Significance
The conserved PCH2 gene in baker’s yeast regulates meiotic double-strand break repair outcomes, helps establish a proper meiotic chromosome structure, and is important for the progression of meiotic recombination. Its mouse homolog is required for fertility. However, the molecular mechanism of how PCH2 regulates these diverse functions is not known. In this study, we show that Pch2 is an AAA+ family ATPase (ATPases associated with diverse cellular activities) that oligomerizes into single hexameric rings. In the presence of ATP, Pch2 binds to and remodels Hop1, an important component of the synaptonemal complex, and displaces it from DNA. Based on these and previous observations, we suggest that Pch2 impacts meiotic chromosome organization by directly regulating Hop1 binding to DNA.
Keywords: meiosis, hexameric ATPase, AAA proteins
Abstract
In budding yeast the pachytene checkpoint 2 (Pch2) protein regulates meiotic chromosome axis structure by maintaining the domain-like organization of the synaptonemal complex proteins homolog pairing 1 (Hop1) and molecular zipper 1 (Zip1). Pch2 has also been shown to modulate meiotic double-strand break repair outcomes to favor recombination between homologs, play an important role in the progression of meiotic recombination, and maintain ribosomal DNA stability. Pch2 homologs are present in fruit flies, worms, and mammals, however the molecular mechanism of Pch2 function is unknown. In this study we provide a unique and detailed biochemical analysis of Pch2. We find that purified Pch2 is an AAA+ (ATPases associated with diverse cellular activities) protein that oligomerizes into single hexameric rings in the presence of nucleotides. In addition, we show Pch2 binds to Hop1, a critical axial component of the synaptonemal complex that establishes interhomolog repair bias, in a nucleotide-dependent fashion. Importantly, we demonstrate that Pch2 displaces Hop1 from large DNA substrates and that both ATP binding and hydrolysis by Pch2 are required for Pch2–Hop1 transactions. Based on these and previous cell biological observations, we suggest that Pch2 impacts meiotic chromosome function by directly regulating Hop1 localization.
During meiosis, diploid cells programmed to become haploid gametes complete a single round of DNA replication followed by reductional and equational divisions. During the reductional division, homologous chromosomes segregate away from each other. In most organisms, the accurate segregation of chromosomes during the reductional division requires at least one cross-over (CO) between each homolog pair. Crossing over between homologous chromosomes provides physical linkages that promote their proper positioning at metaphase I. Defects in crossing over can lead to widespread nondisjunction of homolog pairs, resulting in aneuploid gametes that are typically inviable (1).
In the budding yeast Saccharomyces cerevisiae, CO formation is initiated early in meiotic prophase by the induction of genome-wide double-strand breaks (DSBs) that are subsequently repaired to form COs and non-COs. Several meiotic regulatory mechanisms have been identified in yeast and other organisms that act in coordination to ensure that the repair of DSBs results in at least one CO between each homolog pair. These include (i) interhomolog bias, a process in which DSBs are preferentially repaired using a homolog instead of a sister; (ii) CO interference, a mechanism that promotes the formation of widely spaced COs; and (iii) CO homeostasis, a regulatory process that maintains CO levels at the expense of non-COs under conditions where DSB levels are limiting (1–11).
The PCH2 gene in baker’s yeast has received significant attention because studies have suggested that it participates in at least a subset of the above regulatory mechanisms. Also, homologs of PCH2 have been identified in fruit flies, worms, and mammals (Fig. 1A), and mutational analyses in these organisms have suggested that the PCH2 homologs have both common and unique functions (12–17). PCH2 was first identified as a meiotic checkpoint factor, with subsequent studies showing that it interacts with DNA damage response proteins to promote checkpoint signaling triggered by unprocessed DSBs (18, 19). Recent genetic, physical, and cytological assays showed that pch2Δ mutants are defective in interhomolog repair bias and the timely progression of recombination. pch2 mutants also display elevated CO levels and defects in CO interference, as well as synthetic spore viability defects in mutant backgrounds where meiotic DSB levels have been reduced. Together these data suggest that Pch2 regulates CO outcomes in meiosis (6, 19–23).
Fig. 1.
Purification of Pch2 and mutants. (A) Alignment of Pch2 amino acid sequences from S. cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens. Walker A and Walker B motifs are highlighted (shading). The alignment was generated using T-Coffee (67, 68). (B) Purification of wild-type and mutant Pch2, His6–Hop1, and His6–Dmc1. Dmc1, His6–Dmc1; EQ, GST–Pch2–E399Q; GA, GST–Pch2–G319A; Hop1, His6–Hop1; Pch2, untagged Pch2 after removal of GST tag; WT, wild-type GST–Pch2. See Materials and Methods and SI Materials and Methods for details.
Very little is known about how Pch2 acts at the mechanistic level. However, hints about its function have been obtained from molecular and cytological studies. For example, Ho and Burgess (19) showed that during meiosis Pch2 and the DNA damage response factor telomere maintenance 1 (Tel1) promote activation of the meiotic kinase 1 (Mek1) kinase; this activation, which is hypothesized to occur through Hop1-dependent autophosphorylation of Mek1, is required to achieve interhomolog bias during meiotic recombination (6, 8, 19, 24, 25). Pch2 was shown to localize to individual chromosomes as well as to the nucleolus (18, 21). More specifically, Pch2 forms foci on chromosomes at the pachytene stage of meiosis where chromosomes are fully paired within the context of the synaptonemal complex (SC), a tripartite structure that is thought to ensure an accurate reductional segregation of homologous chromosomes by promoting and maintaining homolog pairing, and by regulating DSB repair and CO placement (reviewed in ref. 1). Pch2 foci formation is dependent on the presence of the SC central element component Zip1, and colocalizes extensively with Zip3, which marks future CO sites (18, 21).
The SC is a tripartite structure consisting of a central element and two lateral elements that contain the homolog axes. SC formation occurs in temporal and spatial coordination with genetic recombination (26). In budding yeast the Zip1 protein forms a transverse filament that is part of the central element, and Hop1 associates with the chromosome axes. Hop1 is a DNA-binding protein that contains a zinc finger domain, a Hop1p, Rev7p and MAD2 (HORMA) domain associated with oligomerization, and [S/T]Q motifs phosphorylated by the mitosis entry checkpoint 1 (Mec1) and Tel1 kinases in response to meiotic DSBs (8, 24). During the leptotene stage of meiosis, Hop1 is loaded onto chromosomes in a discontinuous manner and few Zip1 foci are observed. At zygotene, Hop1 and Zip1 display a domain-like organization that becomes even more apparent in pachytene when the SC is fully formed (20). Based on these and physical analyses of recombination intermediates in meiosis, Börner et al. (20) suggested that Zip1 is loaded in response to a preexisting Hop1 pattern, and that the Hop1/Zip1 pattern may be dictated by CO placement. Curiously, Hop1 and Zip1 largely colocalize in pch2Δ strains in pachytene (18, 20, 21). Börner et al. (20) also suggested that Pch2 acts as a stringency factor to prevent aberrant loading of Zip1 and additional loading of Hop1. Such a function is likely to be critical to establish interhomolog repair bias, and could thus explain the meiotic defects seen in pch2Δ mutants.
Pch2 has a conserved AAA+ (the abbreviation for “ATPases associated with diverse cellular activities”) module that contains canonical Walker A and B motifs (27). Pch2 does not contain other known functional domains. AAA+ proteins are known to couple ATP binding and/or ATP hydrolysis to conformational changes on macromolecular substrates (28). AAA+ proteins are implicated in a wide range of cellular processes, including DNA replication, membrane fusion, protein degradation, and the regulation of gene expression (28, 29). We purified Pch2 and found that it displays an intrinsic ATPase activity. Both ATP binding and hydrolysis are critical for its function in vivo, and a mutation in the Pch2 Walker B domain confers a dominant negative phenotype. We show, using electron microscopy and size-exclusion chromatography (SEC), that in the presence of nucleotide, Pch2 oligomerizes into single hexameric rings with a central pore. Pch2 binds Hop1 in vitro, and displaces it from DNA. Based on these observations we propose that Pch2 in an ATP-bound state binds to Hop1, inducing a conformational change in Hop1 upon ATP hydrolysis. These data support a model in which Pch2 remodels Hop1 to restrict its localization to specific chromosomal regions, setting up a chromosomal organization that promotes interhomolog repair at CO designation sites.
Results
Pch2 Forms a Sixfold Symmetrical Single-Ring Oligomer.
A hallmark of many AAA+ proteins is that they assemble into hexameric rings with a central pore, and that this structure is critical for function. For example, RuvA/B proteins, which act in genetic recombination in bacteria, form a ring complex and thread DNA through a central pore (30, 31). Pch2, which contains domains homologous to AAA+ proteins, has been extensively studied genetically and cytologically, but little is known about its biochemical activities. We first performed a yeast two-hybrid analysis to test whether Pch2 self-interacts. In this experiment, one copy of PCH2 was fused to a LexA DNA-binding domain and a second copy of the gene was fused to a GAL4-activation domain. High levels of β-galactosidase expression (450 units) were detected only when both fusion proteins were expressed in the same cell, suggesting that Pch2 self-interacts in vivo.
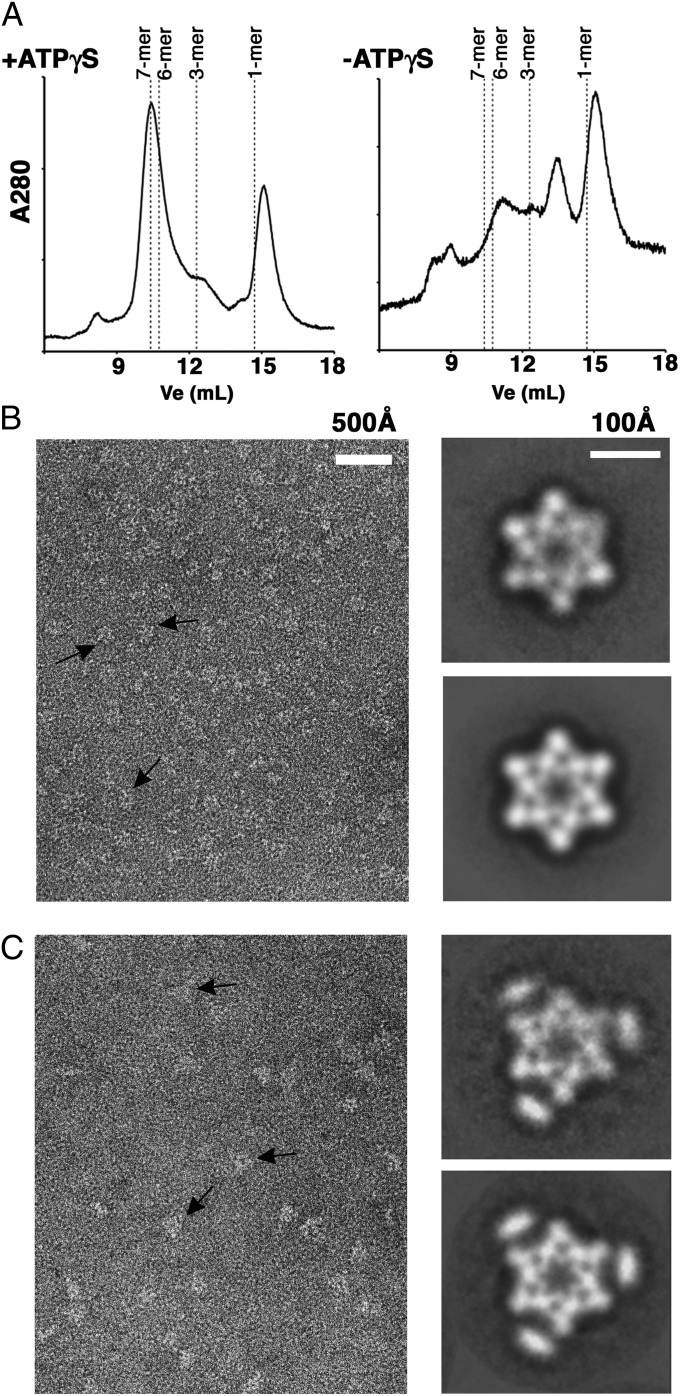
We then expressed and purified Pch2 as a fusion with a thrombin-cleavable N-terminal GST tag. This tag can be cleaved to yield the native protein (Materials and Methods and Fig. 1B). Untagged Pch2 was then analyzed by SEC (Fig. 2A). Pch2 displayed a major peak at the 14.9-mL elution volume, which corresponds to the monomer, and minor peaks at other volumes. However, in the presence of adenosine 5′-[γ-thio]triphosphate (ATPγS), the elution profile changed significantly; a minor peak was observed at the 14.9-mL elution volume, with a predominant peak seen at 10.4 mL (Fig. 2A, Left). These results suggest that ATPγS is important for Pch2 to form a higher order oligomer, as was seen for many AAA+ proteins (32).
Fig. 2.
Pch2 and GST–Pch2 forms hexameric rings in the presence of nucleotides. (A) Elution profiles of the Pch2 protein from a Superdex 200 size-exclusion column in the absence and presence of 2 mM ATPγS. The expected elution volumes (Ve) for different oligomeric forms of Pch2 are indicated by dashed lines. (B, Left) Representative negative staining electron micrograph of Pch2 in the presence of ATPγS. Black arrows indicate ring-shaped particles representing top views of Pch2 hexameric rings. (B, Upper and Lower Right) The projection structure of Pch2 hexameric rings in the presence of ATPγS. Upper Right shows the 2D averages and Lower Right the same average is displayed after sixfold rotational symmetry has been imposed. (C, Left) Representative negative-staining electron micrograph of GST–Pch2 in the presence of ATPγS. Black arrows indicate ring-shaped particles representing top views of GST–Pch2 hexameric rings. (C, Upper and Lower Right) The projection structure of GST–Pch2 hexameric rings in the presence of ATPγS. Upper Right shows the 2D averages and Lower Right the same average is displayed after sixfold rotational symmetry has been imposed. Densities at the vertices of the triangle represent GST dimers.
To visualize the Pch2 oligomer, Pch2 was incubated in the absence and presence of ADP, ATP, or ATPγS, and was imaged by negative staining electron microscopy (Fig. 2B and Fig. S1C). Micrographs of Pch2 incubated in the presence of nucleotides contained ring-shaped particles ∼150 Å in diameter with a stain-penetrated central pore. No ring-like particles were observed in samples containing Pch2 in the absence of nucleotide. Using comparable amounts of protein in the assembly reactions, the concentration of the ring-shaped particles was highest in samples containing ATPγS and lowest in samples containing ADP. We concluded that the higher concentration of ring-shaped particles observed in the presence of ATPγS was due to a more efficient assembly of Pch2 into ring-shaped particles.
Groups of between 200–250 ring-shaped particles from micrographs of Pch2 incubated with ADP, ATP, or ATPγS were selected and analyzed using rotational symmetry algorithms (33). In all three conditions, sixfold symmetry was detected at a radius of 73 Å, where the outer edge of the ring-shaped particle is located. Table 1 lists results of the Student t test and spectral ratio product (33) for Pch2. No other order of symmetry was found to be statistically significant in any of the samples.
Table 1.
Rotational symmetry analysis of Pch2 and GST–Pch2
| Nucleotide | No. of particles | Symmetry detected | Radius, Å | t test and significance level | Spectral ratio product |
| Pch2 | |||||
| ADP | 250 | 6 | 73 | P < 0.000001 | 9.93 × 1034 |
| ATP | 238 | 6 | 73 | P < 0.000001 | 2.51 × 1051 |
| ATPγS | 219 | 6 | 73 | P < 0.000001 | 2.15 × 1045 |
| GST–Pch2 | |||||
| ATPγS | 240 | 6 | 73 | P < 0.000001 | 7.99 × 1041 |
| 3 | 95 | P < 0.000001 | 1.03 × 1087 |
AAA+ proteins typically assemble into single hexameric rings (34), but examples exist where such proteins form double-hexameric ring structures (35–37). The SEC analysis suggested that Pch2 assembles into a single hexameric ring. To confirm this, Pch2 was incubated in the absence and presence of ADP, ATP, or ATPγS and the oligomeric state was assessed by Blue Native (BN) PAGE. In the presence of ADP, ATP, or ATPγS, Pch2 appeared primarily as a single band with mobility similar to the 480-kDa molecular-mass marker, and as a lower smeared band with mobility slightly slower than the 66-kDa marker. The lower molecular-weight band was the prominent band observed when the protein was incubated in the absence of nucleotide (Fig. S1A). Considering that the theoretical molecular masses of the Pch2 monomer and hexamer are 64.1 and 385 kDa, respectively, we inferred that the high molecular band in the gel represented the hexameric ring form of Pch2 and the lower band the monomeric form. The small differences between the observed mobility of Pch2 in BN PAGE and the expected mobility considering its molecular mass can be reconciled because migration in native gels depends both on the molecular mass of the protein and the shape of the complex. Importantly, no additional higher molecular bands that could represent a dodecameric or higher-order complex of Pch2 were observed. As a control, we loaded a mixture of RuvB-like (Rvb)1 and Rvb2 AAA+ proteins under conditions where they oligomerize both as single (∼300 kDa) and double (∼600 kDa) hexameric ring structures (36). These oligomers have theoretical molecular masses similar to the expected mass of Pch2 hexamers and dodecamers. In contrast to the Pch2 samples, the Rvb protein mixture produced two prominent bands representing the hexameric and the dodecameric oligomeric forms of the Rvb1–Rvb2 complex (Fig. S1A). Together, these results suggest that Pch2 assembles as a single hexameric ring.
Projection Structure of the Pch2 Hexameric Rings in Multiple Nucleotide States.
Previous work showed that some AAA+ superfamily members undergo nucleotide-dependent conformational changes (38, 39). We tested whether the Pch2 hexameric ring structure changes in the presence of different nucleotides. Top-view particles from electron micrographs of Pch2 samples in the presence of ADP, ATP, or ATPγS were selected, extracted, and aligned using correlation averaging to produce a projection structure for each nucleotide state. As shown in Fig. S1C, the projection structures were similar in the presence of ATP and ATPγS. The projection structure in the ADP state did not differ significantly from the ATP and ATPγS averages, except the central pore was ∼43 Å in diameter. This difference may not necessarily be intrinsic to the structure and may be caused by variability in stain penetration between samples. Thus, we concluded that the Pch2 hexameric ring did not undergo a substantial conformational change in the presence of different nucleotides.
Influence of the GST Tag on the Oligomeric State and Functionality of Pch2.
Assembly of the hexameric ring structure is typically required for AAA+ proteins to perform their function (34). Because affinity tags may affect the oligomeric state of AAA+ proteins (36), we analyzed the structure of the GST–Pch2 fusion protein. GST–Pch2 protein was incubated in the presence of ATPγS and deposited on electron microscopy grids and visualized using negative staining. Electron micrographs showed primarily triangular-shaped particles (Fig. 2C). Rotational symmetry analysis detected two statistically significant orders of symmetry: threefold at a radius of ∼95 Å and sixfold at radius of ∼73 Å (Table 1). This result is consistent with the projection structure obtained from these particles. The 2D average of the GST–Pch2 oligomer showed a six-vertex regular star polygon structure similar to that observed for Pch2; however, this average featured three globular densities at the periphery positioned around a central threefold symmetry axis, conferring the overall shape of an equilateral triangle (Fig. 2C). The GST protein forms a dimer in solution (reviewed in ref. 40), thus we suggest that the three globular densities at the periphery of the average are GST dimers.
The GST–Pch2 fusion protein was also incubated in the presence of ADP, ATP, or ATPγS and resolved by BN PAGE (Fig. S1B). All samples produced a band with similar mobility to the 720-kDa molecular-mass marker and an additional less prominent band of slightly faster mobility than the 480-kDa molecular-mass marker. Considering that the GST tag adds ∼26 kDa to the Pch2 protein, the high molecular and low molecular weight bands likely represent hexamers (theoretical molecular weight ∼544 kDa) and dimers (theoretical molecular weight ∼181 kDa) of GST–Pch2, respectively. We did not observe protein bands suggestive of a GST–Pch2 dodecamer. Interestingly, the presence of the GST tag removed the dependency of the nucleotide for oligomer formation. Together, these data suggest that the presence of a GST tag fused to the N-terminal end of Pch2 did not prevent the protein from assembling into single hexameric rings and did not induce stacking of the hexameric rings into dodecamers or higher-order oligomers. More importantly, the orientation of the Pch2 monomers in the hexamer can be modeled because the N-terminal GST tag in the GST–Pch2 fusion appears to be located on the exterior of the hexameric ring.
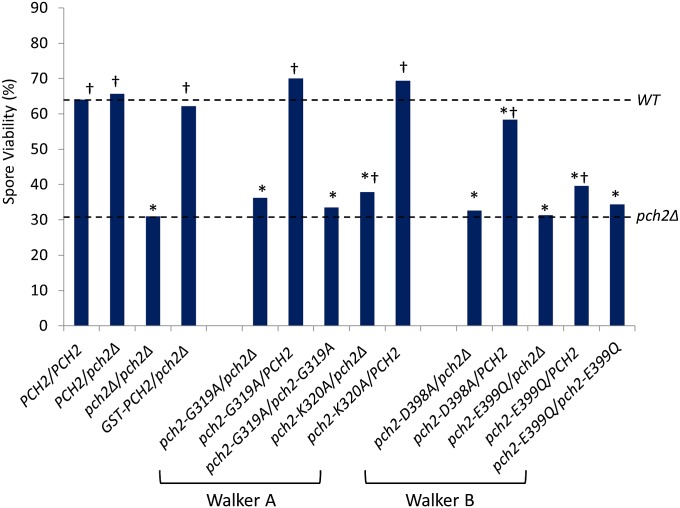
We performed a complementation analysis to assess GST–Pch2 function in vivo. The pch2Δ mutation does not confer a meiotic spore viability defect in the SK1 strain background (22), but confers a synthetic spore viability defect in chromosome segregation in meiosis 4 deletion (csm4Δ) strains. Strains bearing null mutations in CSM4 show ∼64% spore viability, but pch2Δ csm4Δ strains display much lower spore viability (∼31%) (41–43). We believe that this synthetic phenotype is due to the pch2Δ mutation suppressing delays in the meiotic prophase that are required to overcome defects in chromosome motion and recombination progression in csm4Δ strains. As shown in Fig. 3 and Table 2, GST–PCH2 csm4Δ strains displayed spore viability that was indistinguishable from csm4Δ, suggesting that the GST–Pch2 fusion is functional in vivo. The data are also consistent with GST–Pch2 forming hexameric rings. Importantly, these data indicate that GST–Pch2 is appropriate to use in the substrate interaction and ATPase assays performed in sections Pch2 is an ATPase, Pch2 Binds to Hop1, and Pch2 Can Displace Hop1–DNA Complexes.
Fig. 3.
Spore viability of pch2 mutants in the csm4Δ/csm4Δ background. Dashed lines indicate the spore viabilities of csm4Δ/csm4Δ PCH2/PCH2 (64%) and csm4Δ/csm4Δ pch2Δ/pch2Δ (31%) (42, 43). *P < 0.05 compared with csm4Δ/csm4Δ PCH2/PCH2; †P < 0.05 compared with csm4Δ/csm4Δ pch2Δ/pch2Δ. See Table 2 for details.
Table 2.
Spore viability of csm4Δ pch2 strains
| PCH2 allele in csm4Δ/csm4Δ | % spore viability (n) | Significance compared with PCH2/PCH2 | Significance compared with pch2Δ/pch2Δ |
| PCH2/PCH2 | 64.0 (1,164)* | NA | ND |
| PCH2/pch2Δ | 65.7 (78) | − | ND |
| pch2Δ/pch2Δ | 31.0 (200) † | ++ | NA |
| GST–PCH2/pch2Δ | 62.2 (80) | − | ++ |
| pch2–G319A/pch2Δ | 36.3 (82) | ++ | − |
| pch2–G319A/PCH2 | 70.0 (40) | − | ++ |
| pch2–K320A/pch2Δ | 37.9 (120) | ++ | + |
| pch2–K320A/PCH2 | 69.4 (40) | − | ++ |
| pch2–D398A/pch2Δ | 32.7 (78) | ++ | − |
| pch2–D398A/PCH2 | 58.3 (120) | + | ++ |
| pch2–E399Q/pch2Δ | 31.3 (79) | ++ | − |
| pch2–E399Q/PCH2 | 39.6 (120) | ++ | ++ |
| pch2–T428A/pch2Δ | 35.3 (39) | ++ | − |
| pch2–T428E/pch2Δ | 28.3 (38) | ++ | − |
| pch2–G319A/pch2–G319A | 33.5 (100) | ++ | − |
| pch2–E399Q/pch2–E399Q | 34.4 (101) | ++ | − |
Pch2 Is an ATPase.
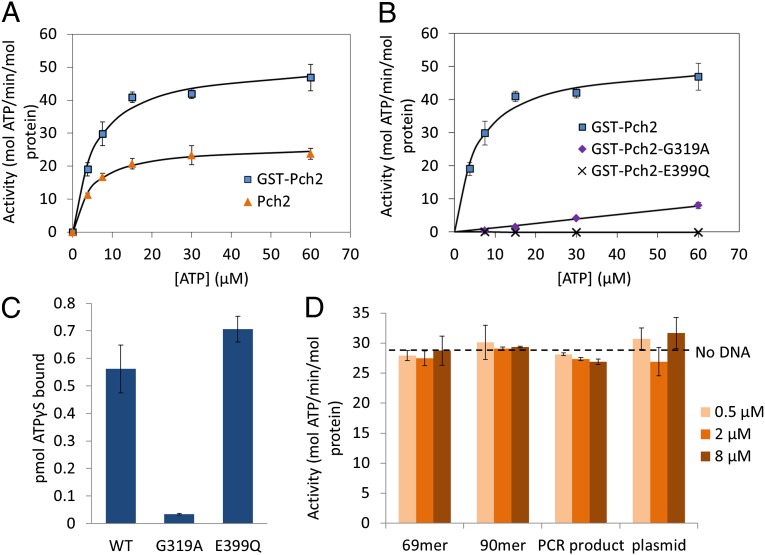
We performed a steady-state kinetic analysis of the ATPase activity of Pch2 and GST–Pch2 proteins. As shown in Fig. 4A and Table 3, Pch2 displayed a Km = 4.5 µM and kcat = 26.4 min−1. Surprisingly, GST–Pch2 displayed an even higher kcat (51.8 min−1) compared with untagged Pch2 (Fig. 4A and Table 3), but the Km values of the two proteins were similar (4.5 µM for Pch2 vs. 5.6 µM for GST–Pch2), indicating that the GST tag stimulates the hydrolysis activity of Pch2, but not its affinity to ATP. Because Pch2 forms hexamers and GST can form dimers, it is likely that dimerization of GST facilitates the formation of Pch2 into hexameric rings (see electron microscopy analysis in section Influence of the GST Tag in the Oligomeric State and Functionality of Pch2), thus stimulating its ATPase activity. We showed by thin layer chromatography (TLC) that GST–Pch2 hydrolyzes ATP into ADP (Fig. S2).
Fig. 4.
ATPase and ATPγS binding activity of Pch2 and mutants. (A) ATPase activity of GST–Pch2 and Pch2. Proteins were present at 6 nM and error bars represent SDs from four experiments. The Km and Kcat values are listed in Table 3. (B) ATPase activity of GST–Pch2, GST–Pch2–G319A, and GST–Pch2–E399Q. GST–Pch2 was present at 6 nM, and GST–Pch2–G319A and GST–Pch2–E399Q were present at 40 nM. Error bars represent SDs obtained from two to four experiments. (C) ATPγS binding activity of Pch2 and mutants. E399Q, GST–Pch2–E399Q; G319A, GST–Pch2–G319A; WT, GST–Pch2. All reactions contained 100 nM wild-type or mutant Pch2 and 12 µM 35S-labeled ATPγS. Error bars represent SDs from two experiments. See SI Materials and Methods for details. (D) ATPase activity of Pch2 (12 nM) in the presence of 0.5, 2, or 8 µM (concentration in nucleotides) of the indicated DNA substrates. Error bars represent SDs from two experiments.
Table 3.
ATPase activity of Pch2
| Protein | Km, μM | kcat, min−1 | kcat/Km, μM−1⋅min−1 |
| GST–Pch2 | 5.6 ± 1.0 | 51.8 ± 2.6 | 9.4 ± 1.6 |
| Pch2 | 4.5 ± 0.4 | 26.4 ± 2.5 | 5.9 ± 0.2 |
Norit A absorption assays were performed as described in SI Materials and Methods. The average and SD of four experiments are shown.
We then tested the effect of DNA substrates on the ATPase activity of Pch2 (Fig. 4D) because Pch2 localizes to chromosomes in meiosis, and DNA-interacting ATPases such as disrupted meiotic cDNA 1 (Dmc1) display altered ATPase activities in the presence of DNA (6, 18–22, 44). We also pursued this because Pch2 binds weakly to single- and double-strand DNA (ssDNA and dsDNA, respectively) as well as other DNA structures (Fig. S3). The weak DNA-binding activity was specific to Pch2 because the GST protein alone displayed DNA binding at background levels. As shown in Fig. 4D, the ATPase activity of Pch2 was not stimulated by various DNA substrates. Based on these and previous observations (18) we hypothesize that Pch2 functions on meiotic chromosomes require posttranslational modifications and/or interactions with other factors.
To confirm that the Pch2 and GST–Pch2 ATPase activities were intrinsic and not caused by contaminating proteins, we analyzed GST-Pch2 proteins containing mutations in the ATPase domain. GST–Pch2–G319A, containing a glycine-to-alanine mutation in the Walker A motif, and GST–Pch2–E399Q, containing a glutamic acid-to-glutamine mutation in the Walker B motif have severely reduced (GST–Pch2–G319A) or no apparent (GST–Pch2–E399Q) ATPase activities (Figs. 1A and 4B). These data indicate that the ATPase activity described above was specific to Pch2. We note that there is a residual ATPase activity for GST–Pch2–G319A; however, based on complementation tests (see section Both ATP Binding and ATP Hydrolysis Are Important for Pch2 Functions, Fig. 3 and Table 2), this reduced activity does not appear to support the in vivo function of Pch2.
For many Walker A/B ATPases, mutations in the Walker A motif disrupt ATP binding, whereas Walker B mutations disrupt ATP hydrolysis but not binding (28). To test if this was the case for Pch2, we measured ATPγS binding by both wild-type and mutant (Walker A and Walker B) Pch2 proteins using a filter-binding assay. GST–Pch2 and GST–Pch2–E399Q showed similar binding to ATPγS; however, GST–Pch2–G319A was strongly defective (Fig. 4C). These results, in concert with the ATPase assays, indicate that GST–Pch2–E399Q can bind but not hydrolyze ATP, and thus may be in a “locked” ATP bound state.
Both ATP Binding and ATP Hydrolysis Are Important for Pch2 Functions.
We analyzed the phenotype of ATPase pch2 mutants in a spore viability assay. In addition to testing the two mutants described above (G319A and E399Q), we generated two more mutants, one with a lysine to alanine mutation in the Walker A motif, pch2–K320A, and the other with aspartic acid to alanine mutation in the Walker B motif, pch2–D398A. We assessed the functionality of these four pch2 alleles by testing complementation of pch2Δ in a csm4Δ background (for plasmids and strains used, please see Tables S1 and S2). As mentioned in section Influence of the GST Tag in the Oligomeric State and Functionality of Pch2, full complementation restored spore viability to ∼64%, and a null allele conferred a spore viability of ∼31%. pch2–G319A/pch2Δ, pch2–K320A/pch2Δ, pch2–D398A/pch2Δ, and pch2–E399Q/pch2Δ displayed 36%, 38%, 33%, and 31% spore viability, respectively (Fig. 3 and Table 2), indicating that they are loss-of-function mutants (P < 0.05 for all cases compared with PCH2/PCH2). All four alleles except pch2–K320A also conferred spore viabilities indistinguishable from pch2Δ/pch2Δ, indicating they are null alleles, whereas pch2–K320A conferred an intermediate phenotype (P < 0.05 compared with either PCH2 or pch2Δ; Fig. 3 and Table 2), suggesting partial function. Because Pch2–G319A displayed a residual ATPase activity, we tested whether pch2–G319A was partially functional by testing the complementation of homozygous pch2–G319A/pch2–G319A, and as a control, we also tested pch2–E399Q/pch2–E399Q. In the csm4Δ background, pch2–G319A/pch2–G319A and pch2–E399Q/pch2–E399Q displayed 33.5% and 34.4% spore viability, respectively, both indistinguishable from pch2Δ/pch2Δ (31%, P > 0.2 in both cases), suggesting complete loss of function for pch2–G319A and pch2–E399Q. In conclusion, both Walker A and Walker B motif mutants are disrupted for PCH2 function, indicating that both ATP hydrolysis and ATP binding are critical for Pch2 function.
pch2 Walker B Motif Mutants Display a Dominant Negative Phenotype.
Walker B mutants often display dominant negative phenotypes. Such phenotypes can be explained by the mutant protein being in an ATP-bound state that prevents or locks in an interaction with a substrate (45). Because Pch2–E399Q can bind ATP but is defective in ATP hydrolysis, we tested whether pch2–E399Q confers a dominant negative phenotype. Heterozygous diploid strains in the csm4Δ background were generated with one copy of wild-type PCH2 and one copy of mutant alleles of PCH2. pch2Δ/PCH2 showed 66% spore viability (Fig. 3 and Table 2), indicating that pch2Δ is recessive. pch2–G319A/PCH2 and pch2–K320A/PCH2 displayed 70% and 69% spore viability, respectively, indicating that the Walker A mutations were recessive. In contrast, pch2–D398A/PCH2 and pch2–E399Q/PCH2 displayed 58% and 40% spore viability (P < 0.05 compared with wild type), indicating that both Walker B mutations conferred a dominant negative phenotype. One explanation for this phenotype is that the Walker B mutant Pch2 proteins bind substrate continuously and competitively interfere with wild-type Pch2 or form a mixed hexamer that is not functional.
Pch2 Binds to Hop1.
Genetic and molecular studies suggested that Pch2 and its homologs induce conformational changes in specific targets (17, 20, 46). We investigated potential substrates of Pch2 by testing interactions with Dmc1 and Hop1. Dmc1 is a meiotic strand-exchange protein that mediates ssDNA invasion into homologous duplex sequences (reviewed in ref. 2). Because Pch2 acts in interhomolog bias, we speculated that a Pch2 interaction with Dmc1 is important for Dmc1’s role in mediating recombination between homologs. The axial element of the SC, Hop1, was chosen for analysis because (i) Hop1 distribution on meiotic chromosomes was shown to be altered in pch2Δ mutants (18, 20, 21); (ii) Pch2 acts to exclude Hop1 from the nucleolus (18); (iii) HORMA domain containing 1 (HORMAD1), a mammalian protein related to yeast Hop1, was shown to be depleted from mouse meiotic chromosome spreads in a process dependent on thyroid hormone receptor interactor 13 (Trip13), the mouse homolog of Pch2 (17); and (iv) Hop1 levels were elevated in pch2Δ mutants as detected in Western blots (19).
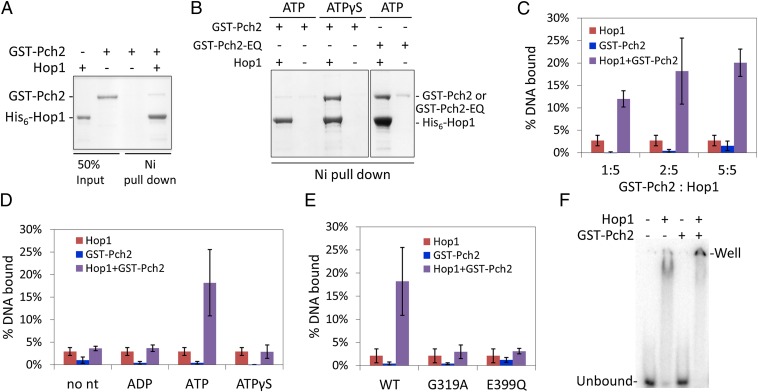
To test whether Dmc1 or Hop1 interacts with Pch2, we purified full-length Dmc1 and Hop1 with His6 tags. We carried out in vitro-binding assays using purified proteins. No interactions between Pch2 and Dmc1 were detected (Fig. S4). However, as shown in Fig. 5 A and B, GST–Pch2 strongly bound Hop1 in the presence of ATPγS, whereas weak binding was seen in the absence of nucleotide or in the presence of ATP. We reasoned that Hop1–Pch2 interactions were transient in the presence of ATP (and would thus be difficult to detect in vivo) but could be stabilized in the presence of ATPγS, where an ATP hydrolysis cycle cannot be completed, and the interaction is in a locked state. If this is correct, then an ATP hydrolysis mutant of Pch2 should also lock the interaction in the presence of ATP. As shown in Fig. 5B, Pch2–E399Q strongly interacted with Hop1 in the presence of ATP. These data are also consistent with the dominant negative phenotype conferred by the pch2–E399Q mutation. It is important to note that in all of our experiments Hop1 remained intact after incubation with Pch2, indicating that protease activity is not associated with Pch2 as has been found for some AAA+ proteins (e.g., ref. 47).
Fig. 5.
Pch2 interacts with Hop1. (A and B) In vitro-binding assays performed with purified Hop1 and GST–Pch2 or GST–Pch2–E399Q in the absence (A) or presence (B) of the indicated nucleotide (200 µM). (C–E). Hop1 (41.7 nM) binding to a 69-bp 32P-dsDNA substrate (1.15 µM, concentration in nucleotides) was determined in filter-binding assays. In C–E, error bars represent SDs from at least three repetitions. (C) Hop1 DNA-binding activity in the presence of different amounts of GST–Pch2 (8.3, 16.7, and 41.7 nM) and 200 µM ATP. (D) Hop1 DNA-binding activity in the presence of GST–Pch2 (16.7 nM) and the indicated nucleotides. (E) Hop1 DNA-binding activity in the presence of indicated GST–Pch2 mutant proteins (16.7 nM) and 200 µM ATP. (F) Acrylamide gel EMSA. Hop1 (110 nM) was incubated with 69-bp 32P-dsDNA (1.38 µM, concentration in nucleotides) in the presence of ATP and in the presence or absence of 25 nM GST–Pch2, and the amount of DNA bound was analyzed by EMSA.
Pch2 Can Displace Hop1–DNA Complexes.
To investigate the functional consequences of a Pch2–Hop1 interaction, we tested whether Pch2 alters the biochemical activities of Hop1. Hop1 cooperatively binds to dsDNA and has been shown to synapse noncontiguous and contiguous dsDNA molecules into complexes that resemble higher-order nucleoprotein structures (48–50). To determine if Pch2 affects Hop1 activity we examined Hop1 binding to 69-bp duplex DNA in the presence of GST–Pch2 or untagged Pch2 in filter-binding assays. Hop1 DNA binding appeared to increase in the presence of ATP and GST–Pch2 or Pch2 (Fig. 5C and Fig. S5A); however this stimulation was not seen if ADP or ATPγS were included instead of ATP. Both GST–Pch2–G319A and GST–Pch2–E399Q failed to stimulate Hop1 DNA-binding activity (Fig. 5E), suggesting that ATP hydrolysis by Pch2 was critical for the stimulation. Pch2 also stimulated Hop1 binding to a 40-bp dsDNA substrate predicted to contain a single Hop1 binding site (49), indicating that multiple Hop1 binding sites were not required for stimulation (Fig. S5B).
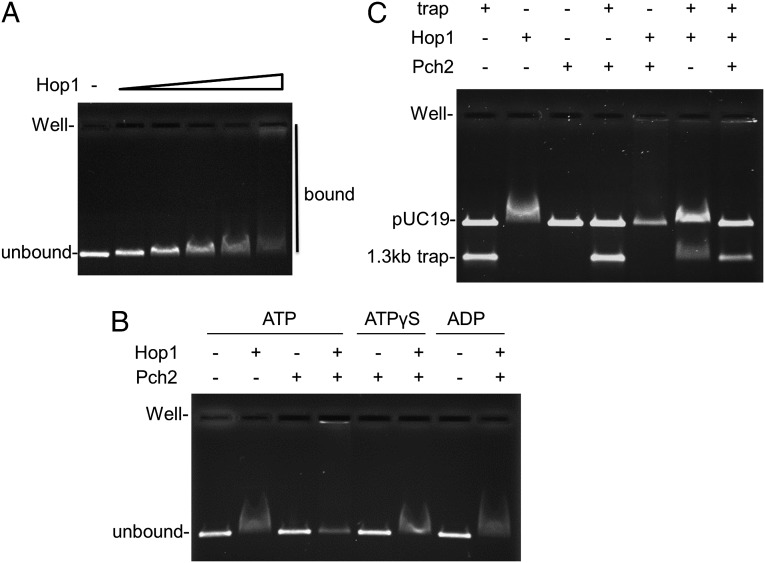
A drawback of filter-binding assays is that it is not possible to characterize the nature of the protein-DNA complexes because they are irreversibly trapped on a filter. To overcome this hurdle we performed gel electrophoretic mobility-shift assays (EMSAs) to examine Hop1 binding to a 69-bp 32P–dsDNA substrate. As shown in Fig. 5F, Hop1 shifted this substrate to a specific position in the gel; however, when Pch2 and ATP were added to the Hop1-binding reaction, the DNA shifted to the well of the gel. We found this result interesting because Hop1 binds cooperatively to large DNA substrates and can form protein–DNA aggregates (49). These observations and cytological observations suggesting that Hop1 forms domains on meiotic chromosomes (18, 20) encouraged us to test whether Pch2 can affect binding of Hop1 to a large DNA substrate. We examined this by performing an agarose gel EMSA using a 2.7-kb linear pUC19 DNA substrate (Fig. 6 and Supporting Information). As shown in Fig. 6A, pUC19 mobility was retarded by Hop1 in a concentration-dependent manner. At 200 nM Hop1, a lower mobility gel shift was observed. At higher concentrations (240 nM and above) the shift became much less discrete and extended to the well of the gel (Fig. 6A).
Fig. 6.
Pch2-mediated dissociation of Hop1 from DNA. (A) Hop1 titration. Reactions (25 μL) in Buffer A [20 mM Tris (pH 7.5), 0.01 mM EDTA, 2 mM MgCl2, 40 µg/mL BSA, 0.1 mM DTT, 75 mM NaCl, 9% glycerol], 60 ng BamHI-digested pUC19 (2.7 kb), and 0, 80, 120, 160, 200, and 240 nM Hop1 were incubated at 30 °C for 20 min, after which they were loaded onto an agarose gel (0.7%) and analyzed as described in SI Materials and Methods. (B) Reactions (25 μL) in Buffer A with 60 ng BamHI-digested pUC19 (2.7 kb) and 200 nM Hop1 were incubated at 30 °C for 10 min, after which 200 nM GST–Pch2 and 300 µM ATP or 50 µM ATPγS or 300 µM ADP were added as indicated. Reactions were then continued for 5 min at 30 °C, and loaded onto a 0.7%-agarose gel. (C) Reactions (25 μL) in Buffer A with 60 ng BamHI-digested pUC19 (2.7 kb) and 200 nM Hop1 were incubated at 30 °C for 10 min, after which 200 nM GST–Pch2 and 300 µM ATP were added as indicated in the presence of 40 ng 1.3-kb trap DNA. Reactions were then continued for 10 min at 30 °C, after which they were loaded onto a 0.7%-agarose gel and analyzed as before.
We then tested whether the addition of GST–Pch2 would affect binding of Hop1 (200 nM) to DNA. As shown in Fig. 6B, the addition of 200 nM GST–Pch2 in the presence of 300 µM ATP resulted in a significant change in the gel shift pattern. Approximately 20% of pUC19 DNA was present at the unbound position, with another portion shifted to the well. This effect is dependent on ATP hydrolysis because the addition of 200 nM GST–Pch2 in the presence or absence of ATPγS (50 µM) or ADP (300 µM) did not alter the Hop1-mediated gel shift (Fig. 6B).
These observations suggest that Pch2 dissociates a pool of Hop1 from large DNA substrates in steps that require ATP hydrolysis. To test this we performed an order of addition experiment in which pUC19 DNA was preincubated with Hop1, after which Pch2 was added in the presence of 300 µM ATP and a 1.3-kb DNA trap. If Pch2 can displace Hop1 from DNA then we would predict that the presence of the DNA trap at the time of Pch2 and ATP addition would increase the amount of unbound pUC19 DNA. As shown in Fig. 6C, the amount of unbound pUC19 10 min after the addition of Pch2, ATP, and trap DNA is significantly higher than the amount seen when only Pch2 and ATP were added. These observations are consistent with Pch2 displacing Hop1 from DNA.
In experiments involving both small (69-bp) and large (2.7-kb) substrates the addition of Pch2 to DNA-binding reactions containing Hop1 and ATP resulted in altered Hop1 DNA binding properties. Such an effect could be due to Pch2 remodeling Hop1 either before or after binding to DNA. We tested the former possibility using a protein cross-linking assay. Previous studies showed that Hop1 forms oligomers in the absence of DNA that can be detected by glutaraldehyde cross-linking and SDS/PAGE electrophoresis, and this property is thought to be important for its cooperative binding to long DNA substrates (49). As shown in Fig. S6, oligomerization of Hop1 in the absence of DNA was not altered in the presence of GST–Pch2. This finding, in conjunction with the gel shift assays described above, suggests that Pch2 remodels Hop1 bound to DNA.
Discussion
In this study we show that Pch2 is an ATPase that assembles in a nucleotide-dependent fashion into a single hexameric ring with a central pore. The diameter of the pore is ∼35 Å, which is comparable to the diameter seen in other AAA oligomeric rings, such as NSF (18–30 Å; refs. 51 and 52). Our study purifies and characterizes a meiosis-specific AAA+ protein that acts as a remodeler of a specific substrate.
Our biochemical studies showed that Pch2 is an active ATPase that displaces Hop1, an axial component of the SC, from DNA. PCH2 homologs are found in worm, fly, mouse, and human and mutations in the corresponding genes confer similar as well as distinct meiotic phenotypes. In mice, the Hop1-like HORMAD1 and HORMAD2 proteins are depleted from chromosome axes by the actions of the PCH2 ortholog Trip13 (17), and the C-terminal domain of Pch2, which likely encodes a substrate-binding site, is highly conserved from yeast to human. Thus, the interaction between Pch2 and Hop1 described here is likely to be conserved.
In pachytene nuclei, Zip1, a central element protein, and Hop1, an axial element component, display an alternating localization pattern along chromosomes that is altered in pch2 mutants. Börner et al. (20) reported that in the early leptotene, before Zip1 loading can be detected, Hop1 is discontinuously loaded onto chromosomes. This pattern is seen in both wild-type and pch2Δ cells, suggesting that the overlapping Zip1/Hop1 pattern observed in pch2Δ is initiated after leptotene. However, the overlapping Zip1/Hop1 pattern seen in pch2Δ becomes prominent during zygotene and persists through pachytene, when Zip1 loading is coupled with SC formation. Based on these observations, Börner et al. (20) propose that the Zip1/Hop1 pattern seen in wild-type meiosis occurs through a differential loading of Zip1 onto regions lacking Hop1. Pch2 is thought to act in this process by preventing aberrant loading of Zip1 and additional loading of Hop1 and/or removing promiscuous loading of Hop1 from the chromosome axis (18, 20, 21). The resulting alternating Zip1/Hop1 localization pattern on chromosomes is thought to be dictated by sites that become COs (18, 20, 21). Similarly, in C. elegans, HTP-1/-2 (Hop1 homologs) and SYP-1 (an SC transverse element protein) are depleted from reciprocal domains on the chromosome, and the boundaries between these domains mark future CO sites (53).
Recently Voelkel-Meiman et al. (54) examined SC dynamics in yeast meiosis by inducing Zip1–GFP expression in cells that have already assembled SC. Under these conditions they observed a nonuniform pattern of Zip1–GFP localization on the SC and concluded that the SC grows continuously in meiotic prophase. Curiously the Zip1–GFP pattern appeared similar in wild type and pch2Δ. Although the above observations may not reflect how Zip1 localization is established, it is consistent with the idea that Pch2 maintains domain-like organization not by interacting with Zip1, but by removing promiscuously loaded Hop1 from the chromosome axis. Such an idea fits with previous studies showing that Pch2 facilitates Hop1 phosphorylation, and that Pch2 and Hop1 are both required for interhomolog bias, a process in which DSBs are preferentially repaired using a homolog instead of a sister (6, 19). This information, in conjunction with studies hypothesizing that Hop1/Zip1 patterns are dictated by CO placement (18, 20, 21), suggests that maintenance of differential Hop1 localization is important for promoting/reinforcing interhomolog repair at CO designation sites.
The Hop1 protein has been shown using a combination of atomic force microscopy, DNase I footprinting, and methylation interference to synapse noncontiguous and contiguous dsDNA molecules into complexes that resemble higher-order nucleoprotein structures (50). In addition, studies have shown that Hop1 can bind to specific structures such as guanine quartets and Holliday junctions and that pairing between two dsDNA sites is facilitated by guanine/cytosine (G/C)-rich sequences (55–57). Based on these observations it is not surprising that Hop1 forms aggregates on DNA when present at high concentrations (49) and that Pch2 can facilitate aggregate formation for a portion of Hop1–DNA complexes (our present study). We find it interesting that Pch2 can partition, in steps likely to require ATP hydrolysis, Hop1–DNA complexes into two different populations: protein–DNA aggregates and unbound DNA. One explanation for this finding is that Pch2 can remove weakly bound Hop1 from DNA but maintains or promotes Hop1 binding to more tightly bound sequences such as those described above. Additional experiments will be required to test this hypothesis.
In pull-down and DNA-binding assays we showed that Pch2 physically interacts with Hop1 and displaces it from DNA. This observation is consistent with a number of studies indicating that Pch2 negatively regulates Hop1 localization on chromosomes (e.g., refs. 17, 18, 20, 54, and 58). We hypothesize that in vivo, after Pch2 binds to promiscuously loaded Hop1 and displaces it, an additional factor or factors are required to denature and degrade Hop1. In support of this idea, Hop1 protein levels are higher in pch2Δ cells, as shown by Western blot (19) and immunostaining (18, 20, 21). A number of AAA+ ATPases display multiple functions when interacting with different substrates and/or cofactors. For example, the AAA+ protein VCP mediates membrane fusion by interacting with p47 and syntaxin5 and activates the nuclear factor κB (NF-κB) by binding to the NF-κB inhibitor nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα); it also regulates the cell cycle by acting on cyclins (reviewed in refs. 59 and 60). We note that Pch2 was shown to interact with origin recognition complex subunit 1 (Orc1), which is an AAA+ protein that appears to act in concert with Pch2 to suppress DSB formation at ribosomal DNA borders in meiosis (46). This function of Orc1 is likely separate from its role in DNA replication—it appears to be mediated through an Orc1 bromo-adjacent homology domain that is required for its chromatin-silencing function (46).
It is also possible that Pch2 requires a posttranslational modification to display full activity toward Hop1. This idea is supported by Ho and Burgess (19), who reported that Pch2 physically interacts with the putative BRCA1 C terminus (BRCT) domain of Xrs2, and BRCT domains are known to bind phosphoproteins (61). Pch2 contains one Thr–Gln site that could be a potential phosphorylation site for Mec1/Tel1, which are yeast homologs of the mammalian ATM/ATR kinases that preferentially phosphorylate their substrates at consensus Ser–Gln/Thr–Gln sites (62). To test this idea we made two mutations at the Thr–Gln site, pch2–T428A (nonphosphorable mutant) and pch2–T428E (phosphomimic mutant). Neither mutation could complement pch2Δ (Table 2), leaving open the possibility that this site could be functionally relevant. Additional studies, which may involve novel binding partners of Pch2, will be needed to test the above ideas.
Materials and Methods
Protein Expression and Purification.
GST–Pch2 was purified as follows: pEAE307 (2µ ampr URA3 leu2-d GAL1-10–GST–PCH2) and its mutant derivatives pEAE323 or pEAE326 (the same as pEAE307 but with GST–pch2–G319A or GST–pch2–E399Q) were transformed into EAY33 (ura3-52, trp1, leu2Δ1, his3Δ200, pep4::HIS3, prb1Δ1.6R, can1, GAL). A single colony of the transformant was inoculated into 160 mL leucine drop-out media, and grown overnight to saturation at 30 °C. Twenty-five milliliters of overnight culture was used to inoculate each of the 6 L synthetic glycerol lactate (SCGL) media. Each liter of SCGL media contains 2% lactic acid, 3% glycerol, 7 g yeast nitrogen base, 0.87 g leucine drop-out mix, and 0.1% glucose. Cultures were grown at 30 °C at 220 rpm (New Brunswick Scientific Incubator Shaker Model G-25) for about 18 h until OD600 reached 0.6. Protein overexpression was induced by adding 40% galactose to a final concentration of 2%. Cells were harvested after 18 h of induction, washed, resuspended in an equal volume of lysis buffer [100 mM Tris (pH 8.0), 1 mM EDTA (pH 8.0), 500 mM NaCl, 10% glycerol, 10 mM β-mercaptoethanol (BME), 1 mM phenylmethylsulfonyl fluoride (PMSF)] and frozen by dropping into liquid nitrogen. Frozen cells were stored at −80 °C.
Frozen cells (10 g) were lysed with dry ice using a Braun coffee grinder run for 2 min, after which dry ice was removed by sublimation at −20 °C. All subsequent steps were performed on ice or at 4 °C. Cell lysates were resuspended in 15 mL lysis buffer, centrifuged at 35,000 × g for 30 min, and the cleared lysate added to 1 mL glutathione resin (Thermo Scientific, catalog no. 16100) and mixed for 2 h. The resin was then transferred into a glass column, washed with 20 mL wash buffer 1 [25 mM Tris (pH 7.5), 1 mM EDTA (pH 8.0), 160 mM NaCl, 10% glycerol, 10 mM BME, 1 mM PMSF], and eluted with 10 mL elution buffer 1 (wash buffer 1 containing 20 mM glutathione). Peak fractions were pooled and loaded onto a 0.6-mL PBE 94 column (Amersham Biosciences), washed using 6 mL wash buffer 2 [25 mM Tris (pH 7.5), 1 mM EDTA (pH 8.0), 250 mM NaCl, 10% glycerol, 10 mM BME, 1 mM PMSF], and eluted with 3 mL elution buffer 2 [25 mM Tris (pH 7.5), 1 mM EDTA (pH 8.0), 500 mM NaCl, 10% glycerol, 10 mM BME]. Aliquots of the eluted protein were frozen in liquid nitrogen and stored at −80 °C. The typical yield was ∼800 µg from 10 g frozen cells for GST–Pch2 and GST–Pch2–G319A, and ∼30 µg from 10 g frozen cells for GST–Pch2–E399Q.
Electron Microscopy and Image Analysis.
Pch2 or GST–Pch2 samples [10 µg/mL in assembly buffer: 50 mM Tris⋅HCl (pH 7.0), 150 mM NaCl, 10 mM MgCl2, 10 mM EDTA, 2 mM BME] were incubated for 10 min in the presence or absence of ADP (10 mM), ATP (10 mM), or ATPγS (2 mM). After incubation, glow-discharged (5 mA for 15 s) 400-mesh electron microscopy grids with a continuous layer of fresh carbon obtained by graphite evaporation were floated into a 5-µL drop of the assembly reaction for 2 min. Excess of sample was blotted with filter paper and the grids were stained with 1% uranyl acetate for 1 min. Images were collected on a JEOL 2010F electron microscope operated at 200 kV at 50,000× magnification with a dose of ∼15 electrons per Å2. Images were recorded on Kodak SO-163 films and scanned on a Nikon Super COOLSCAN 9000 ED at 6.35 µm per pixel. Electron micrographs were binned twofold rendering images with a sampling of 2.54 Å per pixel.
Particles were then picked using Boxer from the EMAN package (63) using 128 × 128-pixel boxes. In the case of Pch2 incubated in the presence of ADP, ATP, or ATPγS, a total of 827, 1,007, and 1,958 particles were manually picked. In the analysis of the GST–Pch2 sample incubated with ATPγS, the number of selected particles was 1,859.
To perform the symmetry analysis, groups of 200–250 particle images were first normalized and then translationally aligned to a circularly symmetrical global average of all of the unaligned particle images in the group. Existence of statistically significant rotational symmetries was determined by comparison with particle images of the micrograph background using spectral ratio product and Student’s t statistical tests as implemented in the Rotastat software (33).
To calculate the 2D averages, normalized particle images were translationally and rotationally aligned using cross-correlation–based methods as implemented in the Xmipp software package (64–66). The reference used for alignments was either a circularly symmetrical global average of all of the unaligned particle images or a reference constructed using a pyramidal combination of a subset of the images (65).
Additional details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the E.E.A. and J.O. laboratories, Michael Lichten, Sarah Zanders, Timothy West, Scott Emr, Michael Goldberg, and Joseph Peters for helpful comments; and Akira Shinohara and Doug Bishop for technical advice. We also thank the Nancy E. Kleckner laboratory for sharing information prior to publication. C.C. and E.E.A. were supported by National Institutes of Health Grant GM53085, J.O. was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-82930, and A.J. was supported by a CIHR Doctoral Research Award.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310755111/-/DCSupplemental.
References
- 1. Hunter N (2007) Meiotic recombination. Molecular Genetics of Recombination: Topics in Current Genetics, eds Aguilera A, Rothstein R (Springer, Berlin), pp 381–442.
- 2.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117(1):9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 3.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117(1):29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 4.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126(2):285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berchowitz LE, Copenhaver GP. Genetic interference: Don’t stand so close to me. Curr Genomics. 2010;11(2):91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanders S, Sonntag Brown M, Chen C, Alani E. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics. 2011;188(3):511–521. doi: 10.1534/genetics.111.129031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90(6):1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 8.Niu H, et al. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16(12):5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingsworth NM. Phosphorylation and the creation of interhomolog bias during meiosis in yeast. Cell Cycle. 2010;9(3):436–437. doi: 10.4161/cc.9.3.10773. [DOI] [PubMed] [Google Scholar]
- 10.Goldfarb T, Lichten M. The sister chromatid is frequently and efficiently used for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454(7203):479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310(5754):1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- 13.Joyce EF, McKim KS. Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics. 2009;181(1):39–51. doi: 10.1534/genetics.108.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce EF, McKim KS. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet. 2010;6(8):e1001059. doi: 10.1371/journal.pgen.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3(8):e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roig I, et al. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 2010;6(8):e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojtasz L, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5(10):e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97(3):313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 19.Ho H-C, Burgess SM. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 2011;7(11):e1002351. doi: 10.1371/journal.pgen.1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Börner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc Natl Acad Sci USA. 2008;105(9):3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi N, Barot A, Jamison C, Börner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5(7):e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanders S, Alani E. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 2009;5(7):e1000571. doi: 10.1371/journal.pgen.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farmer S, et al. Budding yeast Pch2, a widely conserved meiotic protein, is involved in the initiation of meiotic recombination. PLoS ONE. 2012;7(6):e39724. doi: 10.1371/journal.pone.0039724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132(5):758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 25.Terentyev Y, et al. Evidence that MEK1 positively promotes interhomologue double-strand break repair. Nucleic Acids Res. 2010;38(13):4349–4360. doi: 10.1093/nar/gkq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 27.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 29.Tucker PA, Sallai L. The AAA+ superfamily—a myriad of motions. Curr Opin Struct Biol. 2007;17(6):641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Rafferty JB, et al. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996;274(5286):415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, West SC, Egelman EH. Structure and subunit composition of the RuvAB-Holliday junction complex. J Mol Biol. 1997;266(2):217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 32.Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150(1):F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kocsis E, Cerritelli ME, Trus BL, Cheng N, Steven AC. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy. 1995;60(2):219–228. doi: 10.1016/0304-3991(95)00070-2. [DOI] [PubMed] [Google Scholar]
- 34.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure—diverse function. Genes Cells. 2001;6(7):575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 35.Cheung KL, Huen J, Houry WA, Ortega J. Comparison of the multiple oligomeric structures observed for the Rvb1 and Rvb2 proteins. Biochem Cell Biol. 2010a;88(1):77–88. doi: 10.1139/o09-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung KL, Huen J, Kakihara Y, Houry WA, Ortega J. Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J Mol Biol. 2010b;404(3):478–492. doi: 10.1016/j.jmb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Perrote A, Muñoz-Hernández H, Gil D, Llorca O. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012;40(21):11086–11099. doi: 10.1093/nar/gks871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376(5):1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- 39.Rouiller I, et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9(12):950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 40.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360(Pt 1):1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad MN, et al. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133(7):1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 42.Sonntag Brown M, Zanders S, Alani E. Sustained and rapid chromosome movements are critical for chromosome pairing and meiotic progression in budding yeast. Genetics. 2011;188(1):21–32. doi: 10.1534/genetics.110.125575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanat JJ, et al. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4(9):e1000188. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276(45):41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- 45.Brosh RM, Jr, Matson SW. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177(19):5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vader G, et al. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477(7362):115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85(6):875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- 48.Khan K, Vipin Madhavan TP, Muniyappa K. Cloning, overexpression and purification of functionally active Saccharomyces cerevisiae Hop1 protein from Escherichia coli. Protein Expr Purif. 2010;72(1):42–47. doi: 10.1016/j.pep.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Kironmai KM, Muniyappa K, Friedman DB, Hollingsworth NM, Byers B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol Cell Biol. 1998;18(3):1424–1435. doi: 10.1128/mcb.18.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan K, Karthikeyan U, Li Y, Yan J, Muniyappa K. Single-molecule DNA analysis reveals that yeast Hop1 protein promotes DNA folding and synapsis: Implications for condensation of meiotic chromosomes. ACS Nano. 2012;6(12):10658–10666. doi: 10.1021/nn3038258. [DOI] [PubMed] [Google Scholar]
- 51.Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94(4):525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 52.Yu RC, Hanson PI, Jahn R, Brünger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5(9):803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Perez E, et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22(20):2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voelkel-Meiman K, Moustafa SS, Lefrançois P, Villeneuve AM, MacQueen AJ. Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genet. 2012;8(10):e1002993. doi: 10.1371/journal.pgen.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anuradha S, Tripathi P, Mahajan K, Muniyappa K. Meiosis-specific yeast Hop1 protein promotes pairing of double-stranded DNA helices via G/C isochores. Biochem Biophys Res Commun. 2005;336(3):934–941. doi: 10.1016/j.bbrc.2005.08.196. [DOI] [PubMed] [Google Scholar]
- 56.Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol Cell Biol. 2000;20(4):1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tripathi P, Anuradha S, Ghosal G, Muniyappa K. Selective binding of meiosis-specific yeast Hop1 protein to the holliday junctions distorts the DNA structure and its implications for junction migration and resolution. J Mol Biol. 2006;364(4):599–611. doi: 10.1016/j.jmb.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 58.Ontoso D, Acosta I, van Leeuwen F, Freire R, San-Segundo PA. Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. PLoS Genet. 2013;9(1):e1003262. doi: 10.1371/journal.pgen.1003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Song C, Li CC. Molecular perspectives on p97-VCP: Progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146(1-2):44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 60.White SR, Lauring B. AAA+ ATPases: Achieving diversity of function with conserved machinery. Traffic. 2007;8(12):1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 61.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 62.Traven A, Heierhorst J. SQ/TQ cluster domains: Concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27(4):397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- 63.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 64.Marabini R, et al. Xmipp: An Image Processing Package for Electron Microscopy. J Struct Biol. 1996;116(1):237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 65.Scheres SH, Núñez-Ramírez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3(6):977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorzano CO, et al. XMIPP: A new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148(2):194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Di Tommaso P, et al. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39(Web Server issue):W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.