Significance
K+ channels are spontaneously inactivated subsequent to channel opening, a process that is central to their many crucial roles in cell signaling. Several studies suggest that for KcsA inactivation involves ion release. Here we demonstrate that K+ expulsion from the selectivity filter at neutral pH causes spontaneous opening. This illustrates an unexpected opening mechanism for KcsA, which is normally pH gated, and also provides strong evidence for transmembrane allosteric coupling.
Keywords: C-type inactivation, solid-state NMR, membrane protein, protein dynamics, chemical shift assignments
Abstract
It has been hypothesized that transmembrane allostery is the basis for inactivation of the potassium channel KcsA: opening the intracellular gate is spontaneously followed by ion expulsion at the extracellular selectivity filter. This suggests a corollary: following ion expulsion at neutral pH, a spontaneous global conformation change of the transmembrane helices, similar to the motion involved in opening, is expected. Consequently, both the low potassium state and the low pH state of the system could provide useful models for the inactivated state. Unique NMR studies of full-length KcsA in hydrated bilayers provide strong evidence for such a mutual coupling across the bilayer: namely, upon removing ambient potassium ions, changes are seen in the NMR shifts of carboxylates E118 and E120 in the pH gate in the hinges of the inner transmembrane helix (98–103), and in the selectivity filter, all of which resemble changes seen upon acid-induced opening and inhibition and suggest that ion release can trigger channel helix opening.
Potassium channel activation and inactivation is fundamental to many physiological functions including muscle contraction and the generation of synaptic action potentials (1). KcsA is a 160-residue pH-activated homotetrameric K+ channel isolated from the soil bacterium Streptomyces lividans (2, 3) with high sequence homology and functional similarity to mammalian potassium channels (4). It has provided an excellent model for studies of ion-conduction by X-ray crystallography (3, 5, 6), electrophysiology (7, 8), and NMR (9–21). Like many potassium channels, it exhibits (4, 6, 22, 23) slow, spontaneous inactivation involving the residues near the extracellular selectivity filter subsequent to channel activation. Recent results from X-ray crystallography and molecular dynamics suggest that the gates are coupled and that inactivation is prompted by channel opening, mediated via a series of intrasubunit steric contacts involving F103 with T74, T75, and M96 and an intersubunit contact with the neighboring I100 side chain (4–6, 24, 25). In separate experiments, the extracellular gate has been observed to respond directly to ambient [K+]: at high [K+] it exists in a conductive form, and at low K+ it collapses into a nonconductive state (3). Our NMR studies suggest that the low [K+] state and the low pH inactivated state may be similar; this conclusion is supported by the effect of the mutation E71A and the pattern of chemical shift perturbations in the selectivity filter when the ion is depleted (9, 19). Meanwhile, X-ray crystallography studies suggest that mutants (E71A) unable to undergo inactivation are also unable to expel ions (26).
An established similarity of the low pH and the low [K+] states would clarify the importance of allosteric coupling and have the practical consequence that the well-behaved low K+ state could serve as a useful structural proxy for the otherwise fleeting inactivated state. For these reasons we tested this correspondence using NMR experiments. If the low K+ state is similar to the inactivated state of KcsA achieved by lowering the pH, it is expected that structural changes indicative of channel opening observed at low [K+] would occur not only in the selectivity filter but also in the pH gate and the hinge region. However, some studies imply that these two gates might be uncoupled or weakly coupled. For example, X-ray crystallographic studies of KcsA, where K+ sensitivity was largely isolated to the selectivity filter (3). In this work, we asked whether, by contrast, full-length wild-type KcsA (160 aa) reconstituted into hydrated lipid bilayers exhibits global structural changes upon ion expulsion suggestive of channel opening. To accomplish this, nearly complete 13C and 15N chemical shift assignments were obtained for the transmembrane and loop regions from four-dimensional (4D) solid-state nuclear magnetic resonance (SSNMR) (27), providing numerous reporters for conformational change during ion binding. In low [K+] ion conditions at neutral pH, not only does KcsA expel the K+ ions from the inner selectivity filter sites, but the channel also exhibits chemical shift perturbations at the pH gate and the hinge of the inner transmembrane helix, suggesting features akin to the inhibited state that is present at low pH and high [K+]. That these two distinct conditions result in a nearly identical state of the channel offers strong evidence for transmembrane allostery in the inactivation process.
Results
Four-Dimensional Chemical Shift Assignments.
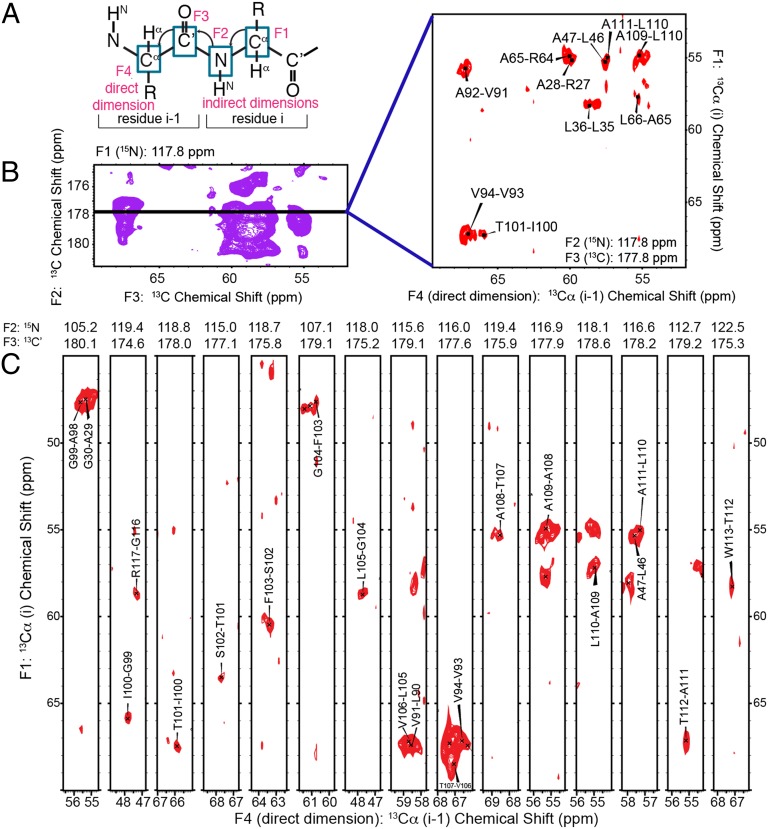
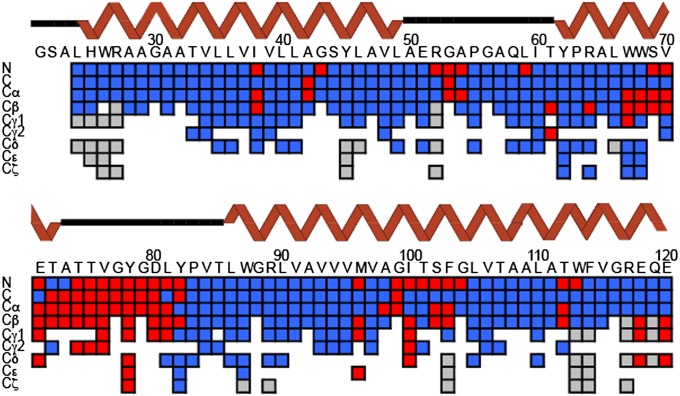
Because KcsA is highly α-helical, it exhibits significant chemical shift overlap, especially in the 15N resonances often required to establish interresidue sequential assignments. We used 3D and 4D spectra to resolve numerous peaks in uniformly 15N,13C–enriched samples of KcsA, which were reconstituted into liposomes composed of 9:1 ratio of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE):1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) (with a 1:1 mass lipid–protein ratio). Unique backbone correlations were identified using 4D CANCOCA (Cα-N-C’-Cα) spectra, which were acquired at a sample temperature of ∼5°. A schematic of the backbone connectivities provided by the CANCOCA experiment is illustrated in Fig. 1A. 3D NCACX (N-Cα-Ci,sidechain), NCOCX (N-C’-Ci-1,sidechain), and CANcoCX (Cα-N-c’- Ci-1,sidechain) spectra (sites indicated in lowercase were used in a magnetization transfer-pathway, without measurement of their chemical shift) were used to aid residue-type assignments for 4D analysis and to assign side-chain resonances. Roughly 42 unique correlations were assigned from the 3D spectra alone, but many regions of the NCACX and NCOCX spectra were still difficult to interpret, especially in planes corresponding to 15N shifts of 115–122 ppm (Fig. S1). The power of 4D spectroscopy is illustrated in Fig. 1B, where resonances appearing along a slice of the 3D spectrum indicated by the black bar are cleanly resolved in the corresponding plane from the CANCOCA spectrum. Examples of an extended 4D backbone walk through the hinge region of KcsA is provided in Fig. 1C, where resonances from W113 to A98 are presented. This is a region of particular interest, often referred to as a hinge, whose “bending” has been assumed to be essential for opening and conduction (6, 28). A separate CANCOCA backbone walk is provided in Fig. S2. A scheme summarizing the atoms assigned in this study (and their completeness) is presented in Fig. 2. Colored tiles indicate a unique resonance assignment. Resonances highlighted in red indicate a K+ concentration-dependent chemical shift perturbation greater than 0.3 ppm at the site, which represents a value greater than the largest chemical shift uncertainty. Predicted secondary structure for each site determined by TALOS+ (29, 30) is presented above the tiles in the wire diagram format. The secondary structure we determined from these assigned shifts largely conforms to that observed in X-ray crystallography (3, 31), including longer transmembrane helices compared with solution NMR studies in detergent micelles. Although several sites along the N-terminal amphipathic helix and the C-terminal four-helical bundle are tentatively assigned, they are not presented because they lack redundant confirmation. Confirming these shifts proved difficult because many sites in these regions of the protein appear to be dynamic and are either absent or significantly broadened in the acquired spectra.
Fig. 1.
Reduction of overlap in spectra of KcsA using 4D CANCOCA (Cα-N-C′-Cα). (A) CANCOCA polarization transfer scheme. In this experiment, the Cα and 15N resonances of the i residue are correlated to the C′ and Cα resonances of the i-1 residue. Here, F4, the directly detected dimension, corresponds to Cαi-1. (B) C′i-1-Cαi-1 plane from a NCOCX 3D experiment at a 15N chemical shift of 117.8 ppm. Spectrum was acquired on a 750 MHz Bruker Avance spectrometer (Bruker Biospin). The Cαi-Cαi-1 4D plane on the right corresponds to the black line at an amide chemical shift of 117.8 ppm and C′ shift of 177.8 ppm. By correlating these sites to the Cαi resonance, 10 overlapped peaks were cleanly resolved. The 4D spectrum was acquired on a 600 MHz Infinity Plus SSNMR spectrometer (Agilent). (C) Cαi to Cαi-1 backbone walk from 4D CANCOCA is shown. The directly detected dimension (Cαi-1, F4) is displayed with the first indirect dimension (Cαi, F1) in 2D strips occurring in the spectra (resolved at the F2 and F3 frequencies listed above each 2D strip). The backbone walk is from W113 to A98, including the hinge region of TM 2 in KcsA.
Fig. 2.
Graphical illustration of assignment completeness and K+ dependence. Backbone assignments are complete from L24 to E120. Most side-chain resonances are also assigned for the high K+ state over this span. The chemical shift-based secondary structure prediction from TALOS+ is depicted in the wire diagram above the assignment grid. Sites observed to move with [K+] (greater than 0.3 ppm) are illustrated in red. Unassigned resonances are depicted in gray. Data probing K+ perturbations are incomplete because for low [K+] samples, only 2D spectra were used.
Global Conformation Change in Response to [K+].
We analyzed chemical shift changes that occurred when the ambient potassium ion concentration was lowered by comparing two otherwise identical samples of KcsA both at pH 7.5, the first prepared with 50 mM [K+] and the second at 0.2 μM [K+] (9). Previously we reported on NMR spectra of full-length KcsA in bilayers at low [K+], and using NMR reporters near the selectivity filter that are well dispersed spectrally, we documented a conformational change upon ion binding and an ion affinity in the low micromolar range (9). Unique chemical shift assignments obtained from the present study attest to global conformational effects of K+ binding. Low K+ spectra are overlaid onto the previously described 50 mM [K+] spectra in Figs. 3 and 4. K+ binding at neutral pH induces changes clustered in functionally important regions, discussed below.
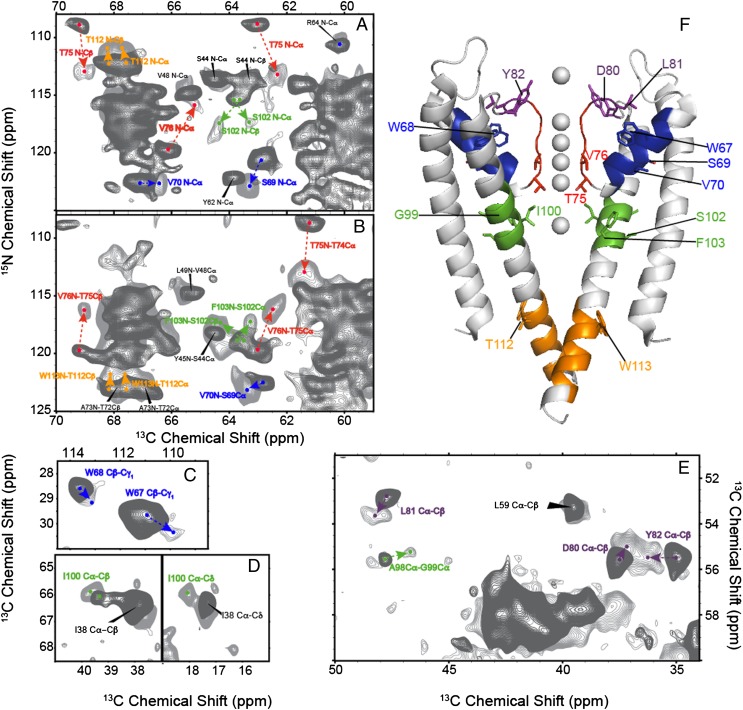
Fig. 3.
Chemical shift perturbations observed when K+ is evacuated from the selectivity filter are shown. Significant perturbations are observed in several key regions across the entire structure including the K+ chelating selectivity filter residues (red), the pore helix (blue), the hinge region (green), the pore loop residues (purple), and the intracellular pH gate region (orange). Overlays of the spectra collected at high K+ (50 mM, dark gray) and low K+ (0.2 μM, light gray) are shown in A–E. Heteronuclear 15N-13C correlation spectra are shown in A and B, and homonuclear 13C-13C correlation spectra are shown in C–E. Chemical shift changes for key residues are highlighted with arrows in the spectra and mapped onto the structure of KcsA (from PDB ID code 1K4C) in F. The changes suggest a global conformational change in response to K+ binding.
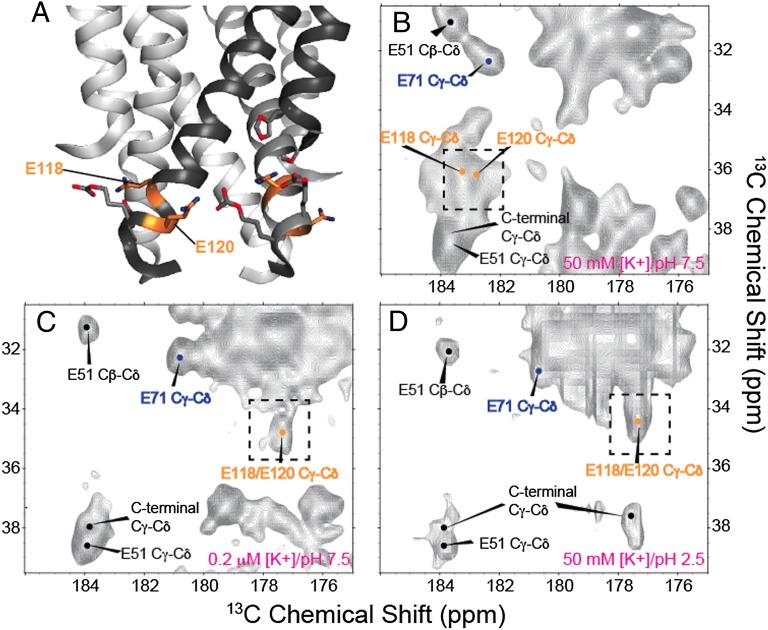
Fig. 4.
The effect of [K+] and pH on the glutamic acid residues in the intracellular activation gate of KcsA is shown, highlighting the similarities between the low K+ and low pH structures. A shows the molecular structure of this gate in the closed (high pH) form based on PDB ID code 1K4C, including key residues E118 and E120 highlighted in orange. B–D show 13C-13C 2D correlation spectra of the glutamate Cγ-Cδ region under different conditions. B shows spectra of samples at high [K+] (50 mM) and neutral pH (7.5). C shows spectra at low [K+] (0.2 μM) and neutral pH (7.5). D shows spectra at high [K+] (50 mM) and low pH (3.5). E118 and E120 exhibited a comparable change in Cγ-Cδ chemical shifts at both low K+ (C) and low pH (D). The shift similarity suggests the local environment of this portion of the inner pH sensitive gate is similar under both the low [K+]/high pH and high [K+]/low pH conditions However, in the low [K+]/high pH spectra (C), the E118/E120 peak appears as a doublet in the Cγ dimension and is replaced by a narrower singlet peak in the high [K+]/low pH spectrum (D). This effect is likely the result of a small shift in the side-chain rotameric state of either E118 or E120 shifting the Cγ resonance by ∼0.5 ppm.
Large chemical shift changes are observed in the pH gate of KcsA (Fig. 4): E118 and E120 resonances shift significantly upfield by ∼5.7 ppm (Fig. 4B). The pH gate of KcsA is composed of these two carboxylate side chains and a network of hydrogen bonds to neighboring arginines and H25 (8). Unfortunately the side chains of H25 were not uniquely assigned in the low K+ spectra. On the basis of these striking changes it is likely that the protonation states and pKa values of E118 and E120 change when the ambient K+ level is lowered, or alternatively that there is a change in their hydrogen bonding environment.
The hinge region of TM2 exhibits changes (residues 96–103 and to a lesser extent, 112–113) in response to ambient potassium concentration. Because they contact selectivity filter sites on adjacent strands, residues M96 and F103 are presumed to be directly sensitive to the ion occupancy of the selectivity filter and have been shown through mutation to have an important role in the allostery of this system (6, 25). It has been hypothesized that steric effects from the side chain of F103 are important in inactivation, both for KcsA and possibly analogously for eukaryotic homologs (32). Changes in the M96 and F103 15N backbone and 13C side-chain resonances were observed, suggesting structural shifts in this region upon K+ binding. The I100 spin system (Fig. 3D) is relatively weak at high [K+], while being strong and well-resolved at low [K+], suggesting that this site rigidifies with loss of ion. Chemical shift perturbations were observed at G99 (Cα correlated A98 Cα; Fig. 3E), I100 (complete aliphatic spin system; Fig. 3D), S102 (Cα, Cβ, 15N; Fig. 3A), F103 (Cα, Cβ, 15N; Fig. 3B), A111 (Cβ), T112 (15N; Fig. 3A), and W113 (15N; Fig. 3B).
In addition to the selectivity filter, pH gate, and hinge region, residues exhibiting aggregate chemical shift perturbations greater than 1 ppm (defined as the sum of the absolute chemical shift difference of all resonances in each residue) were found in several other key regions of the protein including parts of the S1 helix (I38, V39, G43), the pore helix (W67, S69, V70, E71), and residues along the loop connecting the selectivity filter to the S2 helix (D80, L81, Y82). By contrast, much of the first transmembrane helix and the C-terminal section of the inner transmembrane helix show no notable changes in NMR spectra upon ion binding. The relative magnitudes of the changes are shown overlaid on the structure of KcsA [from Protein Data Bank (PDB) ID code 1K4C] (3) in Fig. 5A. We conclude that K+ binding in bilayers induces global changes beyond the vicinity of the ion binding sites.
Fig. 5.
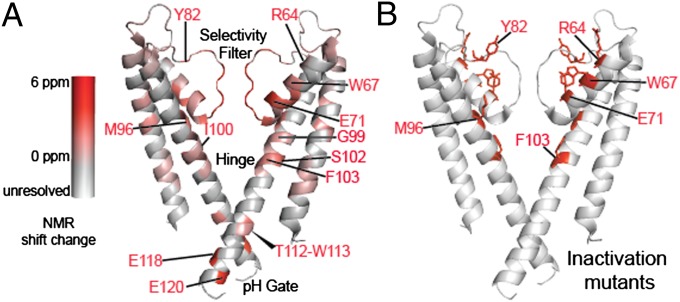
Residues showing large K+-dependent chemical shift differences map well to mutation sites known to promote or suppress channel inactivation. (A) The absolute deviation of chemical shifts (defined as the sum of the absolute chemical shift changes of all resolved resonances for each residue) of residues in KcsA in response to the ambient potassium level is mapped onto the 1K4C crystal structure in a scale ranging from gray (no change) to red (6 ppm summed over N, CA, CB, CO, and any assigned side-chain resonances). (B) Residues at which mutations are known to impact inactivation are highlighted in red (6, 22, 23, 25, 33). The mutations lie in regions that register distal NMR effects of the ambient potassium level. The chemical shifts are provided in Table S3.
Mutations in a number of locations in the channel have a strong effect on inactivation (Fig. 5B) (6, 22, 23, 25, 33). The amino acids affected by K+ binding are compared with those that have been implicated in inactivation of the channel in Table S1 and Fig. 5 A and B. Overall there is a remarkable correspondence between the locations where NMR shift changes are seen upon ion binding and the locations that are implicated in inactivation based on mutation functional data.
Similarity of pH Gate at Low [K+] to Low pH Conditions.
We further examined the changes observed as a function of [K+] by comparing spectra to a third sample of KcsA prepared at 50 mM [K+], but with a pH of 3.5, well below the reported pH change for channel opening. Our spectra of KcsA at low pH, where “opening” of the channel is expected to occur followed by inactivation, are in many ways similar to the studies presented previously (12, 20, 34). Although the line-widths and sensitivity are not as favorable for low pH samples as for low [K+] samples, possibly because of sample disorder present in the low pH sample, the comparison suggests that the low [K+] and low pH states of KcsA are very similar in several regions (Fig. S3). Previous work from our group (19) and the group of Shimada (20) has suggested a similarity based on markers in the selectivity filter. Here we show that this similarity pervades the entire structure of KcsA—including the hinge and the pH gate. The carboxylate resonances of E118 and 120 (Fig. 4) are particularly striking because they move upfield by ∼5.7 ppm for both the low pH and low [K+] conditions relative to neutral pH and high ambient [K+]. The groups of Nimigean (8) and Perozo (35) have reported detailed mutational analysis and shown that when E118 and E120 are mutated to Alanine, the pKa of the activation is shifted by 2–3 units, indicating the importance of the carboxylate functionality in setting the activation pKa for KcsA. Our results show that the pKas of these critical carboxylates are sensitive to K+ binding on the opposite side of the protein, which highlights the importance of a strong allosteric coupling network in controlling the function of KcsA. We conclude that the state produced at low pH is the same or highly similar to the state produced at low ambient potassium ion concentration. The group of Baldus has previously assigned the side-chain carboxyl groups of E71, E118, and E120 (34). The results from the Baldus group showed partial ionization dependent upon [K+], finding that at high [K+], E118 and E120 are 20% protonated at pH 7.5 and 40% protonated at pH 4, whereas at low K+, they are ∼10% protonated at pH 7.5 and 80% protonated at pH 4. Although our assignments at high K+ and neutral pH are in good general agreement with theirs, we observe chemical shifts consistent with near complete protonation of E118 and E120 at both low pH and low [K+] within the experimental signal-to-noise (S/N). The discrepancy is perhaps explained by further work from the Baldus group showing the importance of anionic nonannular lipids near the selectivity filter of KcsA (36). In the KcsA–Kv1.3 chimera used in the Baldus studies, R64 is mutated to an aspartate, and there is reduced binding to these anionic lipids. This lipid binding is present in our samples and is likely required for the strong channel allostery we observe.
Discussion
Implications for Channel Gating and Inactivation.
We show that extensive regions of KcsA quite distal to ion binding exhibit NMR spectral shifts in response to low potassium concentration. Moreover, the loci of these changes are well correlated with regions that are hypothesized from crystal structures to be perturbed as the channel opens at low pH (5, 6, 24) or correlated to regions where mutations affect inactivation kinetics (6, 22, 23, 25, 33). Evidence shown here for changes specifically in the hinge region and the pH sensor lead to the suggestion that not only is the ion expelled but that the global conformational change is likely to be the opening of the channel, akin to the changes during activation at low pH.
The many spectral similarities between the state produced at low pH and that produced at low ambient potassium ion concentration suggest that the inactivated state can be produced either by proton loading followed by spontaneous ion ejection at high ambient ion concentration or by ion removal, followed by spontaneous proton binding at neutral pH. This surprising conclusion further suggests that the pKas of E118 and 120 may be perturbed by several units as a function of the ambient K+ level, and that the ion affinity is strongly influenced by the pH gate, implying strong allosteric coupling across the bilayer.
The hypothesis that the low potassium state exhibits opening of the inner transmembrane helices was further explored using available structures of open or semiopen channels (5), by predicting 13Cα chemical shifts (37, 38). 13Cα chemical shifts are known to be strong indicators of backbone dihedral conformation (37–39). Analyzing the low K+ structure (1K4D) (3) and several structures with different degrees of opening [with distances between the Cαs of T112 ranging from 14 Å (3FB5) to 32 Å (3F5W)] (5). The best agreements between prediction and experimental chemical shifts, particularly for the hinge region of the TM2, were observed for structures 3F7Y and 3FB8, which exhibit intermediate openings of 17 Å and 20 Å, respectively (measured at T112). Further discussion of these calculations is provided in SI Text, Fig. S4, and Table S2. It has been suggested that an opening of about 20 Å triggers the appearance of the collapsed state of the selectivity filter (5), in low pH protocols. Perhaps removal of the ion induces a global conformational change and opening or partial opening to a similar extent.
It was previously suggested that upon acidification of the intracellular pH gate, the channel opens and subsequently an ion is spontaneously expelled from the selectivity filter, causing inactivation. Our working model for the processes by which low potassium conditions result in the same state is that, upon washing the ion from the selectivity filter, the inner transmembrane helix and pH gate spontaneously open, due to the same transmembrane allosteric coupling. Remarkably, this coupling allows the binding of a simple ion to influence the large-scale conformation of the potassium channel.
Conclusions
Chemical shifts from high-resolution NMR studies of the K+ channel KcsA in hydrated bilayer environment show changes as a function of potassium ion concentration in a number of functionally important regions: the extracellular selectivity filter, the hinge of the inner transmembrane helix, and the intracellular pH gate, consistent with the global conformational change that occurs accompanying ion release at neutral pH. Moreover, chemical shift similarities indicate that the dominant state of KcsA at low pH and high [K+] is strikingly similar to the state at low [K+] and neutral pH. We conclude that both states are good structural models for the inactivated state known from electrophysiology experiments, and that their equivalence occurs because of an unusual degree of allosteric coupling across the bilayer.
Materials and Methods
Expression and Purification of U-15N,13C KcsA.
The complete protocol for optimal SSNMR sample preparation is provided in detail elsewhere (40). However, briefly, five samples were prepared for this study. The first two were prepared using a plasmid that was a gift from the group of Rod Mackinnon at Rockefeller University (New York, NY). The last three samples were prepared from a second plasmid, which was a gift from the group of Crina Nimigean at Weill Cornell Medical School (New York, NY). The first plasmid is optimal with Escherichia coli M15 cells and the second with E. coli JM83 cells.
Transformed cells were grown on LB-Agar plates with the requisite antibiotic (100 μg/mL Amp, 25 μg/mL Kan for plasmid 1 and 100 μg/mL Amp for plasmid 2). The plates were incubated at 37 °C for 6–8 h. Single colonies were picked and transferred into a 4x 10 mL LB preculture with the appropriate antibiotic. The preculture was incubated at 37 °C and 250 rpm shaking for 3–4 h or until the OD600 is ∼0.5. The preculture was then transferred into four flasks with 4 L of LB and incubated at the same conditions of temperature and shaking. Once the OD600 reached ∼0.9, the cells were then harvested by centrifugation and resuspended in 0.5 L of M9 minimum media supplemented with 2 g of 13C-labeled glucose, 0.5 g of 15-labeled NH4Cl, 10 mL of 13C-15N–labeled Bioexpress from Cambridge Isotopes, and 1 g proline in the case of plasmid 2. Cells were allowed to grow for 1 h in the M9 media, and then protein expression was induced using 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for plasmid 1 or 1 mM aTC (from Cayman Chemicals #10009542) for plasmid 2. After induction, the cells were allowed to grow for 12–14 h (overnight) at 37 °C and then harvested by centrifugation. The cells were lysed via French Press and membrane protein extracted using 1 g of Decyl-β-Maltopyranoside (DM, from Affymetrix #D310S) for every 10 g of cells. The unlysed cells and membranes were pelleted by centrifugation and purified via nickel affinity column. After concentration of the purified fractions, the concentrated protein solution was then dialyzed against a 5 mM DM solution to reduce the detergent concentration. Low K+ samples were prepared by dialysis against a buffer containing 50 mM Tris and 50 mM ultrapure NaCl (from Sigma) and adjusted to a pH of 7.5 with HCl. The residual K+ level was quantified by atomic absorption spectroscopy and shown to be 0.2 μM.
The U-[13C]-[15N]-KcsA was reconstituted into 9:1 DOPE/DOPS liposomes. DOPE (18:1 PE, 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine, Avanti # 850725) has a zwitterionic headgroup and DOPS (18:1 PS, 1,2-Dioleoyl-sn-Glycero-3-[Phospho-l-Serine], Avanti # 840035) has an anionic headgroup. The chloroform was evaporated under nitrogen and the lipids were resuspended in 50 mM KCl, 50 mM Tris, 0.01 mM sodium azide, 10 mM DM, pH 7.5 buffer to a total lipid concentration of 10 mg/mL The lipid solution was then added to the desired amount of KcsA in DM such that the protein–lipid weight ratio was 1:1. The vesicle/protein mixture dialyzed against a 50 mM Tris/50 KCl buffer at pH 7.5 or 50 mM Tris/50 mM NaCl (using ultrapure NaCl). The liposome pellet was then harvested. The pellets were then subject to a –80 °C overnight freeze-thaw cycle to remove excess bulk water, which reduced the volume to ∼30 μL.
SSNMR Spectroscopy.
SSNMR experiments were performed either on a 750 MHz (1H frequency) three-channel Bruker Avance spectrometer (Bruker Biospin) or a three-channel Varian/Agilent Infinity Plus spectrometer (Agilent). The liposome pellets were centrifuged into either 3.2 mm Agilent limited speed rotors or 4 mm Bruker rotors. Typically 20–30 μL of the sample were packed into the rotors, which corresponds to 7–10 mg of KcsA. SPECIFIC-CP (41) was used for all transfers between 15N and 13C nuclei unless otherwise noted.
The 750 MHz Bruker Avance spectrometer was equipped with a three-channel 4 mm 1H-13C-15N probe. The magic-angle spinning (MAS) rate was set to 14 kHz and the variable temperature apparatus was set to –30 °C, corresponding to ∼0 °C in this instance. Typical 90° pulse lengths for 1H were ∼2.5 μs. 13C and 15N 90° pulse lengths were ∼5 μs. We used 80–85 KHz of SPINAL64 decoupling (42) on the 1H channel during acquisition. High-resolution 13C–13C correlation experiments using the 2D dipolar assisted rotational resonance (DARR) sequence (43) and a 15 ms mixing time show narrow lines for several resonances. Six experiments were assigned for the sake of assignments. Six 2D spectra were used: a 13C-13C 2D spectrum with 15 ms of DARR mixing, a 13C-13C 2D with 150 ms of DARR mixing, an NcaCX with 20 ms of DARR mixing, an NcaCX with 100 ms of mixing, an NcoCX with 20 ms of DARR mixing, and an NcoCX with 100 ms of DARR mixing. Two 3D spectra were acquired: One NCACX with 25 ms of DARR mixing and an NCOCX also with 25 ms of DARR mixing.
The 600 MHz Infinity Plus (Agilent) spectrometer was equipped with an Agilent 3.2 mm HFXY 4 channel T3 probe operating in 1H-13C-15N mode. Spinning was maintained at 13.333 kHz except for the CANCOCA 4D experiments performed at 11.111 kHz. Typical 90° pulse lengths were 2.0 μs for 1H, 2.5 μs for 13C, and 5 μs for 15N. We used 80–85 KHz of SPINAL64 decoupling (42) on the 1H channel during acquisition. Two 2D spectra were acquired: an NC correlation spectrum where polarization transfer as provided by a short zf-TEDOR period (44) and an NcxCX spectrum acquired with a short zf-TEDOR period followed by 50 ms of DARR mixing. A 3D CAncoCX spectrum with 50 ms DARR mixing was acquired, followed by a CANCOCA 4D experiment (27). The CANCOCA 4D used rotational resonance tickling (R2T) mixing (45) for optimal transfer from carbonyl to Cα.
Supplementary Material
Acknowledgments
We thank Crina Nimigean for many helpful suggestions. This work was supported by grants from the National Institutes of Health (NIH): NIH R01 GM 88724 (to A.E.M.) and NIH NRSA F32 087908 (to B.J.W.). Professor McDermott is a member of the New York Structural Biology Center, a Strategically Targeted Academic Research (STAR) center supported by the New York State Office of Science, Technology, and Academic Research. NMR resources were supported by NIH P41 GM66354.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319577110/-/DCSupplemental.
References
- 1.Tang XD, Santarelli LC, Heinemann SH, Hoshi T. Metabolic regulation of potassium channels. Annu Rev Physiol. 2004;66:131–159. doi: 10.1146/annurev.physiol.66.041002.142720. [DOI] [PubMed] [Google Scholar]
- 2.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Zhou YF, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414(6859):43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 4.McCoy JG, Nimigean CM. Structural correlates of selectivity and inactivation in potassium channels. Biochim Biophys Acta. 2012;1818(2):272–285. doi: 10.1016/j.bbamem.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466(7303):203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuello LG, et al. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466(7303):272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeMasurier M, Heginbotham L, Miller C. KcsA: It’s a potassium channel. J Gen Physiol. 2001;118(3):303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA. 2008;105(19):6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J Mol Biol. 2010;401(2):155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zachariae U, et al. The molecular mechanism of toxin-induced conformational changes in a potassium channel: Relation to C-type inactivation. Structure. 2008;16(5):747–754. doi: 10.1016/j.str.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Schneider R, et al. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. J Am Chem Soc. 2008;130(23):7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- 12.Ader C, et al. A structural link between inactivation and block of a K+ channel. Nat Struct Mol Biol. 2008;15(6):605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 13.Varga K, Tian L, McDermott AE. Solid-state NMR study and assignments of the KcsA potassium ion channel of S. lividans. Biochim Biophys Acta. 2007;1774(12):1604–1613. doi: 10.1016/j.bbapap.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282(20):15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 15.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14(11):1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange A, Giller K, Pongs O, Becker S, Baldus M. Two-dimensional solid-state NMR applied to a chimeric potassium channel. J Recept Signal Transduct Res. 2006;26(5-6):379–393. doi: 10.1080/10799890600932188. [DOI] [PubMed] [Google Scholar]
- 17.Lange A, et al. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 2006;440(7086):959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 18.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15(4):684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhate MP, McDermott AE. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc Natl Acad Sci USA. 2012;109(38):15265–15270. doi: 10.1073/pnas.1211900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai S, Osawa M, Takeuchi K, Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc Natl Acad Sci USA. 2010;107(14):6216–6221. doi: 10.1073/pnas.0911270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu LP, et al. Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex. Biochemistry. 2005;44(48):15834–15841. doi: 10.1021/bi051656d. [DOI] [PubMed] [Google Scholar]
- 22.Cordero-Morales JF, Jogini V, Chakrapani S, Perozo E. A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys J. 2011;100(10):2387–2393. doi: 10.1016/j.bpj.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordero-Morales JF, et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14(11):1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 24.Uysal S, et al. Mechanism of activation gating in the full-length KcsA K+ channel. Proc Natl Acad Sci USA. 2011;108(29):11896–11899. doi: 10.1073/pnas.1105112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockless SW, Zhou M, MacKinnon R. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 2007;5(5):e121. doi: 10.1371/journal.pbio.0050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng WWL, McCoy JG, Thompson AN, Nichols CG, Nimigean CM. Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proc Natl Acad Sci USA. 2011;108(13):5272–5277. doi: 10.1073/pnas.1014186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR. 2007;39(2):107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 28.Jiang YX, et al. The open pore conformation of potassium channels. Nature. 2002;417(6888):523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 29.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 30.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA. 2009;106(16):6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Seebohm G, Sanguinetti MC. Position of aromatic residues in the S6 domain, not inactivation, dictates cisapride sensitivity of HERG and eag potassium channels. Proc Natl Acad Sci USA. 2002;99(19):12461–12466. doi: 10.1073/pnas.192367299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13(4):311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 34.Ader C, et al. Coupling of activation and inactivation gate in a K+-channel: Potassium and ligand sensitivity. EMBO J. 2009;28(18):2825–2834. doi: 10.1038/emboj.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuello LG, Cortes DM, Jogini V, Sompornpisut A, Perozo E. A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 2010;584(6):1126–1132. doi: 10.1016/j.febslet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weingarth M, et al. Structural determinants of specific lipid binding to potassium channels. J Am Chem Soc. 2013;135(10):3983–3988. doi: 10.1021/ja3119114. [DOI] [PubMed] [Google Scholar]
- 37.Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38(4):289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Bax A. SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48(1):13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neal S, Nip AM, Zhang HY, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR. 2003;26(3):215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 40.Bhate MP, et al. Preparation of uniformly isotope labeled KcsA for solid state NMR: Expression, purification, reconstitution into liposomes and functional assay. Protein Expr Purif. 2013;91(2):119–124. doi: 10.1016/j.pep.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95(6):1197–1207. [Google Scholar]
- 42.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson. 2000;142(1):97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 43.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-driven C-13-C-13 recoupling without C-13 rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118(5-6):2325–2341. [Google Scholar]
- 44.Hing AW, Vega S, Schaefer J. Transferred-echo double-resonance NMR. J Magn Reson. 1992;96(1):205–209. [Google Scholar]
- 45.Costa PR, Sun BQ, Griffin RG. Rotational resonance tickling: Accurate internuclear distance measurement in solids. J Am Chem Soc. 1997;119(44):10821–10830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.