Epigenetic marks such as DNA methylation and histone modifications are widely involved in regulating different aspects of developmental and environmental responses (1). Meanwhile, DNA methylation and histone modification are also used constitutively to silence transposable elements and repeat elements (TREs) (2). Such TRE-mediated silencing should necessarily be limited to the intended targets only and not spread to adjacent genes and their regulatory elements. Higher eukaryotic organisms have evolved antisilencing mechanisms to keep the balance between silencing and antisilencing that is required for precise gene expression regulation. Earlier work revealed that repressor of silencing 1 (ROS1) is one such antisilencing gene (3). Recently, a group of unique antisilencing genes has been discovered, including enhanced downy mildew 2 (EDM2) reported both by Lei et al. in PNAS (4) and earlier by Tsuchiya and Eulgem (5), anti-silencing 1 (ASI1) by Wang et al. (6), and increase in bonsai methylation 1 (IBM1) by Saze et al. (7). What distinguishes the latter group of genes from ROS1 is that they seem to be involved in posttranscriptional regulation through alternative polyadenylation (APA).
Polyadenylation is an essential mRNA processing step for almost all genes in eukaryotes (8, 9). The poly(A) site defines the end of the transcript and thus its 3′-UTR. However, an alternate site of processing and poly(A) addition would create a different transcript by an inclusion or exclusion of certain RNA sequences such as the miRNA target site, a recognition site of an mRNA stability factor, or a translational repressor. In recent years, APA has been shown to change the fate of a cell or a developmental process and to alter cellular responses to the environment, including tumorigenesis, flowering time control, and oxidative responses (10–12). Genomewide analyses by deep sequencing have revealed that 70–80% of human and plant genes are subject to APA (13–15). The role of such abundant APAs in cellular functions, however, remains largely unknown.
It was previously suggested that histone modification might influence poly(A) choice through correlation analysis (16). The significance of the work on EDM2 and ASI1 is that they provide direct genetic evidence demonstrating an involvement of epigenetic marks in the selection of APA sites. EDM2, identified as an enhancer of fungal disease resistance (17), specifically recognizes intronic heterochromatin in its target genes presumably through its composite plant homeodomain (PHD) domain (4). This binding of EDM2 somehow blocks the poly(A) site choice of the transcript in the vicinity of the binding location. This is deduced from the fact that in the edm2 mutant, use of intronic poly(A) sites [so called proximal poly(A) sites, in contrast to the full-length distal poly(A) sites] results in the generation of shortened transcripts that encode no or nonfunctional proteins (4, 5). In other words, the normal function of EDM2 is to reduce the production of nonfunctional transcripts and to promote the generation of full-length mRNA ending at the distal poly(A) site. The net result is the avoidance of reduced gene expression caused by silencing due to TREs in the intron (hence antisilencing).
The antisilencing phenomena of EDM2 were clearly demonstrated in the APA of a group of genes with large TRE-containing genes such as the fungal disease resistance gene resistance to Peronospora parasitica (RPP7) (5) and IBM1 (4). Interestingly, another gene (ASI1) was also found to have very similar function of generating APA of IBM1 transcripts (6). Adding an additional layer of complexity to this story is the fact that IBM1 itself is a histone H3K9 demethylase that prevents plant-specific CHG methylation (7). Moreover, genomewide epigenetic modification profiles of all three mutants (asi1, edm2, and ibm1) largely overlap, which indicates that they work together through a similar antisilencing pathway. In fact, it was suggested that both ASI1 and EDM2 function mostly through IBM1 by modulating APA of IBM1 transcripts (4, 5).
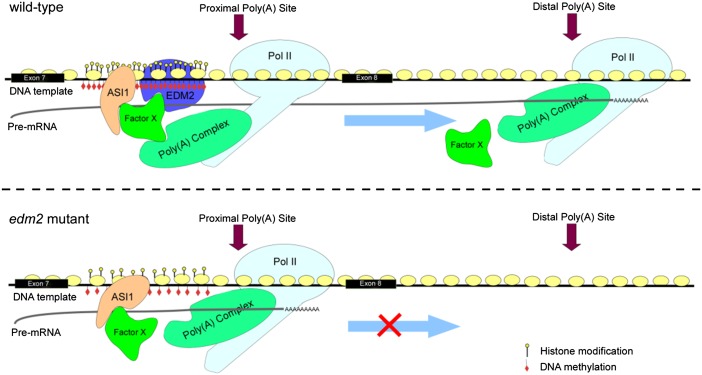
The question is how are these epigenomic marks sensed by the polyadenylation machinery so that a different poly(A) site is chosen? Polyadenylation is a cotranscriptional event where numerous factors affecting transcription would impact polyadenylation (18). The polyadenylation machinery is composed of 20–25 different proteins in plants that recognize a set of poly(A) signals on the pre-mRNA (9). The use of one poly(A) site over the other is likely owing to the relative strength of the polyadenylation signal being perceived by poly(A) machinery, as well as the availability of the site. It is also possible that associated protein(s) may recruit (or repel) the polyadenylation apparatus close to a particular site. It is equally possible that a protein could inhibit the function of the complex. One possibility in the EDM2 or ASI1 cases is that, while binding to epigenetic marks, these proteins also interact with the poly(A) signals, thus blocking the recognition of the proximal poly(A) site. Indeed, ASI1 was found to have an RNA recognition motif and a bromo-adjacent homology domain involved in preventing the genomewide CHG methylation (6). EDM2, on the other hand, possesses an N6-adenine methyltransferase domain (4), providing a potential to methylate adenine residues in RNA. Methylated adenine was shown to affect 3′-end formation (19). Binding of EDM2 to chromatin through its PHD domain could bring the methyltransferase domain to close proximity to the nascent RNA. However, there is no evidence to show a direct interaction between ASI1 and EMD2. Thus, a conceivable model would be that both ASI1 and EDM2 bind to target DNA and to associated nascent RNA, forming a complex through an unidentified factor X. The unknown factor may also mediate the interaction with the polyadenylation complex that is associated to the C-terminal domain of RNA polymerase II (Fig. 1).
Fig. 1.
Hypothetic model for antisilencing regulation of alternative polyadenylation. In WT Arabidopsis, EDM2 and ASI1 bind to the intronic heterochromatin region by recognizing enriched epigenetic marks. With the assistance of an unknown factor X, the three proteins form a complex that potently masks the poly(A) signals from being recognized by the polyadenylation apparatus riding on the C-terminal domain of RNA polymerase II. Consequently, the distal poly(A) site is used. In the edm2 or asi1 mutant, absence of either EDM2 or ASI1 reduces the masking effect on the proximal poly(A) site. The result is an extensive use of the proximal site, producing shortened transcripts encoding nonfunctional proteins, leading to a silencing effect of the target gene.
The analysis of both edm2 and asi1 mutants also implied that the transcription rates of IBM1 and RPP7 do not change in the mutants, because there were no differences in the distribution of pol II along the genes (4–6). Clearly this is a posttranscriptional regulation event. Particularly in RPP7, the degree of disease resistance conferred is tightly associated with its transcript level (5). Could the APA just be a byproduct of other modes of regulation? This possibility is very unlikely. There exist other examples showing that APA is a predominant way to attenuate the expression of
The significance of the work on EDM2 and ASI1 is that they provide direct genetic evidence demonstrating an involvement of epigenetic marks in the selection of APA sites.
genes. One involves the flower control locus A (FCA) gene, a regulator of flower locus C (FLC) expression, and of flowering time in Arabidopsis (12). Another is the oxidative tolerant 6 (OXT6) gene in Arabidopsis that encodes two proteins: a smaller one produced through use of an intronic poly(A) site and a larger one produced using a distal poly(A) site (11). Both FCA and OXT6 genes have large introns where the proximal APA occurs. However, it seems that these APA events use a different mechanism because these introns do not contain TREs (4). Recent work by Duc et al. indicated that a spen family protein FPA might be involved in the selection of some intronic polyadenylation (20). Nonetheless, recent genome-level profiling discovered a significant amount of intronic poly(A) sites in Arabidopsis and rice (14, 15). Further analysis of these intronic sites would broaden our view on the interrelationship of RNA processing and epigenetic regulation.
The connection between gene silencing and polyadenylation was demonstrated by Herr et al. (21) where mutations of poly(A) protein factors led to enhanced silencing of some genes. This work is indicative that normal functions of these polyadenylation factors prevent silencing of the tested genes, an equivalent to an antisilencing effect. While there was not clear demonstration of the involvement of TREs, this is further evidence that motivates investigations to elucidate a potential direct engagement of poly(A) factors in antisilencing.
Meanwhile, many interesting questions remain. What are the specific functions of ASI1 and EDM2 during this process? If any, what are other factors involved? Is binding of ASI1 or EDM2 to intronic heterochromatin region needed to influence poly(A) site choice? Is RNA modification (e.g., methylation) or RNA secondary structure involved? What is the role of the splicing of the large intron in all this? Addressing these questions would ensure a fine-grained understanding of posttranscriptional regulation of genes in this ever-increasing complex world of the epigenome.
Acknowledgments
Research in the authors' laboratory is supported by US National Science Foundation Grant IOS–0817829 (to Q.Q.L.) and US National Institutes of Health Grant 1R15GM94732-1 A1 (to Q.Q.L.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 527.
References
- 1.Chinnusamy V, Zhu J-K. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dooner HK, Weil CF. Give-and-take: Interactions between DNA transposons and their host plant genomes. Curr Opin Genet Dev. 2007;17(6):486–492. doi: 10.1016/j.gde.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Agius F, Kapoor A, Zhu J-K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103(31):11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lei M, et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci USA. 2014;111:527–532. doi: 10.1073/pnas.1320106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchiya T, Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci USA. 2013;110(37):E3535–E3543. doi: 10.1073/pnas.1312545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, et al. RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc Natl Acad Sci USA. 2013;110(38):15467–15472. doi: 10.1073/pnas.1315399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319(5862):462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 8.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 9.Xing D, Li QQ. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA. 2011;2(3):445–458. doi: 10.1002/wrna.59. [DOI] [PubMed] [Google Scholar]
- 10.Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE. 2008;3(6):e2410. doi: 10.1371/journal.pone.0002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113(6):777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y. Alternative polyadenylation: New insights from global analyses. RNA. 2012;18(12):2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108(30):12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Y, et al. Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res. 2011;21(9):1478–1486. doi: 10.1101/gr.114744.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaladkar M, Smyda M, Hannenhalli S. Epigenomic and RNA structural correlates of polyadenylation. RNA Biol. 2011;8(3):529–537. doi: 10.4161/rna.8.3.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eulgem T, et al. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49(5):829–839. doi: 10.1111/j.1365-313X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 18.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108(4):501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 19.Bodi Z, et al. Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front Plant Sci. 2012;3:48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG. Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet. 2013;9(10):e1003867. doi: 10.1371/journal.pgen.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herr AJ, Molnar A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA. 2006;103(41):14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]