) (Fig. 7F). The results demonstrated the complete absence of the mutated protein in the nucleus, suggesting that the heme-binding domain of Cygb is critical to the ability of Cygb to localize to the nucleus of a cell.
Collectively, our gain-of-function in vitro studies further support the role of Cygb in promoting the viability of C2C12 myoblasts under a variety of oxidative-stress conditions. Of equal importance, the mutational studies identified the heme-binding domain as essential for Cygb nuclear localization.
Discussion
This study provides several insights into the role of Cygb during skeletal muscle regeneration. First, we demonstrate that Cygb is localized to the nucleus of MPCs and that its expression is induced transiently during muscle regeneration in vivo. Thus, Cygb is in an appropriate spatial and temporal location to influence MPC biology. Second, we show that Cygb protects myoblasts in culture from ROS-mediated apoptotic cell death and that an intact heme-binding domain is essential for this property. Third, isolated MPCs lacking Cygb have impaired proliferative and differentiative capacity, resulting in the inability to generate fully mature myotubes. Finally, and most importantly, we show in vivo that muscle repair and regeneration is severely impaired in mice with skeletal muscle-specific deletion of Cygb.
Regulation of the transcriptional machinery governing myogenesis and muscle regeneration has been investigated extensively during the past 25 y (5, 10, 22, 47). Although the importance of myogenic-specific transcription factors (i.e., Foxk1, Pax3, Pax7, and the MRF family) in regulating MPC proliferation and differentiation is well established, the understanding of the cellular environment required for the proper execution of the regenerative process is limited. Emerging data support the concept that the cellular redox milieu influences the capacity for proliferation and differentiation of diverse pools of tissue-specific progenitor cells, including MPCs (48–50). Our studies demonstrate that Cygb’s expression pattern, temporal activation, and cytoprotective properties are consistent with its having a role in regulating the proliferative and differentiative capacities of MPCs.
As a member of the globin superfamily of proteins, Cygb is exquisitely designed to participate in a variety of reactions that influence the redox environment of a cell (27, 51, 52). In vitro studies indicate that Cygb is able to bind to various ligands (e.g., oxygen, nitric oxide, and other free radicals) and has peroxidase activity, as does its closest relative Mb (27, 46, 53–58). However, these studies suggest that Cygb may be more important in scavenging free radicals (i.e., reactive oxygen and nitrogen species) and in modulating cellular redox signaling than in modulating O2 metabolism. Our in vitro loss- and gain-of-function studies using C2C12 myoblasts reflect Cygb’s ability to influence cellular redox signaling, with depletion of Cygb increasing ROS generation and sensitizing the cells to stress-induced apoptosis. These findings are consistent with similar loss- and gain-of-function studies demonstrating the ability of Cygb, Mb, or their neuron-specific relative neuroglobin to provide cytoprotection under conditions of oxidative stress (29–34, 59–68). Mutation of the heme-binding domain of Cygb disrupted its ability to confer protection in vitro, demonstrating that Cygb function in this context depends on this core protein structure, a feature common to all globin proteins, and not on a domain unique to Cygb. However, unlike the other globin proteins, Cygb can localize to the nucleus, and the nuclear localization of Cygb requires an intact heme-binding domain.
Defining the contribution of Cygb to MPC function and muscle regeneration in vivo is challenging. A number of studies suggest that the redox state of MPCs is a major determinant of the regenerative capacity of injured skeletal muscle (69–74) as well as of the proliferation and differentiation of MPCs in vitro (69, 73, 74). These studies suggest that in an oxidizing milieu MPCs are more responsive to signaling pathways that promote cell differentiation and cell death, whereas in reducing environments MPCs are more responsive to signals that promote cellular proliferation and survival.
It is likely that a primary mechanism through which Cygb affects MPC biology is through its ability to bind and traffic reactive oxygen and nitrogen species, thus influencing the balance of the cellular redox state. However, Cygb localizes to the nuclei of MPCs, and this characteristic also requires an intact heme-binding domain. Cygb is distinctive among the globins in that it is found both in the nucleus and cytoplasm [all other members of the family are localized exclusively in the cytoplasm (35–38)]. In addition, our Leptomyocin B and the mutational studies suggested that Cygb’s expression within the cytosol and nucleus is dynamic and can be altered. The distinct nuclear, subcellular localization suggests that Cygb has unique nuclear targets not shared by the other globins. This notion is strengthened by our observation that myoglobin, a closely related globin protein with a structure similar to Cygb, fails to overcome the inability of Cygb-depleted MPCs to differentiate into fully mature myotubes. Our expression-profiling studies identified changes in the transcript levels for oxidative stress- and apoptosis-related genes when Cygb was depleted from C2C12 myoblasts. However, whether nuclear Cygb can influence transcription directly or whether these changes are indirect, secondary to influences on the nuclear redox state, remains to be determined.
It is intriguing that we observed colocalization of Cygb with MyoD in the nucleus of activated MPCs during the regenerative phase of muscle injury. MyoD-dependent transcription drives subsequent expression of myogenin and other members of the MRF family (75). Although MyoD was expressed at the appropriate time during regeneration after CTX-induced skeletal muscle injury in Cygb−/− mice, subsequent expression of myogenin was largely absent in the regenerating Cygb−/− skeletal muscle.
Skeletal muscle injury resulting from ischemia, hypoxia, or extreme exercise triggers the generation of ROS (13, 74, 76, 77). Likewise, in muscular dystrophies, such as Duchenne muscular dystrophy, instability in the myocyte cytoskeleton structure is associated with oxidative stress-induced damage, leading to a continuous cycle of damage and regeneration that ultimately depletes MPCs (78–80). In mouse models of Duchenne muscular dystrophy, chemical scavengers of ROS (e.g., N-acetylcysteine) are able to break the repetitive cycle of degeneration/regeneration and improve muscle function, demonstrating the protective benefit of antioxidant mechanisms (81). Based on our studies, we propose that Cygb plays such a role in MPCs during adult skeletal muscle regeneration. Its protective properties may be linked exclusively to its capacity to maintain an appropriate redox state directly. However, additional mechanisms related to direct or indirect effects on transcription also could play a role by virtue of Cygb’s localization within the nuclei of MPCs.
In summary, Cygb is a stress-responsive hemoprotein that promotes the viability of MPCs and thus serves a critical role in myogenesis and skeletal muscle regeneration (Fig. 7G). Future studies will need to focus on the molecular mechanism by which Cygb exerts a positive influence on the viability and the proliferative and differentiative capacities of activated MPCs under oxidative stress. Ultimately, an enhanced understanding of the cellular function of Cygb within activated MPCs and skeletal myoblasts will provide opportunities for the development of therapeutic measures for the treatment of patients with a variety of muscle injuries and disorders, including burn patients and muscular dystrophy patients.
Materials and Methods
Murine Model of CTX-Induced Muscle Injury.
Control and experimental C57BL/6 adult mice (2–4 mo of age) were handled in accordance with National Institutes of Health and the University of Texas Southwestern Medical Center’s (UTSW) Institutional Guidelines. Induction of CTX-induced muscle injury was undertaken by injecting 100 μL of 10 μM CTX (Sigma) i.m. into the hindlimb (i.e., gastrocnemius/plantaris/soleus) muscles of adult male mice, and the injured skeletal muscles were harvested at various time points (40, 42–44).
Generation of a Conditional Cygb-Knockout Mouse Line.
Using the Cre/loxP system, the Mammen laboratory engineered a conditional Cygb-knockout mouse model, which was backcrossed into a pure C57BL/6J background strain. Details related to the construction of this unique mouse model are outlined in SI Materials and Methods.
Histological and IHC Studies.
Tissues for histological and IHC studies were prepared as previously described (68) and as outlined in SI Materials and Methods. TUNEL staining for apoptotic cells was performed on tissue sections as previously described (82).
C2C12 Myoblast Differentiation and Isolation of MPCs.
C2C12 myoblasts were plated in equal numbers under normoxic conditions as previously described (24). MPCs were isolated and cultured from the hindlimbs of 8-wk-old male mice as previously described (83). Further details regarding these methods are outlined in SI Materials and Methods.
BrdU Incorporation Assay.
In the BrdU incorporation assay MPCs isolated from the hindlimb muscles of Cygb+/+ and Cygb−/− mice were pulsed with 10 μM BrdU for 6 h. The assay was undertaken using a standardized protocol and as previously described (41).
Western Blot Analysis, RNA Isolation, and qRT-PCR.
Proteins from cell lysates, cellular fractions (i.e., nuclear and cytosolic), or whole-muscle tissue were isolated from control and experimental groups. Western blot analysis was performed as previously described (24, 28, 68). Total RNA, RT-PCR, and qRT-PCR were performed as previously described (24) These experiments are described in more detail in SI Materials and Methods. The primer sets used in the various qRT-PCR reactions are listed in Tables S1 and S2.
Knockdown and Overexpression of Cygb in Cell Culture.
Knockdown of Cygb expression was achieved by using murine-specific siRNA targeting Cygb (siCygb) that was obtained from Dharmacon. Cells initially were seeded at 70% (vol/vol) confluency and were maintained in DMEM containing 20% FBS at 37 °C under normoxic conditions. These cells were transfected with siCygb (80 nM) or a scrambled siRNA oligo (control, 20 nM) using HiPerFect (QIAGEN) and low-serum OptiMEM transfection medium (Invitrogen) for 6 h. Subsequently, the medium was changed back to DMEM, and the cells were cultured for another 24 h before the in vitro experiments were initiated.
The full-length murine Cygb gene was cloned into the pEGFP-N1 plasmid (BD Biosciences Clontech), which was capable of generating a Cygb-GFP fusion protein. Using Lipofectamine (Invitrogen), we achieved Cygb overexpression by transiently transfecting cells with the Cygb-GFP plasmid (24). The control cells were transiently transfected with the empty pEGFP-N1 plasmid vector. Finally, a mutant form of Cygb with an unstable heme-binding domain was generated by introducing base-specific mutations into the Cygb-GFP plasmid using the QuikChange Mutagenesis Kit (Stratagene). Specifically, the two conserved histidines at positions 81 and 113 were changed to glycine, and the conserved cysteine at position 83 was changed to serine in two successive rounds of mutation. Expression of the full-length mutated Cygb protein was undertaken by transiently transfecting cells with the mutant Cygb plasmid (Mut-Cygb).
Induction and Modulation of Oxidative Stress.
Cells were placed in an in vitro hypoxic chamber and exposed to very low oxygen tension (0.1% O2, 4.99% CO2, and 95% N2) for 12 h (24). Induction of oxidative stress with generation of ROS was initiated by exposing cells to menadione (40 μM) or etoposide (50 μM) (Sigma) for 12 h. ROS scavenging or inhibition of caspase-mediated apoptosis was undertaken by exposing cells to DMTU (2 mM) (Sigma) or ZVAD-fmk (100 μM) (Calbiochem), respectively, for 60 min.
Cell-Death Assays: TUNEL and Flow Cytometry.
Control or experimental samples (tissues or cells) were assessed for apoptotic cell death using the DeadEnd Fluorometric Kit (Promega). Cell death in cell culture was quantified using FACS analysis as described previously (84). Briefly, the cells were washed twice with PBS and fixed in 80% methanol for 1 h and then were rinsed in PBS, stained with propidium iodide (50 μg/mL) (Calbiochem), sodium citrate (0.1%), and RNase (1 μg/mL) and were analyzed on a FACScan (Becton Dickinson). The population of cells with reduced DNA content (i.e., showing a reduced G1 peak) was identified as undergoing apoptosis.
Measurement of ROS Levels.
Intracellular levels of ROS were estimated by using the fluorescent dye H2DCFDA (Invitrogen). Control and experimental cells were plated in equal numbers and then were exposed to various forms of oxidative stress. After a given amount of time, the cells were incubated with 10 μM of H2DCFDA for 45 min, washed in PBS, and then placed in growth medium for ∼90 min. The fluorescence of the cells in each group was assessed at an excitation of 490 nm and emission of 520 nm. The fluorescent reading for the experimental group was normalized to the fluorescence of untreated cells loaded with the dye.
Detection of Apoptosis.
Induction of apoptosis in control and stressed cells was measured in vitro by measuring the mitochondrial transmembrane potential and caspase 3, 4, and 7 activity and assessing cytosolic cytochrome c levels. The mitochondrial transmembrane potential was measured by assessing the degree of retention of the dye 3, 3′-dihexyloxacarbocyanine [DiOC6(3)] (Molecular Probes) (85). Briefly, control and experimental C2C12 myoblasts were incubated at various time points poststress with 50 nM DiOC6(3) at 37 °C for 30 min. Subsequently, the cells underwent repeated PBS washes and then were resuspended in 500 μL of PBS. The intensity of the cells was measured using FACS analysis, and cells with low fluorescent intensity were identified as having depolarized mitochondrial membranes.
The combined activities of caspase 3, 4, and 7 were assayed in control and experimental cell extracts by measuring the proteolytic cleavage of the fluorogenic substrate Ac-VAD-AFC(Calbiochem) (84). Finally, subcellular fractionation was undertaken on control and experimental cells, and the protein was prepared for Western blot analysis (85). The amount of cytochrome c released from the mitochondria into the cytosol was assessed by Western blot analysis.
Statistical Analysis.
Data are reported as the mean ± SEM. Statistical significance was assessed by performing a two-tailed Student t test or one-way ANOVA between groups.
Supplementary Material
Acknowledgments
The J.W.S. and J.A.R. laboratories provided key resources and personnel to assist in completing this project. This study was supported by American Heart Association (AHA)-South Central Affiliate Postdoctoral Research Fellowship 09POST2260759 (to S.S.); AHA-South Central Affiliate Postdoctoral Research Fellowship 11POST7620124 (to D.C.C.); AHA-South Central Affiliate Beginning Grant-in-Aid (BGIA) 0655202Y and National Institutes of Health (NIH) Research Grants R01 HL-072016 and R01 HL-097768 (to B.A.R.); AHA-South Central Affiliate BGIA 0565085Y and Grant-in-Aid 09GRNT2250797 (to P.P.A.M.); by the Donald W. Reynolds Clinical Cardiovascular Research Center at University of Texas Southwestern; by the GlaxoSmithKline Research Foundation (to P.P.A.M.); and by NIH Research Grants K08/R01 HL-076440 and HL-102478 (to P.P.A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314962111/-/DCSupplemental.
References
- 1.Deconinck N, Dan B. Pathophysiology of duchenne muscular dystrophy: Current hypotheses. Pediatr Neurol. 2007;36(1):1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Heslop L, Morgan JE, Partridge TA. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J Cell Sci. 2000;113(Pt 12):2299–2308. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- 3.Yokota T, et al. Expansion of revertant fibers in dystrophic mdx muscles reflects activity of muscle precursor cells and serves as an index of muscle regeneration. J Cell Sci. 2006;119(Pt 13):2679–2687. doi: 10.1242/jcs.03000. [DOI] [PubMed] [Google Scholar]
- 4.Hawke TJ, Garry DJ. Myogenic satellite cells: Physiology to molecular biology. J Appl Physiol (1985) 2001;91(2):534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20(13):1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 6.Li J, et al. Evidence of heterogeneity within bovine satellite cells isolated from young and adult animals. J Anim Sci. 2011;89(6):1751–1757. doi: 10.2527/jas.2010-3568. [DOI] [PubMed] [Google Scholar]
- 7.Shefer G, Rauner G, Yablonka-Reuveni Z, Benayahu D. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS ONE. 2010;5(10):e13307. doi: 10.1371/journal.pone.0013307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev Biol. 2010;340(2):330–343. doi: 10.1016/j.ydbio.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scimè A, et al. Transcriptional profiling of skeletal muscle reveals factors that are necessary to maintain satellite cell integrity during ageing. Mech Ageing Dev. 2010;131(1):9–20. doi: 10.1016/j.mad.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Holterman CE, Rudnicki MA. Molecular regulation of satellite cell function. Semin Cell Dev Biol. 2005;16(4-5):575–584. doi: 10.1016/j.semcdb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Cossu G, Biressi S. Satellite cells, myoblasts and other occasional myogenic progenitors: Possible origin, phenotypic features and role in muscle regeneration. Semin Cell Dev Biol. 2005;16(4-5):623–631. doi: 10.1016/j.semcdb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15(12):666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Irintchev A, Wernig A. Muscle damage and repair in voluntarily running mice: Strain and muscle differences. Cell Tissue Res. 1987;249(3):509–521. doi: 10.1007/BF00217322. [DOI] [PubMed] [Google Scholar]
- 14.Wernig A, Irintchev A, Weisshaupt P. Muscle injury, cross-sectional area and fibre type distribution in mouse soleus after intermittent wheel-running. J Physiol. 1990;428:639–652. doi: 10.1113/jphysiol.1990.sp018232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kihlström M, Salminen A, Vihko V. Food deprivation decreases the exertion-induced acid hydrolase response in mouse skeletal muscle. Eur J Appl Physiol Occup Physiol. 1988;57(2):177–180. doi: 10.1007/BF00640659. [DOI] [PubMed] [Google Scholar]
- 16.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 17.Schultz E, Jaryszak DL. Effects of skeletal muscle regeneration on the proliferation potential of satellite cells. Mech Ageing Dev. 1985;30(1):63–72. doi: 10.1016/0047-6374(85)90059-4. [DOI] [PubMed] [Google Scholar]
- 18.Schultz E, Jaryszak DL, Valliere CR. Response of satellite cells to focal skeletal muscle injury. Muscle Nerve. 1985;8(3):217–222. doi: 10.1002/mus.880080307. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan SM, Allen RE. Skeletal muscle satellite cell proliferation in response to members of the fibroblast growth factor family and hepatocyte growth factor. J Cell Physiol. 1999;181(3):499–506. doi: 10.1002/(SICI)1097-4652(199912)181:3<499::AID-JCP14>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Schultz E. Satellite cells in normal, regenerating and dystrophic muscle. Adv Exp Med Biol. 1985;182:73–84. doi: 10.1007/978-1-4684-4907-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Schultz E. Satellite cell proliferative compartments in growing skeletal muscles. Dev Biol. 1996;175(1):84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- 22.Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84(1):209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 23.Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170(4):421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- 24.Singh S, et al. Calcineurin activates cytoglobin transcription in hypoxic myocytes. J Biol Chem. 2009;284(16):10409–10421. doi: 10.1074/jbc.M809572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270(5639):725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 26.Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: A novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol. 2002;19(4):416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 27.Trent JT, 3rd, Hargrove MS. A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem. 2002;277(22):19538–19545. doi: 10.1074/jbc.M201934200. [DOI] [PubMed] [Google Scholar]
- 28.Mammen PP, et al. Cytoglobin is a stress-responsive hemoprotein expressed in the developing and adult brain. J Histochem Cytochem. 2006;54(12):1349–1361. doi: 10.1369/jhc.6A7008.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fordel E, Thijs L, Moens L, Dewilde S. Neuroglobin and cytoglobin expression in mice. Evidence for a correlation with reactive oxygen species scavenging. FEBS J. 2007;274(5):1312–1317. doi: 10.1111/j.1742-4658.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 30.Fordel E, et al. Neuroglobin and cytoglobin overexpression protects human SH-SY5Y neuroblastoma cells against oxidative stress-induced cell death. Neurosci Lett. 2006;410(2):146–151. doi: 10.1016/j.neulet.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 31.Hodges NJ, Innocent N, Dhanda S, Graham M. Cellular protection from oxidative DNA damage by over-expression of the novel globin cytoglobin in vitro. Mutagenesis. 2008;23(4):293–298. doi: 10.1093/mutage/gen013. [DOI] [PubMed] [Google Scholar]
- 32.Li D, et al. Cytoglobin up-regulated by hydrogen peroxide plays a protective role in oxidative stress. Neurochem Res. 2007;32(8):1375–1380. doi: 10.1007/s11064-007-9317-x. [DOI] [PubMed] [Google Scholar]
- 33.Xu R, et al. Cytoglobin overexpression protects against damage-induced fibrosis. Mol Ther. 2006;13(6):1093–1100. doi: 10.1016/j.ymthe.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 34.Stagner JI, Seelan RS, Parthasarathy RN, White K. Reduction of ischemic cell death in cultured Islets of Langerhans by the induction of cytoglobin. Islets. 2009;1(1):50–54. doi: 10.4161/isl.1.1.8936. [DOI] [PubMed] [Google Scholar]
- 35.Geuens E, et al. A globin in the nucleus! J Biol Chem. 2003;278(33):30417–30420. doi: 10.1074/jbc.C300203200. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt M, et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279(9):8063–8069. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- 37.Garry DJ, et al. Postnatal development and plasticity of specialized muscle fiber characteristics in the hindlimb. Dev Genet. 1996;19(2):146–156. doi: 10.1002/(SICI)1520-6408(1996)19:2<146::AID-DVG6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Ordway GA, Garry DJ. Myoglobin: An essential hemoprotein in striated muscle. J Exp Biol. 2004;207(Pt 20):3441–3446. doi: 10.1242/jeb.01172. [DOI] [PubMed] [Google Scholar]
- 39.Garry DJ, et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci USA. 2000;97(10):5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawke TJ, et al. Xin, an actin binding protein, is expressed within muscle satellite cells and newly regenerated skeletal muscle fibers. Am J Physiol Cell Physiol. 2007;293(5):C1636–C1644. doi: 10.1152/ajpcell.00124.2007. [DOI] [PubMed] [Google Scholar]
- 41.Hawke TJ, Jiang N, Garry DJ. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J Biol Chem. 2003;278(6):4015–4020. doi: 10.1074/jbc.M209200200. [DOI] [PubMed] [Google Scholar]
- 42.Hawke TJ, Kanatous SB, Martin CM, Goetsch SC, Garry DJ. Rad is temporally regulated within myogenic progenitor cells during skeletal muscle regeneration. Am J Physiol Cell Physiol. 2006;290(2):C379–C387. doi: 10.1152/ajpcell.00270.2005. [DOI] [PubMed] [Google Scholar]
- 43.Yan Z, et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278(10):8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 44.Hawke TJ, et al. p21 is essential for normal myogenic progenitor cell function in regenerating skeletal muscle. Am J Physiol Cell Physiol. 2003;285(5):C1019–C1027. doi: 10.1152/ajpcell.00055.2003. [DOI] [PubMed] [Google Scholar]
- 45.Li S, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA. 2005;102(4):1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiland TR, Kundu S, Trent JT, 3rd, Hoy JA, Hargrove MS. Bis-histidyl hexacoordination in hemoglobins facilitates heme reduction kinetics. J Am Chem Soc. 2004;126(38):11930–11935. doi: 10.1021/ja046990w. [DOI] [PubMed] [Google Scholar]
- 47.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 48.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 49.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15(2):247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 50.Finkel T. Radical medicine: Treating ageing to cure disease. Nat Rev Mol Cell Biol. 2005;6(12):971–976. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- 51.Reeder BJ. The redox activity of hemoglobins: From physiologic functions to pathologic mechanisms. Antioxid Redox Signal. 2010;13(7):1087–1123. doi: 10.1089/ars.2009.2974. [DOI] [PubMed] [Google Scholar]
- 52.Petersen MG, Dewilde S, Fago A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J Inorg Biochem. 2008;102(9):1777–1782. doi: 10.1016/j.jinorgbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Kiger L, et al. Electron transfer function versus oxygen delivery: A comparative study for several hexacoordinated globins across the animal kingdom. PLoS ONE. 2011;6(6):e20478. doi: 10.1371/journal.pone.0020478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeder BJ, Svistunenko DA, Wilson MT. Lipid binding to cytoglobin leads to a change in haem co-ordination: A role for cytoglobin in lipid signalling of oxidative stress. Biochem J. 2011;434(3):483–492. doi: 10.1042/BJ20101136. [DOI] [PubMed] [Google Scholar]
- 55.Gardner AM, Cook MR, Gardner PR. Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes. J Biol Chem. 2010;285(31):23850–23857. doi: 10.1074/jbc.M110.132340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halligan KE, Jourd’heuil FL, Jourd’heuil D. Cytoglobin is expressed in the vasculature and regulates cell respiration and proliferation via nitric oxide dioxygenation. J Biol Chem. 2009;284(13):8539–8547. doi: 10.1074/jbc.M808231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fago A, et al. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J Biol Chem. 2004;279(43):44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 58.Hamdane D, et al. The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J Biol Chem. 2003;278(51):51713–51721. doi: 10.1074/jbc.M309396200. [DOI] [PubMed] [Google Scholar]
- 59.Fang J, Ma I, Allalunis-Turner J. Knockdown of cytoglobin expression sensitizes human glioma cells to radiation and oxidative stress. Radiat Res. 2011;176(2):198–207. doi: 10.1667/rr2517.1. [DOI] [PubMed] [Google Scholar]
- 60.Fordel E, et al. Anoxia or oxygen and glucose deprivation in SH-SY5Y cells: A step closer to the unraveling of neuroglobin and cytoglobin functions. Gene. 2007;398(1-2):114–122. doi: 10.1016/j.gene.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 61.Stagner JI, Parthasarathy SN, Wyler K, Parthasarathy RN. Protection from ischemic cell death by the induction of cytoglobin. Transplant Proc. 2005;37(8):3452–3453. doi: 10.1016/j.transproceed.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Sun Y, et al. Neuroglobin protects the brain from experimental stroke in vivo. Proc Natl Acad Sci USA. 2003;100(6):3497–3500. doi: 10.1073/pnas.0637726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendgen-Cotta UB, et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2008;105(29):10256–10261. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li RC, Guo SZ, Lee SK, Gozal D. Neuroglobin protects neurons against oxidative stress in global ischemia. J Cereb Blood Flow Metab. 2010;30(11):1874–1882. doi: 10.1038/jcbfm.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raychaudhuri S, Skommer J, Henty K, Birch N, Brittain T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis. 2010;15(4):401–411. doi: 10.1007/s10495-009-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, et al. Effects of neuroglobin overexpression on acute brain injury and long-term outcomes after focal cerebral ischemia. Stroke. 2008;39(6):1869–1874. doi: 10.1161/STROKEAHA.107.506022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li RC, et al. Neuroglobin protects PC12 cells against oxidative stress. Brain Res. 2008;1190:159–166. doi: 10.1016/j.brainres.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mammen PP, et al. Hypoxia-induced left ventricular dysfunction in myoglobin-deficient mice. Am J Physiol Heart Circ Physiol. 2003;285(5):H2132–H2141. doi: 10.1152/ajpheart.00147.2003. [DOI] [PubMed] [Google Scholar]
- 69.Csete M, et al. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J Cell Physiol. 2001;189(2):189–196. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- 70.Franco AA, Odom RS, Rando TA. Regulation of antioxidant enzyme gene expression in response to oxidative stress and during differentiation of mouse skeletal muscle. Free Radic Biol Med. 1999;27(9-10):1122–1132. doi: 10.1016/s0891-5849(99)00166-5. [DOI] [PubMed] [Google Scholar]
- 71.Hansen JM, Klass M, Harris C, Csete M. A reducing redox environment promotes C2C12 myogenesis: Implications for regeneration in aged muscle. Cell Biol Int. 2007;31(6):546–553. doi: 10.1016/j.cellbi.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S, et al. Glutathione-peroxidase-1 null muscle progenitor cells are globally defective. Free Radic Biol Med. 2006;41(7):1174–1184. doi: 10.1016/j.freeradbiomed.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 73.Urish KL, et al. Antioxidant levels represent a major determinant in the regenerative capacity of muscle stem cells. Mol Biol Cell. 2009;20(1):509–520. doi: 10.1091/mbc.E08-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaccagnini G, et al. p66(ShcA) and oxidative stress modulate myogenic differentiation and skeletal muscle regeneration after hind limb ischemia. J Biol Chem. 2007;282(43):31453–31459. doi: 10.1074/jbc.M702511200. [DOI] [PubMed] [Google Scholar]
- 75.Armand AS, et al. Cooperative synergy between NFAT and MyoD regulates myogenin expression and myogenesis. J Biol Chem. 2008;283(43):29004–29010. doi: 10.1074/jbc.M801297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies KJ, Quintanilha AT, Brooks GA, Packer L. Free radicals and tissue damage produced by exercise. Biochem Biophys Res Commun. 1982;107(4):1198–1205. doi: 10.1016/s0006-291x(82)80124-1. [DOI] [PubMed] [Google Scholar]
- 77.Walker PM. Ischemia/reperfusion injury in skeletal muscle. Ann Vasc Surg. 1991;5(4):399–402. doi: 10.1007/BF02015307. [DOI] [PubMed] [Google Scholar]
- 78.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Disatnik MH, et al. Evidence of oxidative stress in mdx mouse muscle: Studies of the pre-necrotic state. J Neurol Sci. 1998;161(1):77–84. doi: 10.1016/s0022-510x(98)00258-5. [DOI] [PubMed] [Google Scholar]
- 80.Rando TA, Disatnik MH, Yu Y, Franco A. Muscle cells from mdx mice have an increased susceptibility to oxidative stress. Neuromuscul Disord. 1998;8(1):14–21. doi: 10.1016/s0960-8966(97)00124-7. [DOI] [PubMed] [Google Scholar]
- 81.Whitehead NP, Pham C, Gervasio OL, Allen DG. N-Acetylcysteine ameliorates skeletal muscle pathophysiology in mdx mice. J Physiol. 2008;586(7):2003–2014. doi: 10.1113/jphysiol.2007.148338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu JR, et al. Control of facial muscle development by MyoR and capsulin. Science. 2002;298(5602):2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- 83.Musarò A, Barberi L. Isolation and culture of mouse satellite cells. Methods Mol Biol. 2010;633:101–111. doi: 10.1007/978-1-59745-019-5_8. [DOI] [PubMed] [Google Scholar]
- 84.Singh S, Khar A. Activation of NFkappaB and Ub-proteasome pathway during apoptosis induced by a serum factor is mediated through the upregulation of the 26S proteasome subunits. Apoptosis. 2006;11(5):845–859. doi: 10.1007/s10495-006-5878-4. [DOI] [PubMed] [Google Scholar]
- 85.Singh S, Khar A. Differential gene expression during apoptosis induced by a serum factor: Role of mitochondrial F0-F1 ATP synthase complex. Apoptosis. 2005;10(6):1469–1482. doi: 10.1007/s10495-005-1394-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.