Significance
Circadian clocks orchestrate daily rhythms in behavior and physiology using temporal regulation of gene expression to control core clock genes and rhythmic output programs. Although transcription regulation was shown to drive extensive diurnal mRNA rhythms, less is known about the proteins. Here, we provide a proteome-wide study of rhythmic protein accumulation in mouse liver, showing that proteins preferentially accumulate in the morning and during the night. About one-half of these rhythmic proteins could not be explained by rhythmic mRNAs, suggesting that translational or posttranslational regulation plays an important role. Moreover, such rhythms involved many secreted proteins and were clock-independent. Hence, these findings indicate that feeding behavior might determine the rhythmic functions of circulating proteins in the blood.
Keywords: circadian rhythm, proteomics, liver metabolism, posttranslational regulation, protein secretion
Abstract
Diurnal oscillations of gene expression controlled by the circadian clock underlie rhythmic physiology across most living organisms. Although such rhythms have been extensively studied at the level of transcription and mRNA accumulation, little is known about the accumulation patterns of proteins. Here, we quantified temporal profiles in the murine hepatic proteome under physiological light–dark conditions using stable isotope labeling by amino acids quantitative MS. Our analysis identified over 5,000 proteins, of which several hundred showed robust diurnal oscillations with peak phases enriched in the morning and during the night and related to core hepatic physiological functions. Combined mathematical modeling of temporal protein and mRNA profiles indicated that proteins accumulate with reduced amplitudes and significant delays, consistent with protein half-life data. Moreover, a group comprising about one-half of the rhythmic proteins showed no corresponding rhythmic mRNAs, indicating significant translational or posttranslational diurnal control. Such rhythms were highly enriched in secreted proteins accumulating tightly during the night. Also, these rhythms persisted in clock-deficient animals subjected to rhythmic feeding, suggesting that food-related entrainment signals influence rhythms in circulating plasma factors.
Light and heat, the principle energy sources for life, are only periodically available with a period of 1 d. Consequently, organisms acquired a timing system to adapt their physiology and anticipate these diurnal variations. In mammals, this circadian clock influences most aspects of physiology and behavior (1). In humans, perturbations of this clock lead to pathologies, including metabolic and vascular diseases. Although the oscillatory clockwork is cell-autonomous, timing on the scale of organisms uses a hierarchal organization: a master clock within the suprachiasmatic nuclei (SCN) of the hypothalamus receives light input through the retina and communicates timing signals to slave oscillators in other peripheral tissues (2).
In mammals, the molecular oscillator uses interconnected transcriptional and translational feedback loops, in which multiple layers of control, including temporal posttranscriptional and posttranslational regulation, play important roles (3). An active area of chronobiology aims at understanding how the temporal signals from the core oscillator are relayed to clock output function. In this context, genome-wide rhythms in mRNA accumulation were characterized in several models. In general, around 10% of the genes, encoding many enzymes involved in different aspects of cellular metabolism, show rhythmic mRNA accumulation, establishing the role of the circadian clock in temporally gating rhythmic physiology (4).
However, comparatively little is known on the temporal accumulation of proteins, despite increasing evidence suggesting that posttranscriptional mechanisms also contribute to circadian rhythms at the protein level (5), including mRNA translation (6–8). Of interest, two medium-scale studies relying on 2D gel electrophoresis to quantify the liver and SCN proteomes in mouse concluded that around 10–20% of expressed proteins accumulate rhythmically during the day (9, 10). Surprisingly, in both cases, many of the rhythmic proteins were encoded by nonrhythmic mRNAs, highlighting that translational or posttranslational circadian regulation underlies the accumulation of the circadian proteome.
To further study the diurnally rhythmic proteome in mouse liver, we used stable isotope labeling by amino acids (SILAC) for quantitative MS in mouse (11) to monitor temporal protein accumulation profiles in WT samples covering 2 full d and clock-deficient animals. Among >5,000 proteins detected, several hundred exhibited a rhythmic pattern of accumulation in WT mice. Although about one-half of these rhythmic proteins was encoded by rhythmic mRNA and showed about a 6-h delay in peak accumulation time, the other one-half was encoded by nonrhythmic mRNA. This latter set was highly enriched in proteins secreted by the liver, covering functions such as apolipoproteins, coagulation factors, complement factors, and serine protease inhibitors. Thus, this finding suggests that protein secretion may be a rhythmic process in the liver, like in the SCN (10). Finally, these rhythms seemed to be clock-independent, because they persisted in clock-deficient Cry1/Cry2 KO mice.
Results
SILAC MS Identifies the Diurnally Rhythmic Proteome in Mouse Liver.
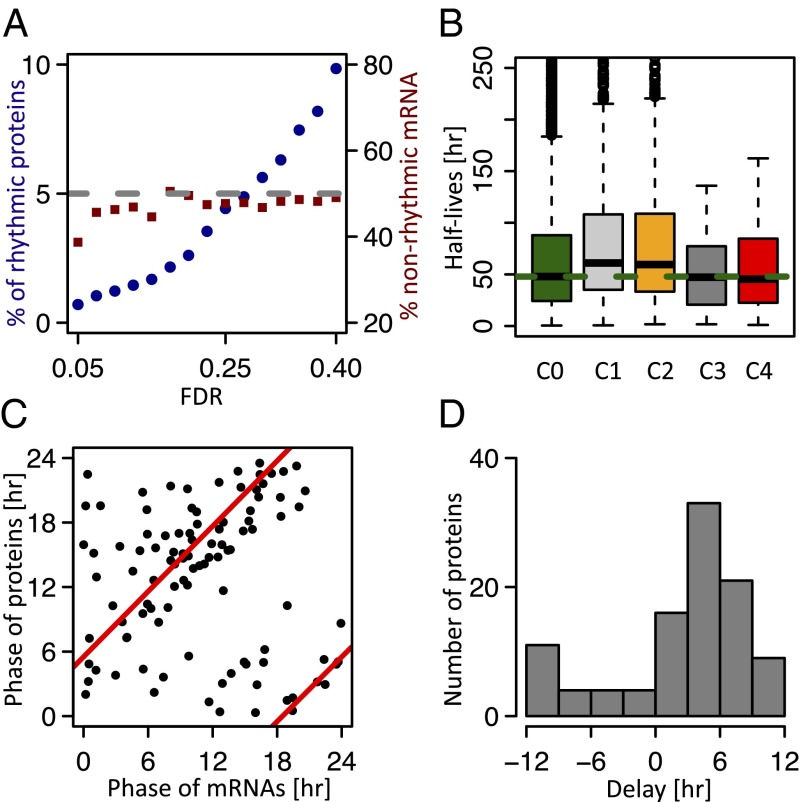
To measure the daily accumulation of proteins, we designed an SILAC MS experiment, in which total protein extracts were harvested from mice every 3 h for 2 d (eight samples per day). Relative protein abundance in each of 16 samples was quantified against a common reference sample labeled using the SILAC method (Fig. S1). Efficiency of the labeling was verified in blood and tail tissues (Fig. S2). The generated mass spectra allowed the identification of a total of 5,827 distinct proteins, of which 70% yielded relative measurements in at least 8 of 16 samples (Fig. S3A). Full data are in Dataset S1. As expected, the detected proteins in the total protein extracts were enriched in abundant cytosolic liver proteins and did not include the lowly expressed core clock transcription factors. Although the number of rhythmic proteins obviously depends on the stringency, we identified 54 [P < 0.0015, harmonic regression, false discovery rate (FDR) = 0.1, Benjamini–Hochberg], 115 (P < 0.0077, FDR = 0.2), and 195 (P < 0.015, FDR = 0.25) rhythmic proteins at increasing FDR thresholds (Fig. S3B). For simplicity, we present the set of 195 proteins and show that our main conclusions do not depend on FDRs. Peak phases of protein rhythms were distributed throughout the day (Fig. 1A) and enriched around the two time points Zeitgeber time 5 (ZT5) and ZT18 (Fig. 1B) independent of FDR (Fig. S3). The peak-to-trough amplitudes of the protein rhythms were, in general, lower than typical amplitudes of rhythmic mRNAs (Fig. 1C and Fig. S4A), with 2.6% of the rhythmic proteins showing amplitudes greater than two. Low amplitudes in protein rhythms are expected considering that the detected proteins tend to have long half-lives, thus dampening circadian amplitudes (see below). Independent validations by Western blots indicated that the phases and amplitudes measured by MS are accurate (Fig. 1 D–F). Although the current proteomics technology prevents from applying very stringent FDRs, lower-stringency examples still show convincing and reproducible rhythms (Fig. S3 E–G). Thus, SILAC MS was able to reliably identify rhythmically accumulating proteins in mouse liver.
Fig. 1.
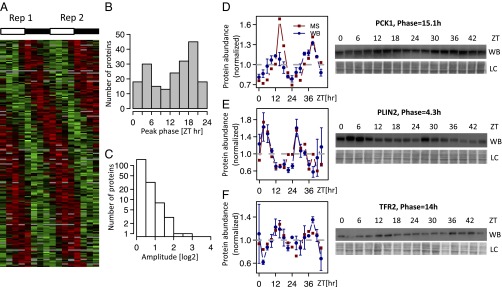
SILAC-based MS identifies the diurnal proteome in mouse liver. (A) Heatmap showing the rhythmic proteome (n = 195, FDR < 0.25); 2 independent d (each comprising eight time points) are juxtaposed; data are standardized by rows, and gray blocks indicate missing data. (B and C) Phase and peak-to-trough amplitude (log2 units) distribution for the rhythmic proteins (FDR < 0.25). (D–F) Examples of rhythmic proteins encoded by the individual genes (D) Phosphoenolpyruvate CarboxyKinase 1 (PCK1; P = 1.3e-05, q = 0.0060), (E) PeriLipIN2 (PLIN2; P = 0.00083, q = 0.068), and (F) TransFerrin Receptor protein 2 (TFR2; P = 6.39e-06, q = 0.0036) confirmed by Western blot (WB) analysis (data are normalized to the temporal mean). The two biological replicates are shown as ZT0–ZT21 (replica 1) and ZT24–ZT45 (replica 2). For the WBs, 16 time points show the mean and SE from two independent biological samples. Naphtol blue-black staining of the membranes was used as a loading control and serves as a reference for normalization of the quantified values. The ZTs at which the animals were killed are indicated. LC, loading control.
Relationship Between mRNA and Protein Accumulation Profiles.
To assess which protein rhythms could be explained from rhythmic mRNA accumulation, we compared the protein data with our previous mRNA expression data under the same light–dark (LD) and feeding conditions (7). After matching mRNA probe sets to proteins identifiers, we selected for each protein the most rhythmic probe set (lowest q value) based on our previous experience (7) that multiple probe sets for the same gene often include nonexpressed isoforms or intronic probes, such that the most rhythmic probe is usually biologically relevant. To define the set of rhythmic mRNAs, we used the standard FDR = 0.05 (12). We found a high proportion of rhythmic mRNA (46.7%) among transcripts encoding the detected proteins. This fraction seemed high but was expected, because MS preferentially detects proteins with high abundances; thus, the corresponding mRNAs are biased to high levels (Fig. S4B). In addition, the fraction of all rhythmic mRNA probe sets changes markedly with mean expression, reaching above 40% in the optimal range (Fig. S4C), and considering the most rhythmic probe set for each protein further mildly increased the expected proportion to match the observed 46.7% (Fig. S4).
Although the percentage of rhythmic proteins was in the range of a few percent and increased (1–10%) with FDR, it is noteworthy that the proportion of rhythmic proteins with nonrhythmic mRNAs was about 50% independent of FDR (Fig. 2A). This number indicated that a large proportion of the rhythmic proteome is regulated at the translational or posttranslational level, which was reported earlier (10). To simplify the discussion, we next considered four classes of genes (protein FDR = 0.25): genes with nonrhythmic mRNA and nonrhythmic protein [class 1 (C1); n = 2,277], rhythmic mRNA and nonrhythmic protein (C2; n = 1,936), rhythmic mRNA and rhythmic protein (C3; n = 102), and nonrhythmic mRNA but rhythmic protein (C4; n = 93) (Fig. S5 A–C). These classes indicated that the proportion of rhythmic proteins among all genes with rhythmic mRNAs was small (4.4%), probably because of long protein half-lives (see below). Despite this limited number of rhythmic proteins, we performed gene ontology analysis on C3 genes separately for day- (ZT0–ZT12) and night-expressed proteins (ZT12–ZT24). Diurnal proteins are enriched in genes encoding for lipid metabolism (PLIN2 and FASN) (13), Acetyl-CoA synthesis (ACSS2 and ACLY), and endoplasmic reticulum (ER) formation (ATL2 and ATL3) (14), whereas night proteins pointed to glucose metabolism (PCK1 and GYG) (15), xenobiotic detoxification (ALAS1, POR, CYP2E1, and CES1D) (16), and protein folding (HSPA8, HSP90AB1, and DNAJA1) (17) (Dataset S2). Meanwhile, C4 genes showed a dominant phase of expression in the middle of the night (around ZT20) and were highly enriched in secreted liver proteins.
Fig. 2.
Relationship between mRNA and protein accumulation profiles show that reduced protein amplitudes originate from long protein half-lives. (A) Percentage of rhythmic proteins (left scale) and fraction of rhythmic proteins (right scale) with nonrhythmic mRNA in function of FDR. (B) Protein half-lives (measured in NIH 3T3 cells) (19) are significantly longer in C1 and C2 (nonrhythmic proteins) compared with C3 and C4 (rhythmic proteins; P < 0.004, Wilcoxon rank sum test). C0 contains all protein half-lives measured in ref. 19. (C) Phase relationships between peak protein and mRNA accumulations (C3). Red lines show circular regression predicting a delay of 5.5 h. (D) Distribution of phase delay between peak protein and mRNA accumulations (C3).
Reduced Protein Amplitudes Originate from Long Protein Half-Lives.
To investigate the reasons for low protein amplitudes (Fig. 1C), we studied the relationship between protein rhythms, protein half-life measurements, and observed delays between peak times in protein and mRNA accumulation. To relate these quantities, we adopted a simple model for protein accumulation, similar to the one that we developed for mRNA accumulation (18) (SI Materials and Methods). On average, the proteins half-lives, taken for mouse fibroblasts (19), are significantly longer (P < 0.004, Wilcoxon test) for the nonrhythmic proteins (C1 and C2; median half-life of 60 h) compared with rhythmic ones (C3 and C4; median half-life of 53 h.) (Fig. 2B). These half-lives are, on average, long compared with the 24-h cycle (Fig. S6A), and our model then predicts that the delay in protein accumulation compared with mRNA should be close to 6 h (assuming constant half-lives) (SI Materials and Methods). To verify this hypothesis, we compared the peak accumulation times for all genes in C3 and found that protein showed an average delay of 5.5 h (Fig. 2 C and D). Individual time traces showing delayed protein accumulation are shown in Fig. S5D. To go one step farther, we tested the predicted relationship between the ratio of relative amplitudes and measured delays (Fig. S5E). Although we find the expected trend of decreased relative amplitude in protein with increased delay, the scatter in the data points is similar to what was obtained for mRNAs (18). This scatter may indicate inaccuracy in the amplitude and phase measurements or reflect temporal posttranslational regulation of protein stability, a mechanism known to play a central role in the control of circadian period length (20–22). Thus, protein half-lives that change over the diurnal cycle could explain protein rhythms showing higher amplitudes compared with their corresponding mRNA (representative examples in Fig. S6B). Finally, because larger amplitudes in mRNA are expected to yield more readily detectable protein rhythms, we compared mRNA amplitudes in C2 and C3 genes, showing that, indeed, C3 transcripts have higher amplitudes compared with C2 (Fig. S5F). Together, the comparison between rhythmic mRNA and protein accumulation profiles showed that our measurements agree, in bulk, with a simple model of protein translation that explains the observed phase and amplitude relationships between mRNA and protein rhythms.
Secreted Proteins Show Synchronous Rhythms in Liver and Plasma.
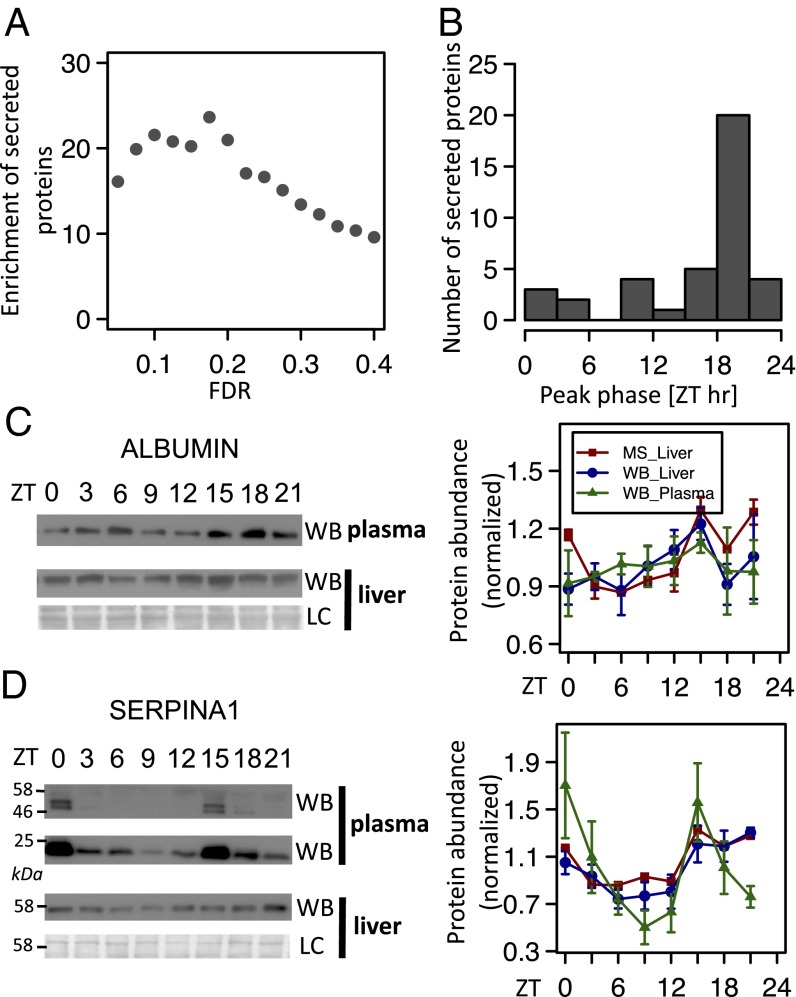
As indicated before, C4 proteins are highly enriched in secreted proteins. In fact, 39 of 53 (74%; P < 10−42, hypergeometric test) (Dataset S3) detected proteins annotated as secreted by the liver present rhythmic accumulation. This high enrichment (up to 25-fold) is independent of FDR (Fig. 3A). Such proteins include serum albumin (ALBUMIN), coagulation factors, complement factors, apolipoproteins, serine protease inhibitors (SERPINs), and other secreted proteins (Dataset S3). As mentioned, these proteins all show a synchronized phase of expression (Fig. 3B) with a peak in the middle of the night and trough in the middle of the day, suggesting that protein secretion may be maximal in antiphase (that is, in the middle of the day). Several of these proteins were described to exhibit a rhythmic pattern of expression in rodent or human serum, which is the case for serum albumin (23, 24), apolipoproteins (25, 26) coagulation factors (27, 28) influencing coagulation time (29, 30), complement factors (31), and SERPIN proteins (27, 31). Consequently, the total concentration of protein in the plasma itself follows a diurnal rhythm (23). We, thus, measured protein expression for two examples (ALBUMIN and SERPINA1) and confirmed the synchronized presence of these proteins in both liver and plasma (Fig. 3 C and D). Interestingly, at least for ALBUMIN, protein synthesis is constant in rodent liver (32), suggesting a potential decrease in liver levels because of the depletion of the cytoplasmic pool during the day when secretion is highest. Secretion of proteins outside the cell requires, among others, the transport of vesicles from the ER to the plasma membrane through the Golgi apparatus. This transport involves the coat protein complex II (COPII) complex and proteins of the ER cargo receptor complex lectin, mannose-binding, 1 (LMAN1) and multiple coagulation factor deficiency 2 (MCFD2) (33, 34). In fact, suppressor of auxin resistance 1 A and B (SAR1A and SAR1B), the required GTPase of the COPII complex, exhibited low-amplitude rhythms in the MS data (Fig. S7), showing maximal accumulation early in the morning (Fig. S7A). These changes are followed by rhythms, albeit of lower significance (Dataset S1), in SEC23B, LMAN1, and MCFD2 (Fig. S7). Although this finding will require additional validations, these rhythms suggest a scenario of maximal protein secretion in the morning.
Fig. 3.
Secreted proteins show synchronous rhythms in liver and plasma. (A) Enrichment (observed over expected numbers) of secreted protein among rhythmic proteins in function of FDR. All points are highly significant (all P < 10−10, hypergeometric test). (B) Phase distribution for the rhythmic secreted proteins (FDR < 0.25) detected in liver shows a tight phase of expression that peaked in the middle of the night (ZT20). (C and D) Temporal levels of ALBUMIN and SERPINA1 in plasma and mouse liver. The graphs present the MS analysis (red lines; P = 0.00073, q = 0.064 for ALBUMIN; P = 0.000036, q = 0.0126 for SERPINA1) as well as the densitometry analyses of the Western blots (WBs) performed with WT mice samples from liver (blue lines) and plasma (green lines). For the WBs, mean and SE from four independent biological samples were represented. In graphs for the SERPINA1 WB in plasma, the 22 kDa band of the blots was quantified. LC, loading control.
WT and Cry1/Cry2 KO Mice Proteomes Show That Diurnal Protein Rhythms Persist in Clock-Deficient Animals for C4 Genes.
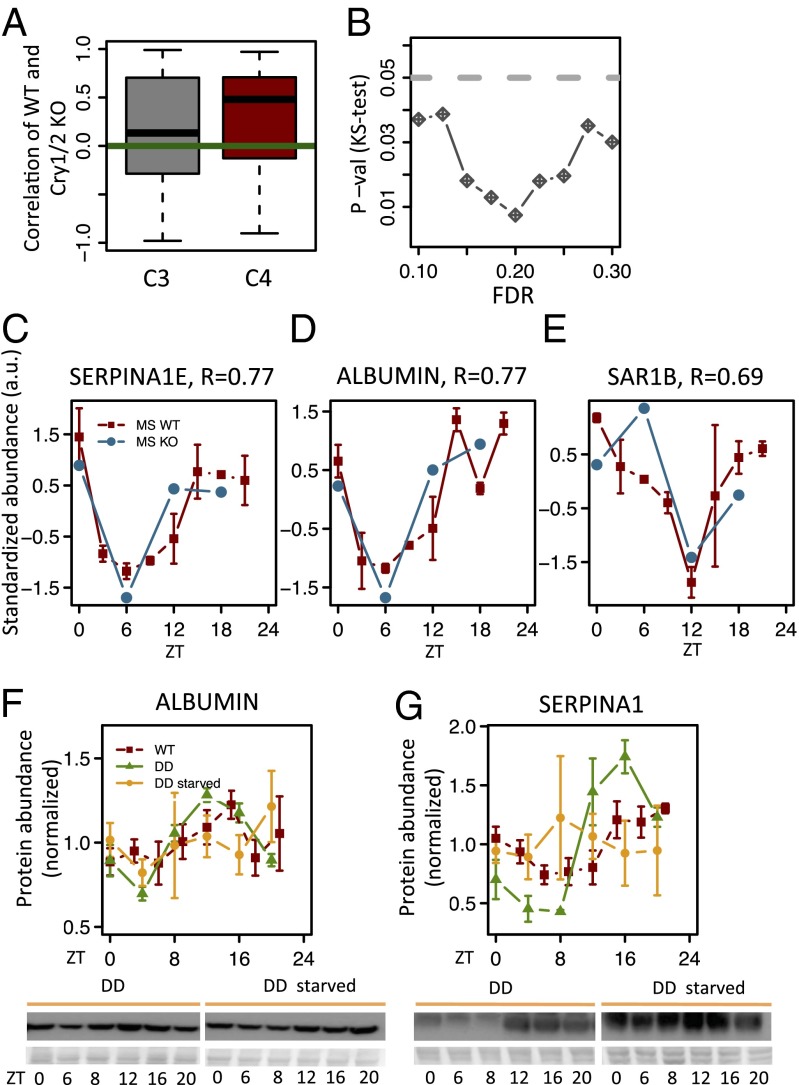
To investigate whether the measured protein rhythms depend on a functional molecular circadian oscillator, we performed MS analysis in Cry1/Cry2 KO arrhythmic animals that were kept under an LD cycle with nighttime feeding to avoid biases from possibly altered feeding behavior in the KO animals. Samples were collected every 6 h for 1 d. Overall, the mean protein abundance for C3 genes in Cry1/Cry2 KO did not differ significantly from WT (Fig. S8A). Interestingly, correlation analysis of the temporal profiles between WT and Cry1/Cry2 KO animals indicated that C4 genes show higher correlations than C3 genes (P < 0.02, Kolmogorov–Smirnov test) persisting over a wide range of FDRs (Fig. 4 A and B). This enrichment indicates that C4 protein rhythms tend to be maintained in arrhythmic but food-entrained animals. This effect concerns many secreted proteins, such as SERPINA1E (Fig. 4C) or ALBUMIN (Fig. 4D), and proteins involved in the regulation of secretion, such as SAR1B, which may possibly explain that secreted proteins also maintain their rhythms in KO animals (Fig. 4E). To test whether feeding rhythms underlie this regulation, we fasted mice placed in constant darkness (DD). Although normally fed mice in DD maintained rhythmic expression of ALBUMIN and SERPINA1 proteins, fasting clearly abolished these rhythms (Fig. 4 F and G). Moreover, although Cry1/Cry2 KO exhibited rhythmic ALBUMIN expression in LD, placing those mice in DD (abolishing rhythmic feeding) showed flat ALBUMIN expression (Fig. S8B). Together, these data indicate that rhythmic feeding, which depends on a functional central circadian clock but not the liver clock, drives rhythmic protein accumulation for C4 proteins.
Fig. 4.
Analysis of WT and Cry1/Cry2 KO proteomes shows that diurnal rhythms in protein abundance persist in clock-deficient animals for C4. (A) Correlations of WT and Cry1/Cry2 KO profiles are significantly higher for C4 compared with C3 genes [Kolgomorov–Smirnov test, P < 0.02; that is, rhythmic proteins with nonrhythmic mRNAs in WT (C4) tend to keep their rhythm in clock-deficient animals under the same conditions (light and feeding), whereas rhythmic proteins with rhythmic mRNAs (C3) do not]. (B) Kolgomorov–Smirnov (KS) tests comparing the temporal correlations between WT and Cry1/Cry2 KO animals for C3 and C4 genes show significant difference over a range of FDRs. (C–E) MS data from WT and Cry1/Cry2 KO mice for individual genes from C4. The two WT replicas are averaged (solid red); tips of error bars indicate the two separate measurements. MS statistics are P = 0.08 and q = 0.20 for SERPINA1E, P = 0.00073 and q = 0.064 for ALBUMIN, and P = 0.0059 and q = 0.19 for SAR1B. (F and G) Temporal levels of ALBUMIN and SERPINA1 in mouse liver. Densitometry analyses of the Western blots performed with liver extracts from WT mice in LD conditions (red; results from Fig. 3 C and D), DD (green), and starvation during constant darkness (yellow; DD starved). Statistics for rhythmicity in DD are P = 2.75 × 10−5 and P = 6.4 × 10−7 for ALBUMIN and SERPINA1, respectively. Statistics for rhythmicity during starvation in DD are P = 0.35 and P = 0.64 for ALBUMIN and SERPINA1, respectively. For the Western blots, mean and SE from three independent biological samples are given.
Discussion
Proteins Show Low-Amplitude Diurnal Rhythms.
Through the use of large-scale quantitative SILAC proteomic analyses, we showed that few percents of liver proteins are rhythmically accumulating in the mouse liver. Although this estimate depends on the applied thresholds (Fig. 2A), the fraction of rhythmic proteins seems to be lower than the fraction of rhythmic mRNAs. Both technical and biological reasons may explain this difference. Although we could identify over 5,000 proteins, sensitivity and accuracy of current MS are still lower compared with the standards for mRNA profiling. In particular, the detection of the proteins is still biased to high-abundance cytosolic proteins, reflected by the absence of core clock transcription factors in our data. Moreover, highly accumulating proteins are expected to be generally more stable, and increased stability dampens oscillatory amplitudes, which was shown by our model. In addition, proteins half-lives are generally much longer than mRNA half-lives (19). Consequently, unless there is significant diurnal posttranslational regulation, protein rhythms are expected to exhibit shallow amplitudes, which is the case for several core circadian regulators. For example, Bmal1 mRNA shows a peak-to-trough amplitude of more than 100-fold in the liver, whereas the change is barely 3-fold for the BMAL1 protein (35). Altogether, low-amplitude and less-accurate measurements imply that it is statistically challenging to identify rhythms in protein abundance, which is the reason that we used lower stringency for proteins than mRNA. These considerations might also explain the low overlap between our analysis and previous results: only 3 proteins identified with 2D gels (P = 0.07, hypergeometric test), a technique certainly not more accurate than MS, are part of our list of 195 rhythmic proteins (9). However, these three peptides (ALBUMIN, APOA4, and SERPINA1D) all correspond to secreted proteins, confirming that rhythmic protein secretion may be a key process in establishing the rhythmic liver proteome. Although it is not excluded that inclusion of lower-abundance proteins would change this picture, the current data suggest that the rhythmic proteome is significantly dampened compared with its mRNA counterpart, with 97% of the protein rhythms showing less than twofold amplitude (Fig. 1C). Accordingly, it becomes intriguing how circadian function can depend on such small amplitudes. However, protein abundance does not necessarily correlate with activity, because for example, newly synthesized proteins are potentially more active than old oxidized proteins (36). Activity may, therefore, exhibit higher amplitude because of a shorter-lived newly synthesized protein pool.
Role of Posttranslational Diurnal Regulation.
Our data confirm previous conclusions on the role of posttranslational control of rhythmic protein accumulation (9, 10). We found that one-half of the rhythmic proteins does not possess a corresponding rhythmic transcript, suggesting that either translation or protein stability might be subject to diurnal regulation. In fact, we previously described that a minority of translation-related genes (7), notably ribosome components, is diurnally regulated at the translational level. Moreover, diurnal regulation of protein stability may be a widespread regulatory mechanism, which was described recently for mRNA processing and stability (18, 37, 38).
Rhythmic Protein Secretion.
Of all identified rhythmic proteins, we found that one-half is encoded by rhythmic mRNAs, with an average delay of 5.5 h (Fig. 2 A, C, and D). The other one-half shows nonrhythmic mRNA and is highly enriched in secreted proteins. Even if a definitive proof of the implication of rhythmically accumulating COPII and ER cargo receptor complexes proteins is missing, our data suggest that this accumulation might provide a possible explanation for maximal rhythmic secretion in the morning. However, this scenario would imply that proteins like ALBUMIN accumulate earlier in the plasma than the liver, which is not observed (Fig. 3C). Diffusion through the liver tissue or posttranslational modifications required for the maturation of secreted proteins, which were suggested for SERPINA1, showing secreted forms in the plasma that are distinct from the forms in the liver (Fig. 3D) may further contribute to the timing of plasma accumulation. Although SAR1 isoforms, the required GTPases of the COPII complex, have been linked to apolipoproteins secretion (39, 40), the ER cargo receptor proteins MCFD2 and LMAN1 seem to mediate the transport of coagulation factors and SERPIN proteins (41–43). Inhibition of both systems induces a protein secretion defect that leads to protein accumulation in the ER and the activation of the unfolded protein response (UPR) (44, 45). Interestingly, we have shown that UPR activation exhibits an ultradian rhythm (with a period of 12 h), with a minimum level of activation that coincides with the minimum level of secreted proteins in mouse liver (46). This periodic UPR activation depends on a functional circadian clock (46, 47), but rescuing a functional clock in the SCN turns these 12-h rhythms into 24-h rhythms, suggesting that signals from the central clock are required for one peak of this ultradian oscillation (47). Because rhythms in secreted proteins persist in animals without functional circadian clocks kept under a nighttime feeding regimen (Fig. 4 and Fig. S8), the second clock-independent peak of oscillation in UPR activation may be caused by the accumulation of secreted proteins in the beginning of the night, when protein secretion potentially reaches its minimum level. Although the nature of the signal controlling rhythmic protein secretion remains to be identified, rhythmic metabolism of food and notably, lipids may underlie this regulation. Indeed, protein secretion is highly regulated by sterol and cholesterol metabolism, at least partly through the activity of acyl-CoA:cholesterol acyltransferase (48, 49). In fact, acyl-CoA:cholesterol acyltransferase inhibitors are used as blockers of vesicular traffic between the ER and Golgi (50), and this enzyme exhibits a diurnal activity (51, 52) that influences sterol synthesis and potentially, protein secretion. Although this hypothesis needs confirmation, it could provide a further link between cell metabolism and the establishment of rhythms in mouse liver physiology.
In conclusion, our high-coverage and temporal MS analyses of the liver proteome showed that many secreted proteins accumulate with a diurnal rhythm. Rhythms in such proteins tended to be maintained in clock-deficient animals kept under a nighttime feeding regimen but vanished under starvation, suggesting that feeding-related signals may determine the rhythmic functions of circulating plasma factors secreted by the liver.
Materials and Methods
Animals.
All animal studies were conducted in accordance with the regulations of the veterinary office of the Canton of Vaud. Cry1/Cry2 double KO mice (53) in the C57BL/6J genetic background are described in ref. 54. Unless noted otherwise, mice were maintained under standard animal housing conditions with free access to food and water and in 12-h LD cycles. However, for all experiments, animals were fed only at night starting 4 d before the experiment to control for genotype-dependent feeding rhythms. SILAC mice were prepared as described in ref. 11 (Fig. S1).
Protein Extraction and Analysis.
Livers were homogenized in a urea lysis buffer containing protease and phosphatase inhibitors. Western blotting on liver samples was performed according to standard procedures. For plasma samples, because total protein concentration changes during the day (23), preventing normalization with an internal loading control, we normalized the loading to the volume of plasma loaded on each well. Antibodies used are given in Table S1.
SILAC MS and Data Analysis.
Equal amounts of proteins extracted from two non-SILAC mice were pooled for 16 time points. A complex and common reference SILAC protein mix (SILAC mix) was prepared from 16 SILAC protein samples. An equivalent procedure was applied for four Cry1/Cry2 KO mice samples, and the same SILAC mix was used as a reference. Samples were analyzed on an LTQ-Orbitrap Velos mass spectrometer as described (SI Materials and Methods). MS data were analyzed and quantified with MaxQuant version 1.3.0.5 (55) and Andromeda (56) to search against UniProt (release 2012_02) restricted to mouse. Protein identifications were filtered at 1% FDR established by MaxQuant against a reversed sequence database (full data in Dataset S1). We assessed rhythmicity in protein expression profiles using standard harmonic regression. Statistical significance was assessed using the F test for multilinear regression. The resulting P values were used to estimate the FDR by the Benjamini–Hochberg method (57). For the comparison of protein accumulation profiles in WT and Cry1/Cry2 KO mice, we used the Pearson coefficient (R) to measure correlations and assessed significance using the Kolgomorov–Smirnov test (Fig. 4 A and B). To compare protein half-lives in different classes, we used the data from NIH 3T3 cells (19). The analysis of protein function is performed using the DAVID Bioinformatics Resources 6.7 at http://david.abcc.ncifcrf.gov/ (Dataset S2).
Supplementary Material
Acknowledgments
Proteome analyses were carried out at the Protein Analysis Facility, Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, and we acknowledge the expert work of Alexandra Potts and Jachen Barblan. The authors thank Prof. Gijsbertus van der Horst for providing Cry1/Cry2 KO mice. This research was supported by the Canton of Vaud, the Ecole Polytechnique Fédérale de Lausanne, European Research Council Starting Grants ERC-2010-StG-260988 (to F.G.) and ERC-2010-StG-260667 (to F.N.), the Leenaards Foundation (F.G. and F.N.), and Swiss National Science Foundation Grant 31-130714 (to F.N).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314066111/-/DCSupplemental.
References
- 1.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 2.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35(1):445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47(2):158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44(1):419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim T-D, et al. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21(7):797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jouffe C, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Deery MJ, et al. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol. 2009;19(23):2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Krüger M, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134(2):353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 12.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimble JM, Sutton GM, Bunnell BA, Ptitsyn AA, Floyd ZE. Prospective influences of circadian clocks in adipose tissue and metabolism. Nat Rev Endocrinol. 2011;7(2):98–107. doi: 10.1038/nrendo.2010.214. [DOI] [PubMed] [Google Scholar]
- 14.Rismanchi N, Soderblom C, Stadler J, Zhu P-P, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17(11):1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gachon F, Firsov D. The role of circadian timing system on drug metabolism and detoxification. Expert Opin Drug Metab Toxicol. 2011;7(2):147–158. doi: 10.1517/17425255.2011.544251. [DOI] [PubMed] [Google Scholar]
- 17.Reinke H, et al. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22(3):331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Martelot G, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10(11):e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 20.Hirano A, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152(5):1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 21.Vanselow K, et al. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20(19):2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo S-H, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152(5):1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheving LE, Pauly JE, Tsai TH. Circadian fluctuation in plasma proteins of the rat. Am J Physiol. 1968;215(5):1096–1101. doi: 10.1152/ajplegacy.1968.215.5.1096. [DOI] [PubMed] [Google Scholar]
- 24.Jubiz W, Canterbury JM, Reiss E, Tyler FH. Circadian rhythm in serum parathyroid hormone concentration in human subjects: Correlation with serum calcium, phosphate, albumin, and growth hormone levels. J Clin Invest. 1972;51(8):2040–2046. doi: 10.1172/JCI107010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mondola P, Gambardella P, Santangelo F, Santillo M, Greco AM. Circadian rhythms of lipid and apolipoprotein pattern in adult fasted rats. Physiol Behav. 1995;58(1):175–180. doi: 10.1016/0031-9384(95)00016-c. [DOI] [PubMed] [Google Scholar]
- 26.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12(2):174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haus E, Cusulos M, Sackett-Lundeen L, Swoyer J. Circadian variations in blood coagulation parameters, alpha-antitrypsin antigen and platelet aggregation and retention in clinically healthy subjects. Chronobiol Int. 1990;7(3):203–216. doi: 10.3109/07420529009056976. [DOI] [PubMed] [Google Scholar]
- 28.Soulban G, Labrecque G. Circadian rhythms of blood clotting time and coagulation factors II, VII, IX and X in rats. Life Sci. 1989;45(25):2485–2489. doi: 10.1016/0024-3205(89)90015-5. [DOI] [PubMed] [Google Scholar]
- 29.Bertolucci C, et al. Circadian rhythms in mouse blood coagulation. J Biol Rhythms. 2005;20(3):219–224. doi: 10.1177/0748730405275654. [DOI] [PubMed] [Google Scholar]
- 30.Kapiotis S, et al. Morning hypercoagulability and hypofibrinolysis. Diurnal variations in circulating activated factor VII, prothrombin fragment F1+2, and plasmin-plasmin inhibitor complex. Circulation. 1997;96(1):19–21. doi: 10.1161/01.cir.96.1.19. [DOI] [PubMed] [Google Scholar]
- 31.Bruguerolle B, et al. [Circadian rhythms of the so-called inflammation proteins in healthy subjects] Rev Rhum Mal Osteoartic. 1986;53(5):313–316. [PubMed] [Google Scholar]
- 32.Peters T, Jr, Peters JC. The biosynthesis of rat serum albumin. VI. Intracellular transport of albumin and rates of albumin and liver protein synthesis in vivo under various physiological conditions. J Biol Chem. 1972;247(12):3858–3863. [PubMed] [Google Scholar]
- 33.Khoriaty R, Vasievich MP, Ginsburg D. The COPII pathway and hematologic disease. Blood. 2012;120(1):31–38. doi: 10.1182/blood-2012-01-292086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14(1):20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Etchegaray J-P, Cagampang FRA, Loudon ASI, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 36.Cui ZJ, Han ZQ, Li ZY. Modulating protein activity and cellular function by methionine residue oxidation. Amino Acids. 2012;43(2):505–517. doi: 10.1007/s00726-011-1175-9. [DOI] [PubMed] [Google Scholar]
- 37.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones B, et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet. 2003;34(1):29–31. doi: 10.1038/ng1145. [DOI] [PubMed] [Google Scholar]
- 40.Levy E, et al. Expression of Sar1b enhances chylomicron assembly and key components of the coat protein complex II system driving vesicle budding. Arterioscler Thromb Vasc Biol. 2011;31(11):2692–2699. doi: 10.1161/ATVBAHA.111.233908. [DOI] [PubMed] [Google Scholar]
- 41.Nichols WC, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93(1):61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 42.Nyfeler B, et al. Identification of ERGIC-53 as an intracellular transport receptor of α1-antitrypsin. J Cell Biol. 2008;180(4):705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34(2):220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 44.Zhang B, et al. Mice deficient in LMAN1 exhibit FV and FVIII deficiencies and liver accumulation of α1-antitrypsin. Blood. 2011;118(12):3384–3391. doi: 10.1182/blood-2011-05-352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao J, et al. SEC23B is required for the maintenance of murine professional secretory tissues. Proc Natl Acad Sci USA. 2012;109(29):E2001–E2009. doi: 10.1073/pnas.1209207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11(1):47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Hughes ME, et al. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8(7):e1002835. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown WJ, Plutner H, Drecktrah D, Judson BL, Balch WE. The lysophospholipid acyltransferase antagonist CI-976 inhibits a late step in COPII vesicle budding. Traffic. 2008;9(5):786–797. doi: 10.1111/j.1600-0854.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runz H, Miura K, Weiss M, Pepperkok R. Sterols regulate ER-export dynamics of secretory cargo protein ts-O45-G. EMBO J. 2006;25(13):2953–2965. doi: 10.1038/sj.emboj.7601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brown WJ, Schmidt JA (2005) Use of acyltransferase inhibitors to block vesicular traffic between the ER and Golgi complex. Methods in Enzymology 404:115–125. [DOI] [PubMed]
- 51.Erickson SK, Shrewsbury MA, Brooks C, Meyer DJ. Rat liver acyl-coenzyme A:cholesterol acyltransferase: Its regulation in vivo and some of its properties in vitro. J Lipid Res. 1980;21(7):930–941. [PubMed] [Google Scholar]
- 52.Szanto A, Ruys J, Balasubramaniam S. Coordinate diurnal regulation of hepatic acyl-coenzyme A: Cholesterol acyltransferase and cellular levels of esterified cholesterol. Biochim Biophys Acta. 1994;1214(1):39–42. doi: 10.1016/0005-2760(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 53.van der Horst GTJ, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 54.Bur IM, et al. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J Biol Chem. 2009;284(14):9066–9073. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.