Significance
Despite the central role that budding yeast has played in telomere biology, structural analysis of the subunits of the yeast telomerase complex has proven to be challenging. We present here the structure of a yeast telomerase protein, Est3, using the resolution-adapted structural recombination Rosetta strategy that combines NMR experimental data with database-derived conformational sampling. A comprehensive in vivo analysis of the experimentally determined Est3 protein surface has identified two functionally important surfaces, opening up the possibility of a similar discovery in the structurally similar human TPP1 protein.
Keywords: RASREC Rosetta, OB-fold protein
Abstract
Telomerase is essential for continuous cellular proliferation. Substantial insights have come from studies of budding yeast telomerase, which consists of a catalytic core in association with two regulatory proteins, ever shorter telomeres 1 and 3 (Est1 and Est3). We report here a high-resolution structure of the Est3 telomerase subunit determined using a recently developed strategy that combines minimal NMR experimental data with Rosetta de novo structure prediction algorithms. Est3 adopts an overall protein fold which is structurally similar to that adopted by the shelterin component TPP1. However, the characteristics of the surface of the experimentally determined Est3 structure are substantially different from those predicted by prior homology-based models of Est3. Structure-guided mutagenesis of the complete surface of the Est3 protein reveals two adjacent patches on a noncanonical face of the protein that differentially mediate telomere function. Mapping these two patches on the Est3 structure defines a set of shared features between Est3 and HsTPP1, suggesting an analogous multifunctional surface on TPP1.
Telomerase is a telomere-dedicated DNA polymerase that is responsible for telomere-length maintenance in most eukaryotes. In cells that lack telomerase, gradual erosion due to incomplete replication of duplex telomeric DNA leads to an eventual block to cellular proliferation. Ectopic expression of telomerase in human cells is sufficient to confer cellular immortality (1) and it is up-regulated in over 90% of tumor biopsies (2). Conversely, reduced telomerase activity is responsible for the age-dependent effects on organs that rely on continual replenishment throughout a normal human life span and can lead to bone marrow failure, pulmonary fibrosis, or aplastic anemia (3). Hence, an increased understanding of the roles of telomerase and its accessory proteins in telomere length homeostasis has the potential to impact several different aspects of human health.
The yeast telomerase holoenzyme is composed of three proteins [the catalytic ever shorter telomere 2 (Est2) subunit, along with the Est1 and Est3 regulatory proteins], which together form a complex with the TLC1 RNA (4, 5). In vivo, telomerase is highly regulated, in that only a subset of telomeres are elongated in each cell cycle (6); however, the mechanism that restricts telomerase to a limited number of substrates is still poorly understood. This deficit stems at least in part from the fact that the surface of yeast telomerase represents a largely unexplored territory. Even though there are numerous interaction surfaces on the three Est proteins with the potential to regulate important interactions, the only well-characterized regulatory step involving Est proteins is the recruitment of telomerase to the telomere through the direct interaction of Est1 and the end-binding protein Cdc13 (7), which was originally uncovered using a labor-intensive genetic approach (8).
High-resolution structural information provides a straightforward route to identifying functionally important surfaces, but obtaining soluble, well-behaved recombinant telomerase proteins in sufficient quantities for biochemical and structural analysis has proven to be particularly challenging. We have overcome this hurdle for the Est3 telomerase protein, allowing us to solve the high-resolution structure of this yeast telomerase subunit. The overall topology of Est3 was revealed to be an OB-fold, a motif that is increasingly common in telomere-associated proteins (9). We have probed this structure with saturation mutagenesis of the complete surface of Est3. This comprehensive approach, which is only possible once structural information is available, allows identification of all functionally relevant residues on the surface of the protein. Strikingly, residues that mediated telomere replication in vivo clustered to a single face of the Est3 protein, which is distinct from the normal ligand-binding surface used by OB-fold proteins. This surface could be divided into two adjacent yet functionally distinct regions. The first is a telomerase interaction surface of Est3 that is shared with its closest structural homolog, HsTPP1 (formerly known as TINT1, PTOP, and PIP1) (10–12), which has been called the “TEL patch.” We find that immediately adjacent to this TEL patch is a second functional surface that is required for yeast telomere replication in vivo. The strong structural similarities between Est3 and HsTPP1 suggest that this second surface might be functional on TPP1. This study illustrates how structure-driven mutagenesis of the surface of a protein, performed at saturation levels, reveals unexpected insights into the function of the protein.
Results
Determination of the Est3 Structure Using a Novel Strategy.
Est3 has been a remarkably elusive structural target. This challenge has stemmed from both poor intrinsic expression as well as the strong tendency of the protein to form both soluble and insoluble aggregates. To overcome this, extensive optimization of the protein construct and sample conditions as well as multiple solubility enhancing tags were investigated (13, 14), with the best results obtained with the fusion of a His10–SUMO tag to the N terminus of the Est3 protein (15). Still, the 15N-HSQC spectrum and heteronuclear NOE (HetNOE) NMR measurement of Est3 suggested the presence of a disordered region within the full-length protein (Fig. S1A). Deletion of 12 N-terminal residues, found to constitute this flexible region, further significantly enhanced protein stability. Additionally, serine-scanning mutagenesis of the putative nonconserved surface cysteine residues (Cys64, Cys76, Cys109, and Cys142) identified Cys142Ser as a mutation that reduced aggregation. Hence, Est3 with a 12-residue N-terminal deletion and a Cys142Ser mutation (hereafter called “Est3∆N”) was used for our structural studies. Overlay of the wild-type and Est3∆N 15N-HSQC spectra (Fig. S1A) confirmed that the optimized Est3∆N protein retained the structural conformation of the full-length protein.
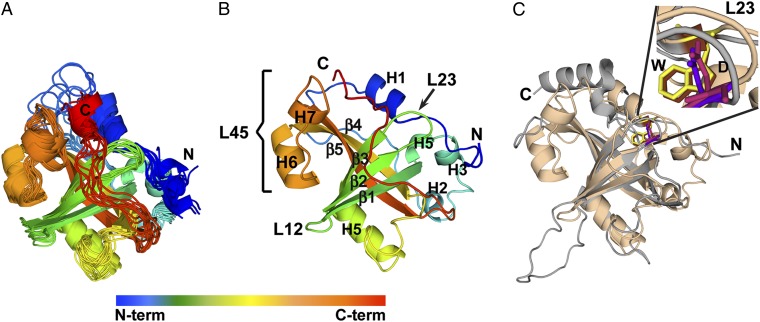
Even with these improvements, the Est3∆N structure could not be obtained using either traditional NMR or crystallographic strategies, due to equilibrium between aggregated and nonaggregated states as well as low sample stability. Instead, we used resolution-adapted structural recombination (RASREC) Rosetta, a novel structure determination strategy combining NMR experimental data with database-derived conformational sampling (16–19), allowing the structures of poorly behaved, larger proteins to be solved (16, 17). To increase sensitivity, NMR data were collected on deuterated samples (20). Nearly complete (97%) backbone resonance assignments were obtained using standard transverse relaxation-optimized spectroscopy–type through-bond triple resonance experiments (21) (Fig. S1B). Long-range restraints were obtained from 37 amide–amide and 91 methyl–methyl NOEs, the latter obtained after selective Ile, Leu, and Val methyl protonation (22), and complete methyl proton assignment (Fig. S1C). Finally, orientational information was obtained from residual dipolar coupling (23) data for 112 N-H bond vectors, collected on an oriented sample. The set of 20 lowest-energy RASREC Rosetta structures using this experimental data generated a well-defined ensemble, with an overall backbone RMSD of 1.5 ± 0.16 Å and greater convergence in the core region (residues 66–163), which exhibits an RMSD of 0.89 ± 0.13 Å (Table S1, 10 lowest energy structures superpositioned in Fig. 1A). The somewhat higher variability for loops outside the central β-barrel is likely due to increased flexibility of the loops, as indicated by the 15N-{1H} heteronuclear NOE NMR measurements and order–parameter random coil index (RCI) S2 values (Fig. S2A) (24).
Fig. 1.
The structure of Est3 is an OB-fold that resembles that of HsTPP1. (A) Ensemble of 10 best-scored Est3∆N structures demonstrates structural convergence. (B) Ribbon representation of the lowest energy Est3∆N structure with secondary structure elements labeled. (C) Superposition of Est3∆N (sand) and HsTPP1-OB (gray) [Protein Data Bank (PDB) ID code 2I46] with the structurally similar W/D motif highlighted. The models were prepared using PyMOL (34).
Several structure validation strategies were used to evaluate the ensemble. Analysis of the final structures showed no residues in disallowed regions, indicating that our structures agreed well with the expected conformational space for residues (Table S1). To ensure that the structures were not adversely biased by individual experimental restraints we also performed several structure calculations with various randomly selected subsets of the NOE constraints (90% and 77% of the final set). All of these calculations produced structure ensembles with good agreement to that of the full calculation (Table S2). Moreover, mapping of the H/D exchange protected residues to mostly the core β-barrel also supported the calculated structure (Fig. S2B). Finally, a Rosetta-independent, external validation was done by using chemical shifts and assigned NOEs as input for a more traditional structure calculation using CYANA-2.1 (25). Although the ensemble RMSD of structures from CYANA-2.1 is increased relative to the RASREC Rosetta structure ensemble and this strategy did not, as expected, yield a well-converged ensemble (Fig. S3A), the overall topology of the structures agrees well with the RASREC Rosetta structure (Fig. S3B), confirming that our observed topology is not biased by the Rosetta input.
Structure of Est3∆N Reveals an OB-Fold Protein.
Est3∆N adopts a classic OB-fold topology (Fig. 1B): a five-stranded β-barrel capped by two helices (H1 and H5) (26). Overall, the region that is N-terminal to the β1-strand of the OB-fold, composed of the first 64 residues of the protein, makes a spiral-shaped structure that caps the top of the β-barrel (Fig. 1B). The β-barrel is formed by two three-stranded sheets composed of β1, β4, and β5 and β1, β2, and β3. The bottom of the β-barrel is capped by a 23-residue stretch between β3 and β4, which forms helix H5 poised at the base of the β-barrel. L12 and L23 are short looped-turns that span five and four residues, respectively, whereas L45 is unusually long, adopting a short helical element. The structure of L45 is supported by chemical shifts that define the helix (H6) and loop secondary structure elements in addition to two intra-L45 long-range NOEs that define the hairpin conformation of the L45 (Fig. S4A). L45 is also highly structured as confirmed by HetNOE and RCI S2 values (Fig. S2A) and it packs against the face of the barrel as defined by four long-range NOEs to the strands β1, β3, and β4 (Fig. S4A). Further, orientational restraints comprising 10 N-H bond vectors in the L45 region validate its calculated orientation in the context of Est3∆N. Following β5, which completes the barrel, is the 19-residue C-terminal tail, which sits over the antiparallel β-sheet formed by β1, β2, and β3. Much of this C tail is dynamic as indicated by the HetNOE NMR measurements and order–parameter RCI S2 values (Fig. S2A).
Even though Est3 has no known sequence homologs in vertebrates, our structure reveals significant similarities to the OB-fold of human TPP1 (HsTPP1-OB) (27) with a Dali (28) Z score of 11.2. Est3 and HsTPP1-OB superimpose with an RMSD of 0.83 Å over all atoms of the OB-fold residues (Fig. 1C and Fig. S4B). Although all hits from the Dali search with a Z score of ≥2.0 are OB-fold proteins, only HsTPP1-OB and telomere end-binding protein β (SnTEBPβ), and to a lesser extent HsRPA70 and archaeal SSB, share several distinguishing structural elements with Est3∆N. First, the N- and C-terminal regions are proximal and antiparallel to each other; second, the C terminus crosses over the antiparallel β-sheet made by β1–β3; and third, helix H1 is adjacent to L23 (Fig. S4B). Furthermore, the interaction between conserved amino acids Trp21 on helix H1 and Asp86 on loop L23 is preserved within the Trp98/Asp148 pair in HsTPP1 and Phe14/Asp54 pair in SnTEBPβ (Fig. S4B). HsTPP1-OB, SnTEBPβ, and Est3∆N also share an H5 positioned at a characteristic angle of ∼40° to the vertical axis of the β-barrel (Fig. S4B). These shared elements further support the structural homology among these proteins, even in the absence of detectable sequence identity.
Structure-Guided Mutagenesis of the Complete Est3 Protein Surface.
A striking feature of the Est3∆N structure is that the long L45 loop occludes the canonical OB-fold ligand-binding surface, suggesting that accessibility to a ligand-binding surface might be regulated in vivo. Surprisingly, however, a strain expressing a variant of Est3 lacking the 19-residue L45 loop exhibited wild-type telomere length (Fig. S5A). Furthermore, missense mutations introduced into this presumed ligand-binding surface (in the variant of EST3 deleted for the occluding L45 loop) did not impair telomere length, providing further support for the dispensability of this face of Est3 in telomere biology.
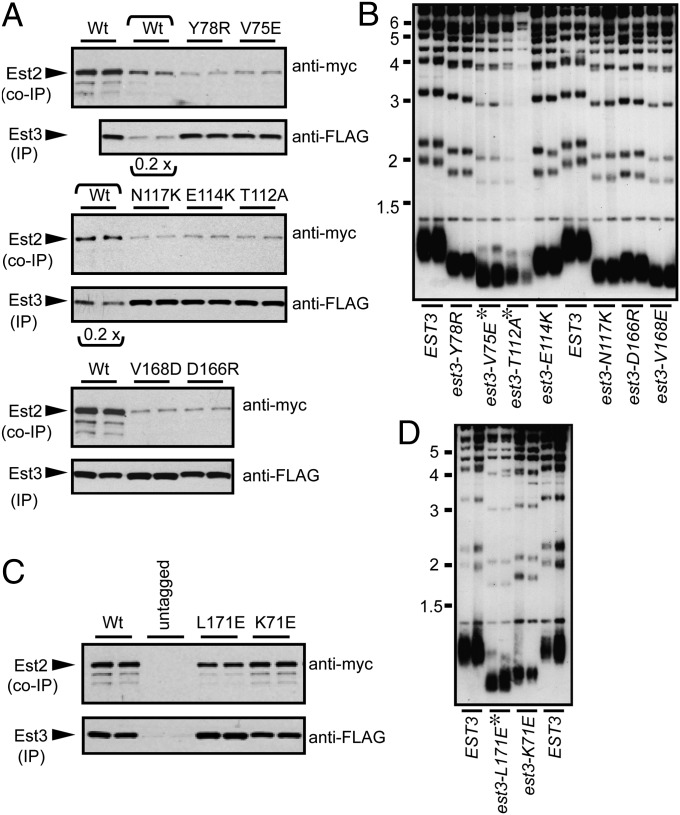
The above results indicated that Est3 employs one or more noncanonical surfaces in telomere replication. We therefore pursued an unbiased comprehensive survey of the entire surface of the Est3 protein by examining the in vivo consequences of mutations introduced into every solvent-accessible surface residue identified in the structure. Analysis of surface exposed residues revealed 112 residues with side chains touching the surface envelope and therefore these were designated as surface-exposed side chains of Est3 (Table S3). These surface residues were mutated by the introduction of a charged amino acid [rather than mutagenesis to alanine, which can often fail to detect functionally important residues (29)], and the resulting mutant collection was examined for effects on telomere replication as previously described (29, 30) (see SI Materials and Methods for more details). Strikingly, this comprehensive analysis revealed that much of the protein surface is dispensable for the functions tested. We clearly identified a total of 15 surface residues which, when mutated, resulted in an inability to maintain telomere length in vivo (29, 30) (Fig. 2 and summarized in Table S3). Notably, these 15 residues map to a noncanonical surface of the Est3 protein which is not commonly used by OB-fold–containing proteins for ligand binding.
Fig. 2.
Two functionally distinct activities on the surface of Est3. (A and C) Coimmunoprecipitation of wild-type (Wt) and mutant Est3 proteins with the catalytic Est2 subunit, bearing (FLAG)3 and (myc)12 epitopes, respectively; the functionality of these two tagged proteins is shown in Fig. S5B. In a subset of the wild-type lanes (indicated by a bracket), 0.2× volume of immunoprecipitate was loaded, to illustrate the detection range. (B and D) Telomere length (assessed after ∼75 generations of growth) of est3-∆ strains transformed with single copy plasmids expressing wild-type EST3 or the indicated est3− missense mutations; mutations that resulted in a telomere maintenance defect severe enough to confer senescence are indicated by an asterisk.
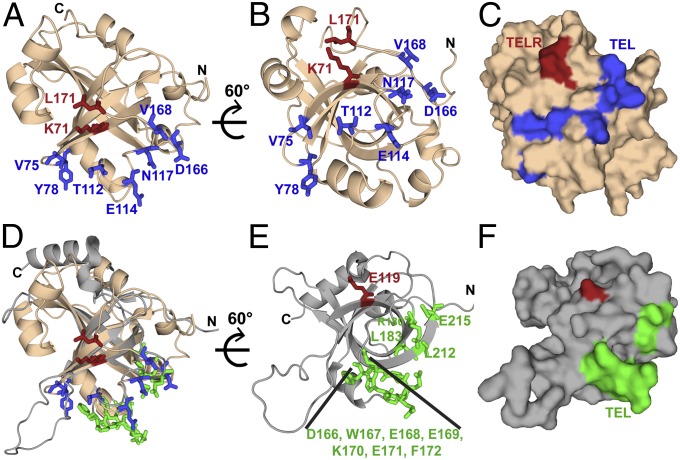
Coimmunoprecipitation of wild-type and mutant Est3 proteins with the Est2 catalytic subunit of telomerase revealed that this collection of surface residues comprised two functionally distinct groups of residues that could be distinguished by their impact on association of Est3 with the telomerase complex (Fig. 2 A and C). Mutations in one set of residues resulted in a greatly reduced ability of Est2 to coimmunoprecipitate with Est3 (Fig. 2A), which was accompanied by a telomere length defect (Fig. 2B), whereas a second set of residues did not impair association of Est3 with telomerase when mutated (Fig. 2C) but nevertheless conferred a profound telomere maintenance defect in vivo (Fig. 2D). Mapping these residues on the surface of Est3 revealed that the residues that mediated association with the telomerase complex defined a narrow contiguous interface, which extended along one face (Fig. 3 A–C) of the Est3 surface along the base of the β-barrel. The second set of residues (Lys71 and Leu171), although distant in sequence, also form a contiguous interface (TELR) located immediately adjacent to this telomerase interaction surface, thereby defining a second, unique function for Est3 (Fig. 3 A–C).
Fig. 3.
The surface of Est3 reveals two distinct contiguous patches. (A) Residues in Est3∆N that mediate binding to telomerase (TEL patch: V75, Y78, T112, E114, N117, D166, and V168) (Fig. 2A) are displayed as sticks in blue, whereas residues not involved in telomerase interaction (TELR patch: K71 and L171) are displayed as sticks in red. (B) Sixty-degree rotation around a horizontal axis shows that the telomerase-interacting residues cluster at the base of the β-barrel. (C) A surface representation demonstrates that the interacting residues form a continuous protein–protein interaction surface. (D) Est3 and TPP1 have a common mode of telomerase association. Superposition of the structure in 3A on HsTPP1-OB (PDB ID code 2I46) with recently identified residues in HsTPP1-OB that mediate binding to telomerase (D166, E168, and K170) (11), (D166-F172, L183 and E215) (10), and (E168, E169, E171, R180, L183, L212, and E215) (12) are shown as sticks in green. The telomerase interaction surface from the two proteins coincides perfectly, indicating structural as well as functional similarity between the two. (E) HsTPP1 structure from D is rotated 60° to show the cluster of telomerase-interacting residues at the base of the β-barrel. Residue E119, identified from structural superposition with K71 of Est3, is displayed as a stick in red. (F) Surface representation of HsTPP1 (same orientation as E), displays two distinct functional patches on its surface. For simplification, the N-term tail has been removed from this view; the structure starts at R96 instead of S90 in the PDB 2I46 structure.
Recent genetic and biochemical studies in human cells have also identified a telomerase interaction surface on the human TPP1 protein, the TEL patch (10–12) (Fig. 3 E and F). Comparison of this surface with the functionally analogous surface on Est3 reveals that these surfaces are essentially completely coincident (Fig. 3D). Furthermore, the second functional Est3 patch (TELR) maps to chemically similar amino acids in TPP1, which predicts that there is a second function for TPP1 that is yet to be elucidated.
Conservation Does Not Fully Predict Functional Interfaces.
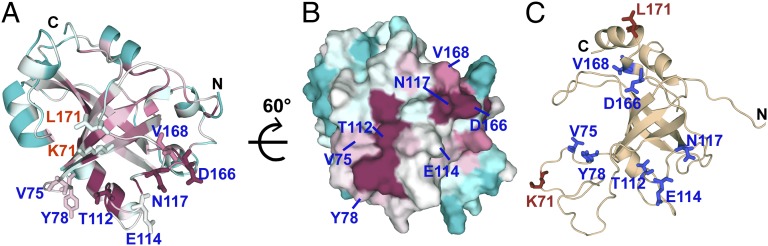
An additional striking conclusion from the surface saturation mutagenesis was that a surprisingly large portion of the Est3 surface appeared to be dispensable for telomere length maintenance in vivo. This opens the possibility that Est3 performs a non–telomere-related function not assessed in our assays. Alternatively, it may be that a large segment of the surface is simply superfluous. As one means of distinguishing between the two possibilities, we compared a map of the functional surface of Est3 with a map of conserved residues on the surface of Est3. Conserved residues (based on a multiple sequence alignment of Est3 proteins from 22 different yeast species) (Fig. S6) were mapped onto the structure of Est3 (Fig. 4 A and B). As expected, most of the highly conserved residues are internal and thus appear to contribute to structure integrity, evident from the sequence conservation in the core OB-fold region (β1–β5 and H5) (Fig. 4A, colored in pink). Indeed, several of these core residues were already identified as being critical to structural integrity, including Trp21, Ile22, and Val157, based on their intolerance to mutation (29). Notably, an assessment of conservation of surface residues indicated that the region of Est3 dispensable for telomere-length regulation displayed a low level of conservation. In contrast, the functional map overlapped, although not precisely, with the region of the surface that displayed the highest degree of conservation. This lack of precise overlap is considered further in Discussion.
Fig. 4.
Conserved surface on Est3 coincides with part of its functional activity. (A) Conserved residues in Est3 (Fig. S6) are mapped on the structure. Residues are color-coded maroon through turquoise indicating conserved through variable residues based on phylogenetic conservation as evaluated by the ConSurf server (35). Residues involved in telomere maintenance are displayed. (B) A view of the structure rotated 60° to match Fig. 3C shows the co-occurrence of the Tel patch with the conserved surface of Est3. (C) The predicted 3D model significantly differs from the calculated structure. The model generated by the PS2 structure prediction server (36) is the same as a recently reported model of Est3 (32). As a result of these differences, functionally important residues (same as A) map to scattered locations on the predicted model’s surface unlike Fig. 3 where a contiguous functional surface was identified.
Discussion
The components of the telomerase enzyme have been elusive structural targets requiring nontraditional approaches. Until recently, Rosetta was restricted to structure predictions of smaller-sized proteins (<150 residues). New improvements in the Rosetta algorithm due to RASREC have provided an effective means to obtain structures of proteins >150 residues through the integration of experimentally obtained long-range and orientation restraints (16, 17). This current study illustrates the power of this approach, especially in experimentally difficult systems.
We found that Est3 adopts an OB-fold with certain distinctive features. The most notable discrepancy between Est3 and other OB-fold proteins is that the canonical OB-fold ligand-binding surface is dispensable for telomere function in Est3, which explains the lack of robust nucleic acid-binding activity exhibited by Est3 (31). Instead, a complete genetic survey of the experimentally defined protein surface revealed two contiguous regions with differential functions in telomere maintenance. Whereas one surface facilitates association with the telomerase holoenzyme, the second serves a separate function in mediating telomerase action. That these surfaces do not fully correspond to those predicted based on conservation alone points to the importance of conducting a comprehensive evaluation of the available surface.
Although a protein fold for Est3 similar to that of TPP1 was accurately predicted on the basis of threading algorithms (30, 32, 33), the topology of the predicted Est3 protein surface was nevertheless strikingly inaccurate. This prior model placed the relevant Est3 surface residues across a wide surface area (Fig. 4C). In contrast, we show here that Est3 mediates its telomere functions through two tightly clustered patches located on a novel face of the OB-fold (Fig. 3C). This illustrates the limitations of homology models, which can be particularly poor at predicting loop conformations in cases where the proteins share scant sequence identity.
Although HsTPP1 and Est3 localize to telomeres through distinct mechanisms, HsTPP1 through the shelterin complex and Est3 as a component of the yeast telomerase holoenzyme, the coincidence of the TEL patches suggests these divergent factors share a common surface for telomerase association. Furthermore, the discovery of a second cluster of functional residues on the surface of Est3 points to the use of additional mechanisms of telomerase regulation by HsTPP1. A full understanding of the activities performed by these surfaces on both Est3 and HsTPP1 may also address whether these two proteins share a common ancestry or instead arose as the result of convergent evolution.
Materials and Methods
Further details can be found in SI Materials and Methods.
Expression and Purification of the Est3∆N Protein.
The Saccharomyces cerevisiae Est3∆N protein was expressed as a His10-Smt3 (Smt3 is yeast SUMO) fusion and first purified by Ni2+-affinity chromatography. Gel filtration was used after Ulp1 cleavage followed by a second round of Ni2+-affinity chromatography. Est3 eluted at >95% purity. Selectively labeled samples were generated similarly using various labeling schemes.
Structure Determination.
All NMR data were collected at 25 °C on 280 μM samples using either a VNMRS 800 or DD2 900 MHz spectrometer equipped with a salt-tolerant proton/carbon/nitrogen cryogenically cooled probe; 97% complete assignment of Est3∆N backbone and chemical shifts were obtained using standard strategies. Sparse distance and orientational restraints were obtained as described in SI Materials and Methods. Structures were generated using RASREC Rosetta (16) as described in SI Materials and Methods.
Telomerase Coimmunoprecipitation Assays.
Wild-type and mutant Est3 proteins, bearing an in-frame (FLAG)3 epitope and under control of the native EST3 promoter, which were expressed from the previously described single-copy plasmid, pVL2076 (30), were introduced into a protease-deficient strain bearing an integrated (myc)12–(Gly)6–Est2 construct. Extract preparation and coimmunoprecipitation of Est3 and Est2 were assessed as previously described (7, 30).
Supplementary Material
Acknowledgments
We thank Chris Lima (Memorial Sloan–Kettering Cancer Center) for the generous gift of the pET-His10-Smt3 vector, Jayakrishnan Nandakumar and Oliver Lange for useful discussions and protocols, David McKay for assistance with the Phoenix protein drop setter for high-throughput buffer screening, and Thayne Dickey for helpful suggestions on the manuscript. We gratefully acknowledge time on the JANUS supercomputer (Research Computing Facility, University of Colorado Boulder) for RASREC Rosetta structure calculations and thank the National Institutes of Health (R01GM059414 to D.S.W., R37AG11728 to V.L., T32GM08759 to T.R., and T32GM007240 to T.M.T.), the National Science Foundation, and a Rose Hills Foundation Fellowship (to T.M.T.) for financial support of this research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates, restraints, and assignment data for the Est3 structure determination have been deposited in the Protein Data Base (PDB ID code 2M9V) and the Biological Magnetic Resonance Bank (accession no. RCSB103391).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316453111/-/DCSupplemental.
References
- 1.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 2.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13(10):693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115(6):413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117(3):323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 7.Tucey TM, Lundblad V. A yeast telomerase complex containing the Est1 recruitment protein is assembled early in the cell cycle. Biochemistry. 2013;52(7):1131–1133. doi: 10.1021/bi3015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144(4):1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31(9):516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287(41):34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong FL, et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150(3):481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandakumar J, et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492(7428):285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17(4):353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhou P, Wagner G. Overcoming the solubility limit with solubility-enhancement tags: Successful applications in biomolecular NMR studies. J Biomol NMR. 2010;46(1):23–31. doi: 10.1007/s10858-009-9371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marblestone JG, et al. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006;15(1):182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange OF, et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci USA. 2012;109(27):10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raman S, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327(5968):1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohl CA. Protein structure estimation from minimal restraints using Rosetta. Methods Enzymol. 2005;394:244–260. doi: 10.1016/S0076-6879(05)94009-3. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009;43(2):63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner KH, Kay LE. In: Modern Techniques in Protein NMR. Krishna NR, Berliner LJ, editors. New York: Plenum; 1998. pp. 27–74. [Google Scholar]
- 21.Fernández C, Wider G. TROSY in NMR studies of the structure and function of large biological macromolecules. Curr Opin Struct Biol. 2003;13(5):570–580. doi: 10.1016/j.sbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125(45):13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 23.Chen K, Tjandra N. The use of residual dipolar coupling in studying proteins by NMR. Top Curr Chem. 2012;326:47–67. doi: 10.1007/128_2011_215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127(43):14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 25.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 26.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445(7127):506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubin JW, Rao T, Mandell EK, Wuttke DS, Lundblad V. Dissecting protein function: An efficient protocol for identifying separation-of-function mutations that encode structurally stable proteins. Genetics. 2013;193(3):715–725. doi: 10.1534/genetics.112.147801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008;15(9):990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Mandell EK, Rao T, Wuttke DS, Lundblad V. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res. 2010;38(7):2279–2290. doi: 10.1093/nar/gkp1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lue NF, Yu EY, Lei M. A popular engagement at the ends. Nat Struct Mol Biol. 2013;20(1):10–12. doi: 10.1038/nsmb.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15(9):985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1.5.0.4. Available at www.pymol.org.
- 35.Glaser F, et al. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 36.Chen C-C, Hwang J-K, Yang J-M. (PS)2: Protein structure prediction server. Nucleic Acids Res. 2006;34(Web Server issue):W152-7. doi: 10.1093/nar/gkl187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.