The viticulture industry has been selectively growing vine cultivars with different traits (grape size, shape, color, flavor, yield of fruit, and so forth) for millennia, and small variations in soil composition, water management, climate, and the aspect of vineyards have long been associated with shifts in these traits. As such, many different clonal varieties of vines exist, even within given grape varieties, such as merlot, pinot noir, and chardonnay. The commensal microbial flora that coexists with the plant may be one of the key factors that influence these traits. To date, the role of microbes has been largely ignored, outside of microbial pathogens, mainly because the technologies did not exist to allow us to look in any real depth or breadth at the community structure of the multitudes of bacterial and fungal species associated with each plant. In PNAS, Bokulich et al. (1) used next-generation sequencing of 16S rRNA and internal transcribed spacer ribosomal sequence to determine the relative abundances of bacteria and fungi, respectively, from grape must (freshly pressed grape juice, containing the skins and seeds) from plants in eight vineyards representing four of the major wine growing regions in California. The authors show that the microbiomes (bacterial and fungal taxonomic structure) associated with this early fermentation stage show defined biogeography, illustrating that different wine-growing regions maintain different microbial communities, with some influences from the grape variety and the year of production.
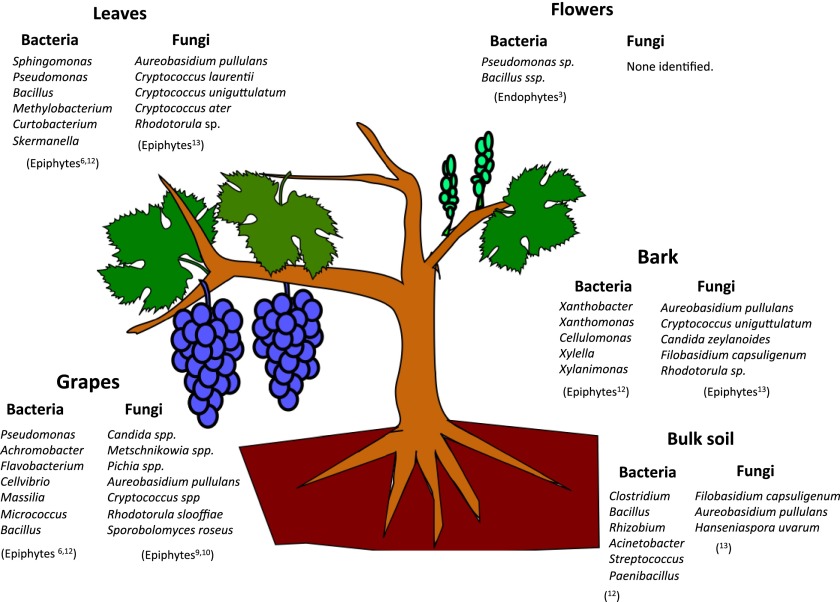
Bacteria and fungi live in complex coassociations with plants and have important roles in shaping the quality of the soil and in promoting the productivity and health of the plant itself. As with the human microbiome (2), the plant microbiome has both direct and indirect relationships with its host, from transforming the availability of organic matter and essential nutrients in the soil, including nitrogen fixation, mitigating the impact of environmental stresses (such as drought or the presence of phytotoxic contaminants), to preventing the growth or activity of plant pathogens through competition for space and nutrients, antibiosis, production of hydrolytic enzymes, inhibition of pathogen-produced enzymes or toxins, and through systemic induction of plant defense mechanisms (3, 4). The most widely studied group of plant-associated microorganisms live in the soil surrounding the roots or inside the roots themselves (endophytes); the interface between roots and soil is often considered the key point of interaction between a plant and its environment. However, microbes colonizing at the root can also migrate through the plant to colonize aerial tissues, either internally or externally (epiphytes). However, the exact way in which microbes make this journey is poorly understood (3). Many microorganisms that actively colonize these tissues have multiple metabolic activities that support plant health, either by promoting growth or suppressing disease-causing pathogens; however, disassociating these two traits and assigning activities to specific taxa is often problematic because of the numerous codependent interactions between the many different species. These beneficial organisms have a very broad phylogeny, but several of them have been well studied, including bacterial genera, such as Azospirillum, Bacillus, etc. (5). In Fig. 1 we highlight some microorganisms that have shown association with the different tissues.
Fig. 1.
Diagrammatic representation of some of characteristic bacteria and fungi known to show associations with the different tissues of Vitis vinifera.
Traditionally, to overcome reduced economic yields caused by disease, viticulturists inoculate the plants with potential antagonists as a biological control for grapevine pathogens (3). However, as our understanding of bacterial and fungal influence on plant characteristics improves, it is possible that microbes could be added to affect other traits. It is possible that different bacteria associated with different tissues could influence the flavor and productivity of grapes, which could impact the organoleptic characteristics of wine. The well-known influence of bacteria and fungi in the wine fermentation, particularly lactic acid bacteria and yeasts, has led to the development of commercial products, including specific fungal taxa to improve wine fermentation processes and flavor. However, the role of prokaryotes was considered to be limited to the malolactic fermentation processes, or even to be only detrimental to fermentation (6). However, metagenomic studies are revealing that grapes can harbor a more diverse microbial community than previously anticipated, for example in botrytized wine ferments (7).
Bokulich et al. (1) focus on the bacteria associated with grapes by directly sampling the initial stage of fermentation, the wine must. Although not previously determined, it stands to reason that the same physical and chemical parameters that determine which plants can grow where (e.g., temperature, humidity, precipitation, soil nutrient concentrations, solar radiation, and so forth) would also have a significant impact on the biogeography of the bacteria and fungi in the ecosystems. The biogeographic regionalization of microbial taxa associated with the grapes begs the question: Where do the taxa come from? For example, is this biogeography based on the native microbial biogeography of the soils or water in the different regions (8), or do the plants themselves select for a different microbiota based on their physiological response to different climatic and soil edaphic variables? Up to now, studies of grape microbiota biogeography have mostly focused on the distribution of yeast, where Saccharomyces cerevisiae populations have been shown to exhibit periodic fluctuation in their presence or absence across different regions, influenced by climatic factors and vineyard age and size (9). Setati et al. (10) studied the spatial distribution of fungal microbial communities within and between vineyards from the same terroir, finding higher yeast heterogeneity on grape samples collected at different points inside individual vineyards than between vineyards with very contrasting farming strategies, which they attribute to myriad microclimates within a vineyard created by differential shading of grapes by leaves, and the aspect of each grape cluster.
Bokulich et al. (1) surveyed 273 grape musts from two vintages of Chardonnay, Zinfandel, and Cabernet Sauvignon, demonstrating categorically that these grape-surface microbial communities were significantly, albeit rather weakly, different between regions. However, the authors also show that the degree of significant differentiation between regions is increased dramatically when they look at the biogeography within a grape variety of vintage. This finding suggests that other factors also play a significant role, including host genotype, and therefore phenotype (grape variety), and local and interannual climate variation (vintage).
Bringing microbial ecology into agriculture presents a golden opportunity to provide mechanistic understanding for observations that farmers and viticulturists have been making for millennia. This is an essential step forward in that it can help to revolutionize how sites for agriculture are chosen, or indeed how they could be manipulated by probiotics designed to establish the “right” bacterial species that can improve soil quality and, hence, crop productivity. This practice can also be used to help improve the wine terroir or even reproduce those terroirs in sites a priori not suitable for generating a wine with such characteristics. However, to be able to exploit the vine microbiome we need to make considerable advances, including: (i) a more comprehensive characterization of the vine-associated microbiota, (ii) an elucidation of the functional potential of these communities, and (iii) a comprehensive analysis of the genomes and regulatory elements associated with each microbe in the context of its interacting role with the plant and in the fermentation stages of wine production. To make these feasible it is necessary to focus both amplicon and metagenomic sequencing on a comprehensive assessment of the microbiota of the epiphytes and endophytes on the grapevine as a whole, including leaves, flowers, roots, stems, and indeed the very soil in which the plants are growing. The microbes associated with grapes are not isolated from the rest of the grapevine niche, and indeed the roots may be the primary source of colonization, and the transmission of these bacteria through the host may be necessary to elicit the favorable wine characteristics that are sought after. Alternatively, it is feasible that the bacteria and fungi only play a role in the fermentate through release of secondary metabolites, and that they have little direct influence on the grape chemistry.
Therefore, as well as soil-derived microbial colonization through root endophytes, there are additional routes for colonization, including soil dust contamination by rain splash, or from people during harvesting, or other agricultural practices; indeed, even the phyllosphere (leaf zone) of nearby plants may transfer microbial taxa through aerial or insect transportation, which suggests even the local biodiversity may impact which taxa are found in which regions (6). Although it is expected that climatic factors could constrain the biogeographic distribution of grape microbiome, the soil characteristics (e.g., availability of nutrients for the plant) and soil structure still has a great impact on constraining the microbiota that could colonize the remaining plant niches (11–14).
The differences in soil microbiota could also generate differential responses in the plant itself, by triggering the expression of different compounds or activating defense mechanisms that could eventually modulate the activity and abundances of other microbes associated with the plant. Elucidating these characteristics is essential to enable more directed vineyard establishment; this is predicated on a better understanding of the mechanisms that select which organisms associate with plant tissues and the environmental factors shaping their complex community composition. Although the biogeography of grape-associated taxa is potentially important (1), the factors that shape endophyte populations in other tissues may be more important, in that these taxa are likely to play a highly significant role in changing the productivity, chemical properties, and pathogen defense for the plant. Although microorganisms associated with grapes are undoubtedly included in the initial fermentation steps, the bacteria or fungi associated with other plant tissues—and indeed the soil—may play a highly significant role in influencing the flavor, color, and quality of the final product; however, this still remains to be shown.
There is a long way to go, but such work provides us tantalizing evidence that the biogeographic characteristics of terrestrial microorganisms may indeed lead to regionalized properties associated with valuable crops. Future work will build on this ecological observation to change the face of agriculture, much as the human microbiome is changing the face of medicine.
Footnotes
The authors declare no conflict of interest.
See companion article on page E139.
References
- 1.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA. 2014;111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A. Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb Ecol. 2011;62(1):188–197. doi: 10.1007/s00248-011-9883-y. [DOI] [PubMed] [Google Scholar]
- 4.Mastretta C, et al. Endophytic bacteria and their potential application to improve the phytoremediation of contaminated environments. Biotechnol Genet Eng Rev. 2006;23:175–207. doi: 10.1080/02648725.2006.10648084. [DOI] [PubMed] [Google Scholar]
- 5.Berg G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84(1):11–8. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 6.Leveau JHJ, Tech JJ. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. Acta Hortic. 2011;905(2):31–42. [Google Scholar]
- 7.Bokulich NA, Joseph CM, Allen G, Benson AK, Mills DA. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS ONE. 2012;7(5):e36357. doi: 10.1371/journal.pone.0036357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martiny JB, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol. 2006;4(2):102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 9.Barata A, Malfeito-Ferreira M, Loureiro V. The microbial ecology of wine grape berries. Int J Food Microbiol. 2012;153(3):243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Setati ME, Jacobson D, Andong UC, Bauer F. The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE. 2012;7(12):e52609. doi: 10.1371/journal.pone.0052609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins G, et al. Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS ONE. 2013;8(8):e73013. doi: 10.1371/journal.pone.0073013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabate J, Cano J, Esteve-Zarzoso B, Guillamón JM. Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol Res. 2002;157(4):267–274. doi: 10.1078/0944-5013-00163. [DOI] [PubMed] [Google Scholar]
- 14.Bulgarelli D, Schlaeppi K, Spaepen S. Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]