Significance
The ability of bacteria to survive antibiotic challenge without mutation (a phenomenon known as “adaptive resistance”) has been traditionally viewed as the consequence of bacterial responses to environmental cues. This study shows that adaptive resistance can also occur in the absence of known environmental stimuli. Cell-to-cell fluctuations in critical physiological traits preadapt certain cells within an isogenic culture to survive lethal selection, and growth of the “lucky” survivors generates bacterial subpopulations with increased antibiotic resistance. Phenotypic heterogeneity must therefore be added to the list of factors that limit the efficacy of antibiotic treatment.
Abstract
Antibiotic-resistant isolates of Salmonella enterica were selected on plates containing lethal concentrations of rifampicin, kanamycin, and nalidixic acid. The stability of the resistance phenotype was scored after nonselective growth. Rifampicin-resistant (Rifr) isolates were stable, suggesting that they had arisen by mutation. Mutations in the rpoB gene were detected indeed in Rifr mutants. In contrast, a fraction of kanamycin-resistant (Kmr) and nalidixic acid-resistant (Nalr) isolates showed reduced resistance after nonselective growth, suggesting that mechanisms other than mutation had contributed to bacterial survival upon lethal selection. Single-cell analysis revealed heterogeneity in expression of the porin gene ompC, and subpopulation separation provided evidence that reduced ompC expression confers adaptive resistance to kanamycin. In the case of Nalr isolates, mutations in the gyrA gene were present in most nalidixic acid-resistant isolates. However, the efflux pump inhibitor Phe-Arg-β-naphtylamide (PAβN) reduced the level of resistance in Nalr mutants, indicating that active efflux contributes to the overall level of nalidixic acid resistance. Heterogeneous efflux pump activity was detected in single cells and colonies, and a correlation between high efflux and increased resistance to nalidixic acid was found. These observations suggest that fluctuations in the expression and the activity of critical functions of the bacterial cell, alone or combined with mutations, can contribute to adaptive resistance to antibiotics.
Except for bacterial species that undergo developmental programs, bacteria have been traditionally viewed as clonal populations of identical cells. In fact, classic genetics and physiology have routinely used batch cultures of bacteria and individual bacterial colonies, assuming that all cells were identical. Because bacterial mutation rates are in the range of 10−10 per base pair per replication (1), most cells in a liquid culture or within a colony are isogenic indeed. However, genetic identity does not necessarily imply phenotypic identity. The existence of spatial organization in Escherichia coli colonies has been known for almost a century (2–4), and the occurrence of diverse gene-expression patterns inside a colony was described 25 years ago (5). These historic examples are not rare exceptions: in the last few decades, single-cell analysis has provided examples of phenotypic variability in bacterial populations made of isogenic cells, both under laboratory conditions and in natural environments (6–12).
Phenotypic heterogeneity can be the consequence of chemical communication, leading to a heterogeneous response at the single-cell level (13). In other cases, however, phenotypic heterogeneity arises either as a programmed epigenetic event or at random, without the involvement of environmental cues. Classic phenomena involving programmed heterogeneity are the bifurcation of a bacterial population into two distinct states or “bistability” (14) and the reversible switching of gene expression or “phase variation” (15). Randomly generated heterogeneity is usually the consequence of noisy gene expression (16, 17). The distinction between programmed and random heterogeneity, albeit useful in practice, is not always clear-cut: quantitative differences caused by noise can become qualitative above a threshold, triggering a programmed response (18). Another source of heterogeneity is gene amplification, which spontaneously occurs in a fraction of cells within a bacterial population (19).
The selective value of phenotypic heterogeneity can be envisioned in certain cases (6, 20). Furthermore, theoretical analysis supports the view that randomly generated phenotypic diversity can increase the chances of survival when bacterial populations are subjected to rapid, severe, or complex environmental fluctuations (21, 22). Such bet hedging strategies imply group selection, which has been considered intrinsically weak in classic population biology studies (23). However, models based on game theory suggest that strategies that generate phenotypic heterogeneity can provide an evolutionary advantage, despite the fact that they lower the immediate fitness of individual organisms (24–26). Models based on information theory also support the view that bet hedging can be advantageous in harsh and changing environments (27).
Coevolution of bacteria with natural antibacterial compounds has fostered the evolution of resistance mechanisms, usually classified into three types: innate resistance, acquired resistance (e.g., by mutation and by horizontal transfer of genetic determinants), and adaptive resistance (28–30). Adaptive resistance typically involves environmentally induced gene-expression changes that increase the ability of a bacterium to survive in the presence of an antibiotic (31–36). In this study, we provide evidence that cell-to-cell fluctuations in the expression and activity of critical cellular functions can induce adaptive resistance to antibiotics in the absence of known environmental stimuli. Physiological differences preadapt certain cells within an isogenic culture to survive lethal selection. If a feedback loop propagates the physiological state that permits survival, growth of the “lucky” survivors generates a bacterial population with increased antibiotic resistance.
Results
Characterization of Antibiotic-Resistant Derivatives of Salmonella enterica SL 1344.
Antibiotic-resistant colonies of S. enterica were selected by plating aliquots (approximately 2–3 × 108 cells) from an S. enterica Luria-Bertani broth (LB) culture on LB agar supplemented with a lethal concentration of kanamycin, nalidixic acid, or rifampicin (25 μg/mL, 10 μg/mL, and 100 μg/mL, respectively). Resistant colonies appeared at frequencies of ≥10−7 mutants per colony-forming-unit, which roughly correspond to mutation rates of ≥10−10 (37). Because the selection was lethal, antibiotic-resistant colonies were expected to derive from “preadapted” antibiotic-resistant cells present in the previous culture. Preadaptation was confirmed by Luria-Delbrück fluctuation analysis (38) (Tables S1–S3).
The stability of antibiotic resistance was scored after nonselective growth. For this purpose, antibiotic-resistant colonies were transferred to LB and grown overnight. The minimal inhibitory concentration (MIC) of each antibiotic for individual isolates was then determined. Results from these experiments (Fig. 1) can be summarized as follows:
Fig. 1.
Stability of the antibiotic-resistant phenotype in independent S. enterica isolates. MICs of rifampicin (A), kanamycin (B), and nalidixic acid (C) for antibiotic-resistant isolates after nonselective growth in LB. Three replicas for each isolate were performed, and the MICs are represented as bars of different colors. Shaded areas indicate the concentration of antibiotic used for selection of antibiotic-resistant isolates (100 μg/mL rifampicin, 25 μg/mL kanamycin, and 10 μg/mL nalidixic acid).
First, all rifampicin-resistant (Rifr) isolates grew in the presence of a high concentration of the antibiotic, and the MICs of rifampicin showed high reproducibility in the replicas (Fig. 1A). Because the degree of resistance of such isolates was stably maintained upon nonselective growth, they were tentatively considered mutants. Because rifampicin resistance is often associated with mutations in the β-subunit of RNA polymerase (39, 40), the rpoB gene of three independent isolates (#12, #13, and #19) was PCR-amplified and sequenced. Nucleotide substitutions that yielded a Q512R amino acid replacement were found.
Second, the MIC of kanamycin showed low reproducibility among individual isolates, with substantial variation between replicas (Fig. 1B). A fraction of kanamycin-resistant (Kmr) isolates showed a level of resistance below the concentration used for selection (Fig. 1B). Hence, Kmr isolates appeared to belong to two classes: (i) stable, putatively carrying mutations that confer kanamycin resistance; and (ii) unstable isolates that lost antibiotic resistance, partially or completely, upon nonselective growth, suggesting that resistance was not mutational. The numbers of stable Kmr isolates showed Luria-Delbrück fluctuation but the numbers of unstable Kmr isolates did not (Table S4).
Third, nalidixic acid-resistant (Nalr) isolates also showed a broad distribution of MIC values but none showed a level of resistance below the concentration used in the initial selection (Fig. 1C). Low reproducibility between replicas was also observed (Fig. 1C). A tentative interpretation was that mutations provided a given level of nalidixic acid resistance but nonmutational resistance contributed to the overall level of resistance in certain isolates.
Contribution of ompC Down-Regulation to Nonmutational Resistance to Kanamycin.
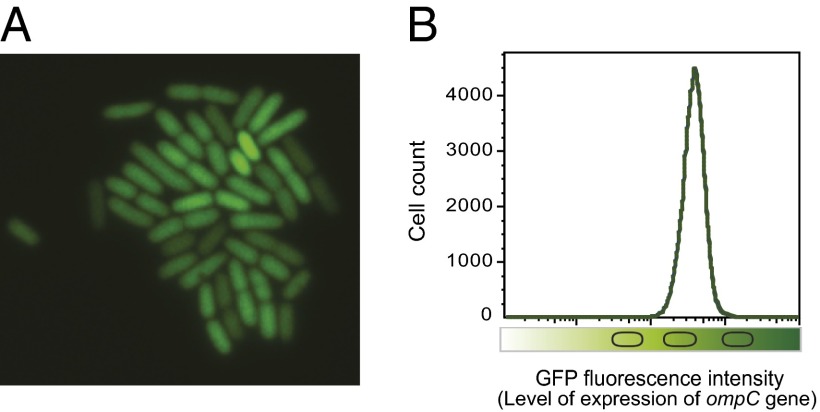
A conceivable explanation for nonmutational resistance to kanamycin was that, in the culture previous to selection, certain cells were in a physiological state that preadapted them to resist killing by the antibiotic. For example, certain cells might be partially or completely refractory to antibiotic uptake. Aminoglycosides diffuse through porin channels in the outer membrane of Gram-negative bacteria, and a low level of aminoglycoside resistance can be acquired by reduced drug uptake (41, 42). Because reduced synthesis of OmpC porin has been shown to contribute to kanamycin resistance (43), we designed experiments to test: (i) whether variations in the level of OmpC synthesis occurred in individual cells of an isogenic population grown in LB broth; and (ii) whether reduced ompC expression yielded Kmr cells. To monitor ompC gene expression in individual S. enterica cells, an ompC::GFP fusion was used. A wide range of fluorescence intensities was detected, thus confirming the occurrence of heterogeneous ompC::GFP expression (Fig. 2).
Fig. 2.
Single-cell analysis of ompC::GFP expression in an isogenic bacterial population. (A) A microcolony of S. enterica strain SV6811 (ompC::GFP) visualized by fluorescence microscopy with a 100× objective. (B) GFP fluorescence intensity distribution in a liquid culture of strain SV6811, monitored by flow cytometry.
In an attempt to correlate ompC expression (fluorescence intensity) and survival (antibiotic resistance), two fractions of the population were sorted in the high and low fluorescence intensity windows (Fig. 3A). The collected cells were plated on LB agar containing kanamycin (25 μg/mL). Kmr colonies were three- to fourfold more abundant in the population that expressed ompC at low levels (Fig. 3B). Therefore, the decrease in ompC expression can be correlated with an increase in antibiotic resistance. A tentative interpretation is that reduced ompC expression may lower the number of porin channels in the outer membrane, thus reducing drug uptake (42, 44). Down-regulation of ompC expression was found to be reversible (Fig. S1), thereby confirming the nongenetic origin of the kanamycin-resistant subpopulation.
Fig. 3.
Correlation between ompC expression and kanamycin resistance. (A) Flow cytometry analysis of GFP fluorescence intensity and percentages of cells within the gates set as high or low GFP fluorescence. Two gates were drawn for each population to include the 10% of cells that expressed GFP at the lower levels, and the 10% of cells expressing GFP at the higher levels. After sorting, aliquots of sorted cells were run again at the cytometer to confirm the efficacy of sorting. (B) Survival of sorted S. enterica cells showing low and high ompC expression. Aliquots from the sorted subpopulations were spread on LB agar and LB agar + kanamycin (25 μg/mL). The absolute numbers of colonies have been normalized to facilitate visual comparison.
Characterization of Nalidixic Acid-Resistant Derivatives of S. enterica: Mutational and Nonmutational Preadaptation.
High levels of resistance to nalidixic acid are often associated with the presence of mutations in the quinolone resistance-determining region (QRDR) of genes that encode gyrase (gyrA and gyrB) and topoisomerase IV (45). Sequencing of the gyrA QRDR region in independent nalidixic acid-resistant isolates revealed mostly amino acid substitutions at codons 81, 83, and 87, as described previously (46, 47). Mutations affecting amino acid 83 caused higher levels of nalidixic acid resistance than those affecting amino acids 87 and 81 (Fig. 4A). However, certain isolates carrying identical mutations showed different MICs (Fig. 4A), and all of the isolates showed high variation between independent replicas (see error bars in Fig. 4A). These variations, we reasoned, might indicate that mutations in the target genes did not thoroughly account for the resistance phenotypes. Hence, we hypothesized that nonmutational mechanisms might be also involved.
Fig. 4.
Characterization of nalidixic acid-resistant isolates of S. enterica. (A) MICs of 46 nalidixic acid-resistant isolates, ordered by average resistance level. Error bars indicate SDs from three MIC replicas. The gyrA QRDR mutations carried by the isolates are indicated. Color codes indicate the GyrA amino acid affected: red, #83; blue, #87; green, # 81; gray, other amino acids. Isolates 19, 34, and 28 did not harbor mutations in gyrA. (B) MICs of nalidixic-resistant isolates of S. enterica carrying gyrA QRDR mutant alleles in the absence and in the presence of the efflux pump inhibitor PAβN. The shaded area indicates the concentration of nalidixic acid used for selection of nalidixic acid-resistant isolates (10 μg/mL).
Although fluoroquinolone resistance has been mainly attributed to mutations (46, 47), resistance can be also mediated by antibiotic efflux (48–51). We tested whether an efflux pump inhibitor, Phe-Arg-β-naphthylamide (PAβN, 20 μg/mL), altered the MIC of nalidixic acid in Nalr isolates showing different and identical mutations in gyrA. In most isolates, addition of PAβN did not fully revert nalidixic acid resistance but decreased the MIC regardless of the types of mutations present in the QRDR of gyrA (Fig. 4B). This effect was especially noticeable among isolates showing low resistance, in which the MIC of nalidixic acid decreased from resistance to sensitivity (in other words, to MICs similar to that of the wild-type). These observations suggest that most amino acid changes in GyrA are unable to confer high level of resistance to nalidixic acid. Even isolates carrying mutations in codon 83, which confer high levels of resistance, showed decreased resistance upon efflux pump inhibition (Fig. 4B). Therefore, high levels of resistance to nalidixic acid seem to result from a combination of mutational and nonmutational mechanisms (QRDR mutation and active efflux, respectively).
Contribution of Heterogeneous Efflux Pump Activity to Adaptive Resistance to Nalidixic Acid.
Evidence for heterogeneous efflux in individual S. enterica cells was obtained using ethidium bromide (EtBr), a common substrate of bacterial efflux pumps (52–54). Heterogeneous EtBr accumulation was observed (Fig. 5A). A priori, cell-to-cell differences in EtBr accumulation may reflect differences in efflux as described in the literature (52), but also cell-to-cell differences in EtBr uptake. Two observations indicate that EtBr accumulation can be used indeed to monitor active efflux: (i) addition of PAβN increased EtBr fluorescence (Fig. 5B); and (ii) in strain SV7371 (∆acrAB), which lacks a major efflux pump, EtBr fluorescence increased to levels similar to those detected in the presence of PAβN (Fig. 5B).
Fig. 5.
Contribution of heterogeneous efflux pump activity to nalidixic acid resistance. (A) Flow cytometry assessment of EtBr accumulation in an isogenic culture of S. enterica. Data are represented by a dot plot [cellular size (forward side) versus fluorescence intensity (EtBr accumulation)]. (B) EtBr fluorescence of a S. enterica culture in the presence of PAβN 20 μg/mL (blue histogram) and in the absence of PAβN (red histogram). As a control, EtBr fluorescence was also monitored in a culture of strain SV7371 (∆acrAB) (green histogram). Because different numbers of cells were counted for each culture, the cell numbers have been normalized to 100. (C) Heterogeneous accumulation of EtBr by S. enterica colonies grown on LB agar supplemented with EtBr (1 μg/mL). EtBr fluorescence was detected under UV illumination. A high level of fluorescence indicates low efflux, and a low level of fluorescence indicates high efflux. (D) Replica-plating of S. enterica colonies grown on LB agar supplemented with EtBr to LB agar supplemented with nalidixic acid (10 μg/mL). (E) Merging of B and C illustrates the correlation between colony color (indicative of efflux pump activity) and nalidixic acid resistance.
Evidence that nalidixic acid resistance can be affected by active efflux was obtained using EtBr-agar, which permits to distinguish colonies with different levels of efflux pump activity (52). When an aliquot from an S. enterica batch culture (small enough to permit formation of isolated colonies) was plated on LB + EtBr, colonies of different colors appeared (Fig. 5C): accumulation of EtBr makes colonies pink and low intracellular levels of EtBr makes colonies white (52). When colonies growing on LB + EtBr were transferred to LB + nalidixic acid by replica-plating (55), colonies exhibiting high fluorescence turned out to be sensitive to nalixidic acid, but nonfluorescent colonies were nalidixic acid-resistant (Fig. 5 D and E).
Because spontaneous gene amplification is a source of heterogeneity (19) and acrAB duplication has been shown to increase active efflux (56), acrAB gene dosage was measured in 17 Nalr colonies (Fig. S2). One case of acrAB amplification was detected, suggesting that spontaneous gene amplification can contribute to nalidixic acid resistance. However, cell-to-cell variations in efflux pump synthesis or activity may be a more frequent cause of heterogeneous nalidixic acid resistance.
Discussion
This study shows that cell-to-cell differences in the expression or activity of critical cell functions can contribute to adaptive resistance to lethal concentrations of antibiotics. Evidence that the antibiotic concentrations used did cause lethality was provided by absence of growth on antibiotic plates, except for antibiotic-resistant colonies that appeared at frequencies typical of spontaneous mutation. As expected, most isolates obtained under such conditions were stable and were considered bona fide mutants (e.g., all of the rifampicin-resistant mutants under study). However, kanamycin-resistant isolates that partially or completely lost resistance after nonselective growth were obtained. Because reversion cannot explain loss of resistance at high frequency, a tentative explanation was that certain cells in the previous culture happened to be in a physiological state that permitted survival upon selection with kanamycin.
Reduced expression of ompC, a gene encoding an outer-membrane porin, has been shown to increase kanamycin resistance (41, 43). Hence, we hypothesized that one cause (perhaps among others) of adaptive resistance to kanamycin might be reduced ompC expression. If such was the case, we reasoned, ompC expression should be heterogeneous in S. enterica cultures, so that only certain cells would be able to survive lethal selection with kanamycin. When ompC expression was monitored at the single-cell level, heterogeneous expression was indeed detected (Fig. 3A), and S. enterica subpopulations exhibiting high and low levels of ompC expression did differ in their levels of kanamycin resistance (Fig. 3B). We thus propose that ompC expression is noisy, and that reduced ompC expression confers adaptive resistance to kanamycin in a fraction of the bacterial population. Because kanamycin down-regulates ompC expression (42), a feedback loop can be expected to propagate the cellular state that initially permitted survival.
Loss of resistance was not detected among nalidixic-resistant isolates, and mutations in the QRDR region of the gyrA gene were found in most Nalr isolates (Fig. 4A). However, several observations suggested that the overall level of nalidixic acid resistance was not solely a result of mutation: (i) isolates carrying identical QRDR mutations showed different MICs (Fig. 4A); (ii) in most Nalr isolates, nonselective growth caused a substantial decrease in the MIC (Fig. 1C); and (iii) variations in the MIC were detected among replicas performed for the same isolate (Figs. 1C and 4). Taken together, these observations led us to hypothesize that resistance to nalidixic acid might have two components, one mutational and another adaptive.
Because active efflux plays a role in quinolone resistance (34), we tested whether inhibition of efflux with PAβN reduced the MIC of isolates carrying QRDR mutations. Reduction of MIC values was obtained for all of the isolates tested, irrespective of the QRDR mutation present in the gyrA gene (Fig. 4B). We thus concluded that the overall level of nalidixic resistance found in most Nalr isolates had two components indeed. One was mutation in the QRDR determinant of the gyrA gene; the other was adaptive resistance caused by active efflux.
Use of EtBr to monitor efflux pump activity confirmed the occurrence of efflux fluctuations: (i) broad distribution of EtBr fluorescence was detected by flow cytometry, indicating heterogeneous efflux pump activity in individual S. enterica cells (Fig. 5 A and B); (ii) heterogeneous EtBr fluorescence was also observed among S. enterica colonies grown on LB agar (Fig. 5C), and replica-plating revealed that colonies with low EtBr fluorescence were nalidixic acid-resistant but colonies with high EtBr fluorescence were nalidixic acid-sensitive (Fig. 5 D and E). Hence, variations in efflux pump activity seem to explain the differences in nalidixic acid resistance observed among S. enterica isolates carrying QRDR mutations and among replicas of MIC assays for a given isolate. In other words, QRDR mutations may confer steady levels of resistance but heterogeneous efflux pump activity may confer unsteady, adaptive resistance.
Although adaptive resistance typically occurs in response to environmental conditions (28), this study provides evidence that mechanisms of adaptive resistance can be activated in the absence of known trigger signals. Spontaneous activation does not occur in the entire bacterial population but in a subpopulation of cells, and may occur at random. This view is in agreement with current notions indicating that isogenic bacterial populations show remarkable levels of phenotypic heterogeneity (18, 57, 58), which is often a consequence of cellular noise (17, 21, 59, 60). Because resistance to stressful conditions can have adverse effects on bacterial physiology (60), subpopulation formation may involve a trade-off between fitness and survival. In analogy with other cell responses that show high levels of heterogeneity, spontaneous activation of mechanisms that confer adaptive resistance may be viewed as a bet-hedging strategy (57, 61, 62). Preadaptation of certain cells to antibiotic challenge will increase the chances that a fraction of the bacterial population survives (63, 64).
Materials and Methods
Bacterial Strains, Media, Culture Conditions, and Chemicals.
Strains of S. enterica serovar Typhimurium derive from SL1344 (65). SV7371 (∆acrAB) belongs to the laboratory collection. LB was used as liquid medium. LB plates contained agar at 15 g/L final concentration. Cultures were grown at 37 °C with shaking. Kanamycin sulfate, nalidixic acid, rifampicin, PAβN, and EtBr were purchased from Sigma Aldrich.
Determination of Minimal Inhibitory Concentrations of Kanamycin, Nalidixic Acid, and Rifampicin.
Exponential cultures in LB broth were prepared, and samples containing around 3 × 102 colony forming units were transferred to polypropylene microtiter plates (Soria Genlab) containing known amounts of antibiotic. After 12-h incubation at 37 °C, growth was visually monitored.
Construction of an ompC::gfp Fusion (Strain SV6811).
A fragment containing the promoterless green fluorescent protein (gfp) gene and the chloramphenicol resistance cassette was amplified from pZEP07 (66) using primers 5′ GCA TCA ACA CCG ACG ACA TCG TAG CGC TGG GTC TGG TTT ACC AGT TCT AAT AAG AAG GAG ATA TAC ATA TGA G 3′, and 5′ TAA GGC ATG AAA AAA GGG CCC GCA GGC CCT TTA GCA ACA TCT TTT GCT GAT TAT CAC TTA TTC AGG CGT A 3′. The 5′ regions of these primers are homologous to the 3′ untranslated region of the S. enterica ompC gene, so that the fusion is formed downstream of the ompC stop codon and the strain is OmpC+. The construct was integrated into the chromosome of S. enterica using the Lambda Red recombination system (67).
Sequencing of gyrA and rpoB Alleles.
The QRDR of the S. enterica gyrA gene was PCR-amplified using primers 5′ GAG ATG GCC TGA AGC CGG TA 3′ and 5′ GGC ATG ACG TCC GGA ATT 3′. The rpoB gene was PCR-amplified using Go-Taq DNA polymerase (Promega) and sets of primers that allowed direct sequencing of PCR products: 5′ CGA GCA AGA TCC TGA AGG GC3 ’, 5′ CGG CTG AAC AAG CTG GAT TC3 ’, 5′ TCA GCG AGC TGA GGA ACC CT3 ’, 5′ GAC TAC GTT GAC GAA TCT AC 3′, 5′ GAA CTC CAA CCT GGA TGA CG 3′, 5′ TTA CTC GTC TTC CAG TTC GA 3′, 5′ TTA CTC GTC TTC CAG TTC GA 3′, 5′ GGA TGA ATC CGG TAT CGT TT 3′, and 5′ ACT TCC ATC TCC CCG AAG CG 3′. PCR products were purified using the MinElute PCR Purification Kit (Qiagen) and sequenced at the facilities of Stab Vida, Caparica, Portugal. DNA sequences were aligned with the gyrA and rpoB sequences of S. enterica SL1344 available at the Web site of the Wellcome Trust Sanger Institute, Hinxton, England.
Flow Cytometry and Cell Sorting.
Exponential cultures were washed and resuspended in PBS to a final concentration of 5 × 106 cells/mL. Cells were sorted using a MoFloTM XDP cytometer (Beckman Coulter). Immediately before sorting, 5 × 106 cells were analyzed for GFP expression. Based on this analysis, gates were drawn to separate the 10% of cells expressing lower GFP levels, and the 10% of cells expressing higher GFP levels. From each gate, 1 × 106 cells were collected into a sterile tube. After sorting, cells were spun at 6,000 rpm for 10 min and FACS buffer was removed. An aliquot of sorted cells was run again at the cytometer to confirm the purity of the preparation. Data were obtained with CXP and WinMDI Software.
Assessment of Efflux Pump Activity by Flow Cytometry.
Flow cytometry was used to monitor accumulation and efflux of EtBr on a real-time basis (52, 53). Data acquisition and analysis were performed using a Cytomics FC500-MPL cytometer (Beckman Coulter). EtBr was excited at 488 nm, and fluorescence was detected using a 585-nm filter. S. enterica strains SL1344 and SV7371 (∆acrAB) were grown in 10 mL of LB broth at 37 °C until an OD600 of 0.3. EtBr was then added (1 μg/mL). When appropriate, PAβN was also added (20 μg/mL). After incubation at 22 °C for 60 min to permit EtBr accumulation, aliquots of 0.5 mL were taken (53). The bacterial suspension was centrifuged for 3 min at 3,400 × g and the pellet was resuspended in PBS for fluorescence measurement. Data were collected for 100,000 events per sample.
Fluorescence Microscopy.
Strain SV6811 (ompC::GFP) was grown at 37 °C in LB, diluted 1:100 in fresh medium, and grown to an OD600 of 0.1–0.2. Samples of 1.5 mL were collected by centrifugation at 3,400 × g for 5 min. Cells were placed on an agarose slab (0.9% agarose/1% LB medium) prewarmed at 37 °C. Images were captured with a Leica DMR fluorescence microscope (Leica Camera).
EtBr–Agar Method to Screen for Nalidixic Acid Susceptibility.
An S. enterica SL1344 culture was swabbed onto LB-agar plates containing EtBr at a concentration of 1 μg/mL, and the plates were incubated at 37 °C in the dark during 16 h. The fluorescence produced upon excitation of EtBr by UV light was detected with a UV transilluminator. Low-fluorescence indicates high efflux and higher fluorescence indicates low efflux (52). EtBr–agar plates containing colonies with diverse levels of fluorescence were replicated to LB agar plus nalidixic acid using replica plating velvets (55).
Supplementary Material
Acknowledgments
We thank Modesto Carballo, Laura Navarro, and Cristina Reyes (Servicio de Biología, Centro de Investigación Tecnología e Innovación, Universidad de Sevilla), and Alberto Álvarez and Pilar Torralbo (Servicio de Técnicas Aplicadas a la Biociencia, Universidad de Extremadura, Badajoz) for help in experiments performed at the facilities; Elena Puerta-Fernández for encouragement, helpful discussions, and suggestions; and Sara Hernández for help in AcrAB experiments. Strain SV7371 vas constructed by Verónica Urdaneta. This work was supported by Grants BIO2010-15023 and CSD2008-00013 from the Ministerio de Economía e Innovación of Spain and the European Regional Fund, and by Grant CVI-5879 from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316084111/-/DCSupplemental.
References
- 1.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legroux R, Magrou J. État organisé des colonies bactériennes. Ann Inst Pasteur (Paris) 1920;34:417–431. [Google Scholar]
- 3.Shapiro JA. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol. 1987;169(1):142–156. doi: 10.1128/jb.169.1.142-156.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho H, et al. Self-organization in high-density bacterial colonies: Efficient crowd control. PLoS Biol. 2007;5(11):e302. doi: 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro JA, Higgins NP. Differential activity of a transposable element in Escherichia coli colonies. J Bacteriol. 1989;171(11):5975–5986. doi: 10.1128/jb.171.11.5975-5986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1(2):117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 7.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61(3):795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 8.Menendez A, et al. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J Infect Dis. 2009;200(11):1703–1713. doi: 10.1086/646608. [DOI] [PubMed] [Google Scholar]
- 9.Helaine S, et al. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci USA. 2010;107(8):3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 11.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10(1):30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 13.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8(5):431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61(3):564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 15.van der Woude MW. Phase variation: How to create and coordinate population diversity. Curr Opin Microbiol. 2011;14(2):205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 17.Silva-Rocha R, de Lorenzo V. Noise and robustness in prokaryotic regulatory networks. Annu Rev Microbiol. 2010;64:257–275. doi: 10.1146/annurev.micro.091208.073229. [DOI] [PubMed] [Google Scholar]
- 18.Casadesús J, Low DA. Programmed heterogeneity: Epigenetic mechanisms in bacteria. J Biol Chem. 2013;288(20):13929–13935. doi: 10.1074/jbc.R113.472274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson RP, Roth JR. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 20.Hernández SB, Cota I, Ducret A, Aussel L, Casadesús J. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet. 2012;8(1):e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167(1):523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309(5743):2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 23.Hull D. Individuality and selection. Annu Rev Ecol Syst. 1980;11:311–332. [Google Scholar]
- 24.Grafen A. Formal Darwinism, the individual-as-maximizing-agent analogy and bet-hedging. Proc Biol Sci. 1999;266(1421):799–803. [Google Scholar]
- 25.Wolf DM, Vazirani VV, Arkin AP. A microbial modified prisoner’s dilemma game: How frequency-dependent selection can lead to random phase variation. J Theor Biol. 2005;234(2):255–262. doi: 10.1016/j.jtbi.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Wolf DM, Vazirani VV, Arkin AP. Diversity in times of adversity: Probabilistic strategies in microbial survival games. J Theor Biol. 2005;234(2):227–253. doi: 10.1016/j.jtbi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462(7269):90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- 28.Fernández L, Hancock REW. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25(4):661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baquero F, Alvarez-Ortega C, Martinez JL. Ecology and evolution of antibiotic resistance. Environ Microbiol Rep. 2009;1(6):469–476. doi: 10.1111/j.1758-2229.2009.00053.x. [DOI] [PubMed] [Google Scholar]
- 30.Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011;35(5):901–911. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 31.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182(12):3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall MN, Silhavy TJ. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146(1):23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 33.Dupont M, Dé E, Chollet R, Chevalier J, Pagès J-M. Enterobacter aerogenes OmpX, a cation-selective channel mar- and osmo-regulated. FEBS Lett. 2004;569(1–3):27–30. doi: 10.1016/j.febslet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido E, Yamaguchi A, Nishino K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem. 2008;283(35):24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikaido E, Shirosaka I, Yamaguchi A, Nishino K. Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology. 2011;157(Pt 3):648–655. doi: 10.1099/mic.0.045757-0. [DOI] [PubMed] [Google Scholar]
- 36.El Garch F, et al. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J Antimicrob Chemother. 2010;65(10):2076–2082. doi: 10.1093/jac/dkq287. [DOI] [PubMed] [Google Scholar]
- 37.Lea D, Coulson C. The distribution of numbers of mutants in bacterial populations. J Genet. 1949;49(3):264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 38.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi H, et al. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996;144(1):103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 40.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 41.Hancock RE. Role of porins in outer membrane permeability. J Bacteriol. 1987;169(3):929–933. doi: 10.1128/jb.169.3.929-933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancock RE, Bell A. Antibiotic uptake into gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988;7(6):713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- 43.Agafitei O, Kim JE, Maguire T, Sheridan J. The role of Escherichia coli porins OmpC and OmpF in antibiotic cross resistance induced by sub-inhibitory concentrations of kanamycin. JEMI. 2010;14:34–39. [Google Scholar]
- 44.Mahendran KR, Kreir M, Weingart H, Fertig N, Winterhalter M. Permeation of antibiotics through Escherichia coli OmpF and OmpC porins: Screening for influx on a single-molecule level. J Biomol Screen. 2010;15(3):302–307. doi: 10.1177/1087057109357791. [DOI] [PubMed] [Google Scholar]
- 45.Giraud E, Brisabois A, Martel JL, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43(9):2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eaves DJ, Liebana E, Woodward MJ, Piddock LJV. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J Clin Microbiol. 2002;40(11):4121–4125. doi: 10.1128/JCM.40.11.4121-4125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randall LP, Coldham NG, Woodward MJ. Detection of mutations in Salmonella enterica gyrA, gyrB, parC and parE genes by denaturing high performance liquid chromatography (DHPLC) using standard HPLC instrumentation. J Antimicrob Chemother. 2005;56(4):619–623. doi: 10.1093/jac/dki293. [DOI] [PubMed] [Google Scholar]
- 48.Hernández A, Sánchez MB, Martínez JL. Quinolone resistance: Much more than predicted. Front Microbiol. 2011;2:22. doi: 10.3389/fmicb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa SS, et al. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Staphylococcus aureus. BMC Microbiol. 2011;11:241. doi: 10.1186/1471-2180-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baucheron S, Chaslus-Dancla E, Cloeckaert A. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J Antimicrob Chemother. 2004;53(4):657–659. doi: 10.1093/jac/dkh122. [DOI] [PubMed] [Google Scholar]
- 51.Lomovskaya O, Watkins WJ. Efflux pumps: Their role in antibacterial drug discovery. Curr Med Chem. 2001;8(14):1699–1711. doi: 10.2174/0929867013371743. [DOI] [PubMed] [Google Scholar]
- 52.Martins M, et al. Management Committee Members of Cost B16 European Commission/European Science Foundation An instrument-free method for the demonstration of efflux pump activity of bacteria. In Vivo. 2006;20(5):657–664. [PubMed] [Google Scholar]
- 53.Viveiros M, et al. Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int J Antimicrob Agents. 2008;31(5):458–462. doi: 10.1016/j.ijantimicag.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Paixão L, et al. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J Biol Eng. 2009;3:18. doi: 10.1186/1754-1611-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lederberg J, Lederberg EM. Replica plating and indirect selection of bacterial mutants. J Bacteriol. 1952;63(3):399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandegren L, Andersson DI. Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7(8):578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 57.Davidson CJ, Surette MG. Individuality in bacteria. Annu Rev Genet. 2008;42:253–268. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- 58.Ni M, et al. Pre-disposition and epigenetics govern variation in bacterial survival upon stress. PLoS Genet. 2012;8(12):e1003148. doi: 10.1371/journal.pgen.1003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silander OK, et al. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLoS Genet. 2012;8(1):e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryall B, Eydallin G, Ferenci T. Culture history and population heterogeneity as determinants of bacterial adaptation: The adaptomics of a single environmental transition. Microbiol Mol Biol Rev. 2012;76(3):597–625. doi: 10.1128/MMBR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veening J-W, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 62.Donaldson-Matasci M, Lachmann M, Bergstrom C. Phenotypic diversity as an adaptation to environmental uncertainty. Evol Ecol Res. 2008;10(4):493–515. [Google Scholar]
- 63.Adam M, Murali B, Glenn NO, Potter SS. Epigenetic inheritance based evolution of antibiotic resistance in bacteria. BMC Evol Biol. 2008;8:52. doi: 10.1186/1471-2148-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baquero F. Epigenetics, epistasis and epidemics. Evolution, Medicine, and Public Health. 2013;2013(1):86–88. doi: 10.1093/emph/eot009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 66.Hautefort I, Proença MJ, Hinton JCD. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol. 2003;69(12):7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.