Abstract
Metallothioneins are proteins that play an essential role in metal homeostasis and detoxification in nearly all organisms studied to date. Yet discrepancies between outcomes of chronic and acute exposure experiments hamper the understanding of the regulatory mechanisms of their isoforms following metal exposure. Here, we investigated transcriptional differences among four identified homologs (mt1–mt4) in Daphnia pulex exposed across time to copper and cadmium relative to a control. Transcriptional upregulation of mt1 and mt3 was detected on day four following exposure to cadmium, whereas that of mt2 and mt4 was detected on day two and day eight following exposure to copper. These results confirm temporal and metal-specific differences in the transcriptional induction of genes encoding metallothionein homologs upon metal exposure which should be considered in ecotoxicological monitoring programs of metal-contaminated water bodies. Indeed, the mRNA expression patterns observed here illustrate the complex regulatory system associated with metallothioneins, as these patterns are not only dependent on the metal, but also on exposure time and the homolog studied. Further phylogenetic analysis and analysis of regulatory elements in upstream promoter regions revealed a high degree of similarity between metallothionein genes of Daphnia pulex and Daphnia magna, a species belonging to the same genus. These findings, combined with a limited amount of available expression data for D. magna metallothionein genes, tentatively suggest a potential generalization of the metallothionein response system between these Daphnia species.
Keywords: metallothioneins, metals, expression, daphnia
1. Introduction
Metallothioneins are metal binding proteins that play a pivotal role in metal homeostasis and detoxification (Amiard, 2006). Since their initial discovery, they have been extensively studied in a variety of organisms ranging from microbes to plants and animals (Roesijadi, 1992). Organisms often possess multiple genes encoding metallothionein homologs with distinct properties, such as varying affinities for different metals, and in many cases different functions (Amiard, 2006; Dallinger et al., 1997; Roesijadi, 1992).
Despite the plethora of available studies, the mechanisms by which metallothionein homologs differ in their regulation remain unclear (Amiard, 2006; Mao et al., 2012). Recent studies have reported differences in expression modulated by heavy metal concentrations and in the exposure period required to induce metallothionein transcription (Amiard, 2006; Roesijadi, 1992; Mao et al., 2012). Two studies differ in their reported responses of the metallothionein homolog 1 (mt1) in Daphnia pulex. Shaw et al. (2007) observed significant induction of RNA levels of the gene encoding mt1when exposing D. pulex for 48 hours to 20 μg Cd L−1, whereas Asselman et al. (2012) observed no elevated levels of mRNA expression for any metallothionein homolog genes when exposing D. pulex for 16 days to 0.5 μg Cd L−1. These experimental results may differ because of a difference in metal concentration, a difference in the elapsed time under stress, or both. Based on studies in other organisms at the protein level (Barka et al., 2001; Bodar et al., 1998; Del Ramo et al., 1995; Martinez et al., 1996), metallothioneins are likely early responders to metal stress. However, too few studies have investigated the time course of expression of the varying homologs in organisms when exposed to metals.
Jenny et al. (2006) observed increasing mRNA levels after 16 and 26 hours exposure to cadmium, copper and zinc in the oyster C. virginica. Soazig and Marc (2003). In the mussel M. edulis, phasic responses of mRNA levels were observed upon exposure to zinc and cadmium for a period of 80 hours. Both studies illustrate the temporal variability of these mRNA expression levels, which may be essential in understanding the mechanisms of metallothionein regulation and their potential use as environmental biomarker (Viarengo, 1999; Valls et al., 2001). However, both studies focus on relatively short exposures of hours, whereas exposures in the environment are more often longer in duration (i.e., chronic). Höckner et al. (2009) investigated the response of a single cadmium mt in two closely related pulmonate species, H. pomatia and C. aspersus. They observed time- and species-dependent regulation of this cadmium mt after exposure to cadmium for 0, 1, 3, 5, and 8 days. Yet, without access to a whole genome sequence it is impossible to know if the primers developed in these three studies were homolog specific. Hence, the mechanisms of differential regulation of metallothionein within a genome remain unknown for aquatic invertebrates. Here, we studied the differential regulation of the genes encoding four metallothionein homologs in D. pulex upon exposure to cadmium and copper. We specifically investigated the hypothesis that exposure duration in addition to exposure concentration can influence metallothionein gene mRNA expression.
In D. pulex, a standard test organism in ecotoxicology and model organism (Ebert, 2011), four homologs of metallothioneins have been identified (Asselman et al., 2012). These homologs have shown distinct responses upon exposure to a variety of environmental stressors, including cadmium and copper (Asselman et al., 2012; Shaw et al., 2007). Studying these homologs at different time points of exposure and environmental stressors can yield a broader understanding of the different roles of each homolog and the overall mechanism regulating their responses.
In the current study, D. pulex were exposed to copper and cadmium over a time-course of sixteen days. During this period, we sampled animals to analyze metallothionein gene transcription at different time points (2, 4, 8 and 16 days) for each of the four individual homologs. We focus on mRNA expression as an indicator of the response to metals. Indeed, metallothioneins are inducible proteins, the synthesis of which is primarily stimulated and regulated at the transcriptional level (Squibb and Cousin, 1977; Andersen and Weser, 1979; Roelofs et al., 2007). Furthermore, the usefulness of mRNA levels as potential biomarkers for metals has already been demonstrated in mussels and fish (Soazig and Marc, 2003; Tom, 1999).
We further related the mRNA expression patterns of all homologs with their upstream regulatory sequences to infer a better mechanistic understanding of the observed expression patterns. Ultimately, we compared these regulatory sequences to those of metallothionein genes in the closely related species Daphnia magna to identify potential similarities. Such similarities or differences may give an indication as to what extent results can be generalized within the Daphnia genus, since these two species are extensively used in regulatory risk assessment. Hence, we aimed to contribute to a better understanding of the presence, function and metal responsiveness of the different metallothionein genes in Daphnia.
2. Material and Methods
2.1 Collection and culturing of the animals
The isolate under study originated from the same isoclonal laboratory culture as the Daphnia pulex isolate used for genome sequencing (Colbourne et al., 2011). Cultures were maintained at a density of 15 adults per liter in no N no P COMBO reconstituted culture water (Shaw et al., 2007) in aerated polyethylene aquaria under constant photoperiod (16:8 light:dark) and temperature (20 ± 1°C). Reconstituted culture water was renewed three times a week. Animals were fed daily with the green algae, Ankistrodesmus falcatus, at a concentration of 1.5 mg C L−1. Neonates (<24h) of the second through seventh brood were used for exposures as well as for setting up a next generation of the maintenance cultures.
2.2 Experimental Design
Neonates were exposed for a period of sixteen days to a cadmium (0.5 μg Cd L−1), copper (5 μg Cu L−1), or control treatment in which no cadmium or copper was added to the media. Concentrations were chosen based on life table and gene expression experiments from Asselman et al. (2012). Stock solutions were made with CdCl2·H2O and CuCl2·H2O (analytical grade, VWR International, Haasrode, Belgium). Each treatment consisted of three replicates, represented by a 4L polyethylene aquarium containing 400 neonates at the beginning of the experiment. Animals were fed daily with A. falcatus (1.5 mg C L−1) under a constant photoperiod (16:8 light:dark) and constant temperature (20 ± 1°C). Every two days, animals were transferred to new reconstituted culture water medium. During the experiment, stocking density (15 adults per liter) was kept constant to avoid crowding effects. After 2, 4, 8 and 16 days animals were randomly sampled from each aquarium for mRNA extraction. The number of animals sampled was 120, 100, 60 and 40 on day 2, 4, 8 and 16, respectively. This sampling scheme ensured enough material for further mRNA extractions (approximately 20–30 mg of Daphnia). Samples for copper and cadmium analysis and for pH measurements were taken at regular intervals. In addition, survival and reproduction were monitored every two days.
2.3 Analysis of population endpoints
Survival and reproduction at every change out were analyzed for effects on these population endpoints. Significant effects on survival and reproduction compared to control at each change out were tested using a t-statistic, after verification of assumptions of normality and homoscedasticity. The effect on reproduction was determined by first calculating the number of neonates per female to exclude effects on survival. Therefore, the age specific reproduction per female was calculated for each biological replicate (i.e. each aquarium). Effects on survival were also determined by using the age specific survival rate.
2.4 mRNA Expression Analysis of Genes
RNA extraction was performed on fresh tissue with the Qiashredder and RNeasy columns (Qiagen, Venlo, Netherlands) following manufacturer’s protocol including the addition of a DNAse treatment (Qiagen, Venlo, Netherlands). RNA Quality and quantity were assessed with a nanodrop spectrophotometer (quality standards were based on the following absorbance ratios: 260/230>2.1 and/or 260/280>1.9). RNA integrity was verified through gel electrophoresis. Afterwards, RNA was aliquoted and stored at −80°C until reverse transcription to cDNA. RNA was reverse transcribed to cDNA with the MessageAmpTM II mRNA Amplification kit (reverse transcription SECTION C only), supplied by Life Technologies (Life Technologies, Carlsbad, CA, USA). cDNA Quantity was assessed with an Oligreen ssDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Samples were subsequently stored at −20°C in nuclease free water until qPCR analysis at a concentration of 2000 ng/μL. Primers for quantitative real-time PCR were developed by Asselman et al. (2012) and purchased from IDT Technologies (IDT Technologies, Coralville, IA, USA) for all four homologs. Although mt1 homolog is duplicated in this daphnia isolate, mt1A and mt1B, it is not possible to distinguish between them with PCR as they differ by only one base. Design and analysis of the quantitative real-time PCR experiment followed Asselman et al. (2012). Briefly, in addition to accurate quantification of template concentrations with an oligreen assay, a reference gene (glyceraldehyde-3-phosphate dehydrogenase) was used to normalize the output data. In particular, for all samples, accurate concentration measurements of cDNA with oligreen were used to confirm that the reference gene was not responsive to the metal stress. This approach has been validated by Lundby et al. (2005) to determine stability and validity of a single reference gene. Primers for the reference gene were developed by Spanier et al. (2010).
qPCR was performed on the Mx3000P qPCR system (Stratagene, Santa Clara, CA, USA). Each 96 well plate included a standard curve per gene, a melting curve per sample, positive controls, no primer and no template controls. Each treatment was represented by three biological replicates and two technical replicates per biological replicate (i.e. each biological replicate is a sample of a different aquarium). SYBR Green Super mix (Quanta Biosciences, Gaithersburg, MD, USA) was used to set up the reactions and rescaled to 25 μL total reaction volume according manufacturer’s protocol. SYBR Green Super mix, primers and H2O were assembled in a mastermix prior to dispensing the appropriate volume to each well with an automated repeater pipette. At the end, 1 μL of cDNA was added in each well and plates were vortexed to homogenize the solution. Amplification consisted of 40 cycles (30s at 95°C, 30s at 60°C, 35s at 72°C), and was preceded by 3 minutes at 95°C.
Quality analysis was performed by analyzing melting curves of all samples and verifying results from positive, no primer and no template controls. Raw fluorescence data were extracted and imported into the software environment R. Data were analyzed according to Asselman et al. (2012), using the qPCR package developed by Ritz and Spiess (2008) and normalizing expression values according to Vandesompele et al. (2002). Significant differences between normalized expression ratios were analyzed statistically with a t-statistic, if assumptions for normality and homoscedasticity were fulfilled. Otherwise, kruskal-wallis test was used as a non-parametric alternative.
2.5 Comparison of D. pulex metallothioneins with those of the closely related species D. magna
Based on protein sequences of D. pulex metallothioneins (Asselman et al., 2012) and two D. magna metallothioneins identified by Poynton et al. (2007), we identified three D. magna metallothioneins in the D. magna genome version 2.4 (unreleased based on a 22x coverage http://server7.wfleabase.org/genome/Daphnia_magna/, Daphnia Genome Consortium: https://wiki.cgb.indiana.edu/display/DGC/Initiatives). We used NCBI protein-protein BLAST with standard settings (Altschul et al., 1990). To further confirm the identity of these metallothioneins, we compared their sequences homology with known metallothioneins in other organisms. Sequences for the latter were accessed through NCBI Genbank (Benson et al., 2011) based on Asselman et al. (2012) and Poynton et al. (2007). Phylogenetic analysis and analysis of regulatory elements in Daphnia magna promoter regions were conducted according to Asselman et al. (2012). In particular, MAFT software (Katoh, 2013) was used to construct a multiple alignment of these protein sequences. A subsequent neighbor-joining tree was constructed using MEGA (Tamura et al., 2011). Finally, results of promoter analysis were compared with results for D. pulex promoter regions of metallothionein genes obtained by Asselman et al. (2012). D. magna sequences were deposited in Genbank: Mta (KF561474), Mtb (KF561476), MtC (KF561475).
3. Results
3.1 Analysis of Population Endpoints
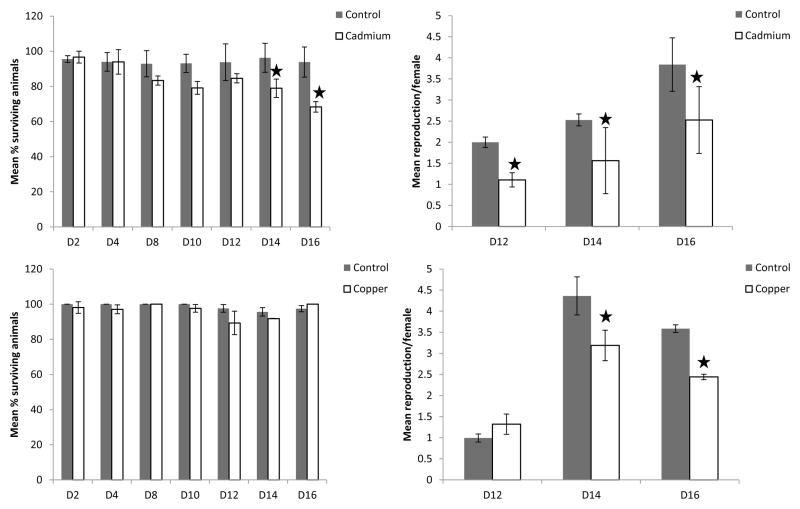
In both copper and cadmium experiments, pH did not vary significantly from control treatments (pH= 7.4 with a standard deviation of 0.1 units). Measured cadmium concentrations were 0.45 ± 0.03 μg Cd L−1 in cadmium treatments and below the detection limit of 0.06 μg Cd L−1 in controls. The measured copper concentrations were 0.88 ± 0.06 μg Cu L−1 in control treatments and 6.25± 0.62 μg Cu L−1 in copper exposures. Different effects on mortality were observed for copper and cadmium exposed organisms. No significant effects on mortality were observed for organisms exposed to copper at any given time point, whereas after 14 and 16 days survival was significantly lower for cadmium exposed organisms compared to their corresponding control treatment (p14=0.04, p16=0.04). (Fig. 1)
Figure 1.
Population characteristics throughout cadmium (Cd) and copper (Cu) experiments are represented after various days (D) of exposure. Ctr denotes control treatments. Left bar charts represent mean % of surviving animals at the given time point. Right bar charts represent mean reproduction per female at the given time point. Error bars represent standard deviations. Stars denote statistically significant differences from control treatments at a significance level of p=0.05.
For both cadmium and copper exposed organisms, significant effects on reproduction were observed. In the cadmium treatment, these effects were significant on the first day on which reproduction was observed (day 12) until the end of the experiment (pday12=0.03, pday14=0.04, pday16=0.04). In the copper treatment, significant effects were observed on the last two time points measured, i.e. day 14 and day 16(pday14=0.04, pday16=0.02) (Fig. 1).
3.2 mRNA expression analysis
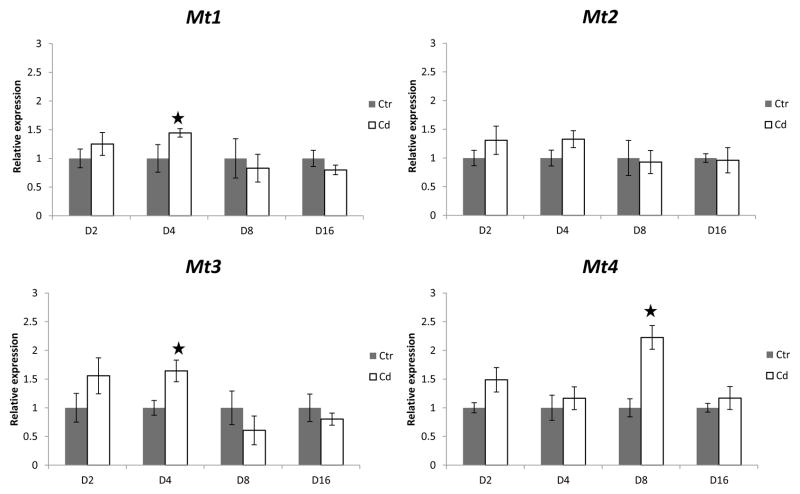
Effect of cadmium on metallothionein transcription was homolog- and time-dependent. Both mt1 and mt3 gene transcription were significantly induced in cadmium exposed animals after four days of exposure (pmt1<0.01, pmt3<0.01) (Fig. 2). This induction was not permanent as after eight and sixteen days of cadmium exposure metallothionein gene transcription of both homologs was not significantly different from control treatment. Mt2 transcription was not responsive to cadmium exposure at any exposure time, whereas mt4 transcription was induced in cadmium-exposed organisms after eight days of exposure (pmt4<0.01) (Fig. 2). Like those of mt1 and mt3, the mRNA expression of mt4 also returned to that of the control treatment after sixteen days of exposure (Fig 2).
Figure 2.
Relative mRNA expression ratios for all four metallothionein homolog genes after 2(D2), 4(D4), 8(D8) and 16(D16) days of exposure to cadmium (Cd) or control (Ctr) treatments after normalization with both oligreen and GAPDH reference gene. Stars denote statistically significant differences from control treatments at a significance level of p=0.05. Error bars represent standard deviations.
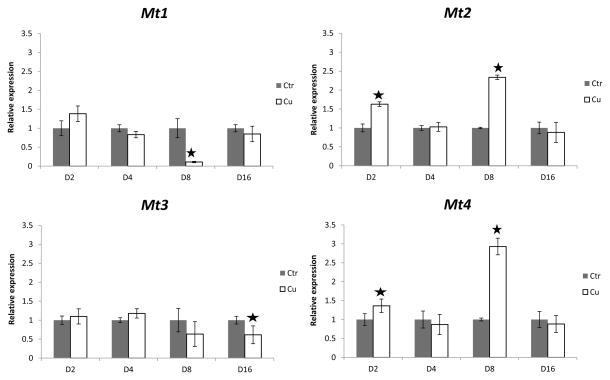
In contrast, the copper treatment induced both mt2 and mt4 transcription after two days (pmt2=0.02, pmt4=0.02) and eight days (pmt2=0.01, pmt4=0.01) of exposure, but not after four and sixteen days of exposure (Fig. 3). In addition, mt1 and mt3 mRNA levels were repressed under copper exposure after eight and sixteen days, respectively (pmt1=0.03, pmt3<0.01) (Fig. 3).
Figure 3.
Relative mRNA expression ratios for all four metallothionein homolog genes after 2(D2), 4(D4), 8(D8) and 16(D16) days of exposure to copper (Cu) or control (Ctr) treatments after normalization with both oligreen and GAPDH reference gene. Stars denote statistically significant differences from control treatments at a significance level of p=0.05. Error bars represent standard deviations.
3.3 Comparison of D. pulex metallothioneins with those of the closely related species D. magna
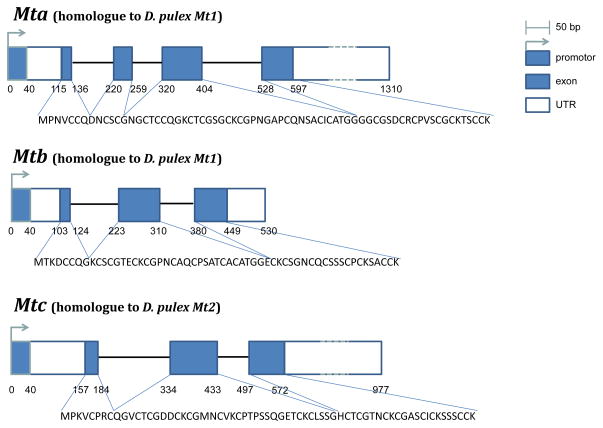
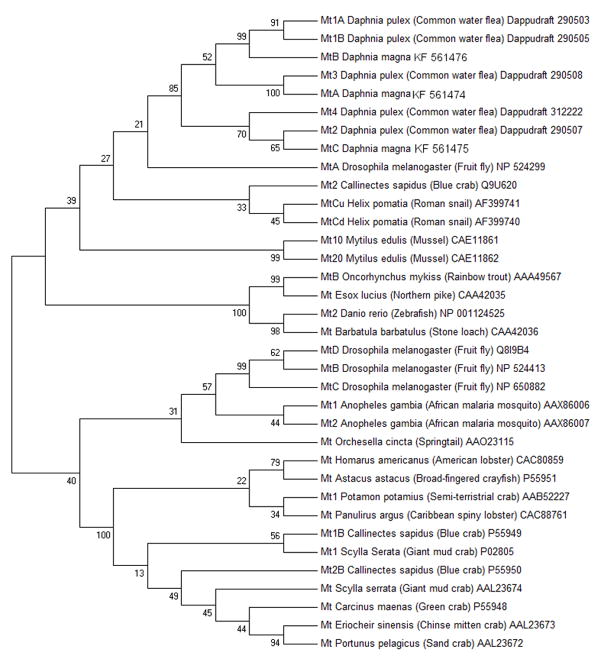
In addition to the two metallothioneins identified by Poynton et al. (2007), we identified a third metallothionein in D. magna (Dma) (Fig. 4). Predicted amino acid sequences of mtA, mtB and mtC gene product contained, respectively, 21, 18 and 18 cysteine residues, which are characteristic for metallothioneins (Fig. 4). These residues were arranged in characteristic motifs (Cys-xaa-yaa-Cys, Cys-x-Cys, and Cys-Cys) typing them as class I metallothioneins (Fowler et al., 1987). Interestingly, Dma mt c contains an unusual histidine residue at position 44(Fig. 4). Histidine residues have been observed in numerous species including the nematode C. elegans (Bofill et al., 2009) and the snail H. pomatia (Berger et al., 1997). The histidine residue has been suggested to play role in metal binding and coordination (Bofill et al., 2009; Capdevila and Atrian, 2011) In addition, amino acid identity scores were high between D. pulex (Dpu) and D. magna metallothioneins (i.e. 91% between Dpu mt1A, Dpu mt1B and Dma mtB; 92% between Dpu mt3 and Dma mtA; 78% between Dpu mt2 and Dma mtC). Phylogenetic analysis revealed strong monophyletic grouping for Daphnia metallothioneins (Fig. 5) indicating them to be quite divergent from other metallothioneins.
Figure 4.
Daphnia magna gene models for mtA, mtB and mtC. These sequence data were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu
Figure 5.
Phylogenetic neighbor-joining (Saitou and Nei, 1987) tree based on multiple alignments of all predicted sequences of 5 different homologs of metallothionein in Daphnia pulex, 3 homologs in Daphnia magna and 27 metallothionein sequences of 18 different species (based on Asselman et al., 2012 and Poynton et al., 2007).). NCBI or wfleabase accession numbers are listed next to each sequence. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Colored dots represent different phylogenetic groups: malacostraca (blue), bony fish (orange), mammals (yellow), insects (purple), molluscs (green), Daphnias (red), nematodes (grey).
Analysis of upstream promoter regions of D. magna was compared with the analysis of D. pulex promoter regions in Asselman et al. (2012) (Fig 6). For the homologs Dpu mt1A, Dpu mt1B and Dma mtB, most distinct difference was the absence of metal regulatory elements in Dma mtB (Table 1). Dpu Mt2 and Dma mtC promoter regions were very similar, both containing a metal regulatory element with the same sequence (Table 2), heat shock factors as well as initiators and 20-hydroxyecdysone responsive elements (Table 1). The same conclusions could be drawn for Dpu mt3 and Dma mtA although mt3’s promoter region did contain three times more heat shock factors than Dpu mtA (Table 1, Table 2).
Figure 6.

Representation of metallothionein (mt) genes in Daphnia pulex (adapted from Asselman et al., 2012) and Daphnia magna. For each gene 3000 bp upstream promoter region with identified regulatory elements (Table 1) is represented. Exons are represented by rectangular blocks, white blocks for non coding exon sequences and blue blocks for coding exon sequences, all connected by triangles representing intron sequences.
Table 1.
List of regulatory elements and their occurrence in promoter region of D. pulex and D. magna metallothionein genes (mt). Data for D. pulex has been obtained from Asselman et al. (2012): Heat shock factors (HSF), heat shock transcription factors (HSTF), ecdysone-responsive key regulator (BR-C_Z4), activator proteins (AP), metal regulatory elements (MRE), initatiors (Inr), anti-oxidant responsive elements (ARE), 20-hydroxyecydsone responsive elements (HERE), heat shock responsive elements (HSP).
| Regulatory Element Metallothionein: | HSF1 | HSTF | BR-C_Z4 | AP-1/4 | HSF2 | MRE | Inr | ARE | HERE | HSP1/2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Mt1a (D. pulex) | 4 | 5 | 1 | 1 | 0 | 1 | 4 | 0 | 0 | 0 |
| Mt1b (D. pulex) | 3 | 6 | 1 | 1 | 2 | 1 | 5 | 0 | 2 | 0 |
| Mtb (D. magna) | 2 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 0 |
| Mt2 (D. pulex) | 2 | 6 | 0 | 1 | 0 | 1 | 2 | 0 | 2 | 2 |
| Mtc (D. magna) | 3 | 6 | 0 | 0 | 0 | 1 | 5 | 0 | 1 | 0 |
| Mt3 (D. pulex) | 1 | 5 | 0 | 0 | 0 | 4 | 4 | 0 | 1 | 1 |
| Mta (D. magna) | 1 | 1 | 0 | 0 | 0 | 3 | 2 | 2 | 2 | 0 |
| Mt4(D. pulex) | 1 | 5 | 0 | 1 | 0 | 1 | 5 | 0 | 0 | 2 |
Table 2.
List of metal regulatory elements (MRE) found in the different D. pulex metallothionein genes based upon the consensus sequence TGCRCNC. If more than one MRE was found, the MRE located the furthest from the transcription start site is denote MRE1, other MREs are then numbered consecutively based upon their location from the transcription start site.
| Consensus: | T | G | C | R | C | N | C |
|---|---|---|---|---|---|---|---|
| Mt1a (D. pulex) | T | G | C | G | C | G | C |
| Mt1b (D. pulex) | T | G | C | G | C | G | C |
| Mt2 (D. pulex) | T | G | C | G | C | A | C |
| Mtc (D. magna) | T | G | C | G | C | A | C |
| Mt3-MRE1 (D. pulex) | T | G | C | A | C | A | C |
| Mt3-MRE2 (D. pulex) | T | G | C | A | C | A | C |
| Mt3-MRE3 (D. pulex) | T | G | C | G | C | A | C |
| Mt3-MRE4 (D. pulex) | T | G | C | G | C | A | C |
| Mta-MRE1 (D. magna) | T | G | C | A | C | G | C |
| Mta-MRE2 (D. magna) | T | G | C | G | C | A | C |
| Mta-MRE3 (D. magna) | T | G | C | G | C | A | C |
| Mt4 (D. pulex) | T | G | C | G | C | A | C |
4. Discussion
4.1 Expression patterns of cadmium exposure
Molecular analysis of the mRNA concentrations of different metallothionein genes extracted at different exposure times confirmed our hypothesis of time dependent induction. Similar to Asselman et al. (2012), we observed no induction of any metallothionein gene after sixteen days of exposure to cadmium. Instead we observed a transient induction of metallothionein gene transcription after four and eight days of exposure (Fig 2). Transient induction of metallothionein genes upon exposure to cadmium has also been observed by Höckner et al. (2009) in Helix pomatia. These authors attributed the transient induction to the presence of a distal MRE overlapping with binding sites for other transcription factors. They also suggested that the latter may act as attenuators or negative regulators of metallothionein induction. However, in D. pulex only Mt3 contains a distal MRE in its promoter region (Fig. 6). Yet, this MRE and also all other MREs in other Mts did not overlap with other transcription factors (Fig. 6). Interestingly, the induction of mt4 differed from the induction of mt1 and mt3 and was only observed after eight days of exposure. Although mt4 encodes a copper metallothionein closely related to that encoded by mt2, it does have-just like mt1 and mt3- a metal responsive element (MRE) within less than 300 basepairs upstream of its starting codon (Asselman et al., 2012), which may explain its induction by cadmium. In contrast, the MRE in the promoter region of mt2 is located relatively far from the transcription start-site (1000 bp upstream). These results show that the choices of both exposure period and homolog are crucial for studying effects on mRNA expression. As a result, the absence of induction at the end of the exposure period in some studies such as Asselman et al. (2012) does not necessarily mean that there has been no induction at all over the entire time course of the exposure. This finding may explain some of the apparent discrepancies between different studies as discussed in Amiard et al. (2006). Our results show that more temporal data on all homologs are needed to adequately incorporate metallothionein mRNA levels into biomonitoring.
Two potential mechanisms may explain the induction of metallothionein genes after four days of metal exposure and the absence of this induction at later time points during the exposure. First, metallothioneins reduce metal toxicity by binding the metal ion and making it non bioreactive (Amiard et al., 2006). As a result, sufficient metallothionein proteins can reduce and potentially eventually eliminate exposure. Second, the absence of induction after longer exposure periods may also be related to the loss of cadmium through continued ecdysis which results in a decreased cadmium burden and potentially decreased metallothionein production (Riddell et al., 2009). Future studies are needed to test this hypothesis for D. pulex by measuring cadmium body burdens and metallothionein protein levels at different time points during the exposure in parallel with transcription levels.
4.2 Expression patterns of copper exposure
In copper exposed organisms, we also observed an early induction. However, as opposed to cadmium, both mt2 and mt4 were transcriptionally induced. Interestingly, this expression pattern demonstrated an oscillating trend defined by induction after two and eight days of exposure and levels not significantly different from control after four and sixteen days of exposure (Fig. 3). Interestingly, both metallothionein genes contain exactly one MRE with the exact same sequence (Table 1), although this MRE is not located at the same distance upstream from the start codon in the two metallothionein genes.
Potential dynamics of metal bound proteins may explain the observed patterns and correlate with proposed mechanisms of reversible induction of metallothionein mRNA expression through metal-responsive transcription factor 1 (Saydam et al., 2002). However, day eight is also related to the onset of maturity and reproduction. In addition, copper metallothioneins have been related to the molting metabolism in blue crabs (Engel and Brouwer, 1987). Hence, the observed pattern may be an interaction between the function of metallothioneins in metal detoxification metabolism and in the molting metabolism.
In contrast to the induction of mt2 and mt4 as a response to copper, mt1 and mt3 expression levels were repressed after eight and sixteen days of exposure respectively. Downregulation of metallothioneins in response to metals has already been observed in other species (Woo et al., 2006). Asselman et al. (2012) proposed a potential inhibition-induction mechanism, similarly to that already known for heatshock proteins, where homologs that are more able to cope with the specific stressor are induced and others are repressed (Franzellitti and Fabbri, 2005). Here, both mt1 and mt3 are responsive to cadmium, whereas mt2 and mt4 are primarily induced by copper. Hence, the proposed induction-inhibition mechanism may explain the differential expression pattern following copper exposure across the different homologs (i.e. induction of mt2 and mt4, repression of mt1 and mt3).
Overall, the differences between the four metallothioneins in mRNA expression patterns across both copper and cadmium exposure reveal a complex regulation of these metallothioneins. Although these metallothioneins are transcriptionally regulated, the differences in their promoter regions cannot fully account for the differences in their expression patterns. Indeed, these metallothioneins differ in the number of MREs (Table 1), the location of these MREs (Fig. 6) and the exact sequence of the MREs (Table 2). Yet, not a single factor (location, number or sequence) can explain all the observed differences in expression patterns. Most likely, the answer lies in complex interaction between the three factors. In addition, protein structure encoded in the genetic sequence of the metallothioneins most likely plays a role in the relative affinity of one metal compared to the other.
4.3 Comparison of D. pulex metallothioneins with those of the closely related species D. magna
Phylogenetic analysis and comparison of regulatory elements in the upstream promoter region of metallothionein genes in D. pulex and D. magna revealed a high degree eof similarity. Although no fourth metallothionein was identified in D. magna, it does not imply the absence of the fourth metallothionein in D. magna, as the reference genome assembly is not yet complete (Daphnia Genome consortium). The observed similarity in predicted protein sequence and regulatory elements needs to be supplemented with similarities in expression patterns prior to generalization. At present, we did not find any studies of metallothionein gene expression in D. magna after chronic exposures and could only compare expression following acute exposures. In addition, Dma mtC has not been identified before and thus, no comparison with expression studies in literature could be made. We did find expression results for acute exposures at varying concentrations for Dma mtA and Dma mtB (Poynton et al., 2008). These results showed a differential regulation of mtB in D. magna, homologous with mt1A and mt1B in D. pulex, after exposure to cadmium and copper at both low and high concentrations (Poynton et al., 2008). Dma mtA, homologous with mt3 in D. pulex, was differentially regulated after exposure to low concentrations of cadmium and after exposure to high concentrations of cadmium and copper (Poynton et al., 2008).
Overall, we observed two clusters of metallothioneins in response to copper and cadmium over time. First, Dpu mt1 and Dpu mt3, homologous with mtB and mtA in D. magna, are induced to cadmium stress after four days of exposure and repressed after eight and sixteen days respectively to copper. Second, Dpu mt2, homologous with mtC in D. magna and mt4 are induced in an oscilating pattern after two and eight days of exposure to copper. These clusters also overlap with their phylogeny and more importantly with the regulatory elements in their promoter regions, underlining the relevance of combining expression data with genomic data.
However, we have in our study only used the isolates of D. magna and D. pulex that were sequenced. Mt gene and promoter sequences may be polymorphic across a range of isolates or populations of these two Daphnia species. As a consequence, care should be taken when extrapolating the reported sequences and expression results obtained here to other isolates. As publically available sequence data grows, these consensus sequences can be improved based upon data for multiple isolates for both species.
5. Conclusion
We observed an early transcriptional induction of metallothionein genes in Daphnia exposed to either cadmium or copper at sublethal concentrations. The expression patterns indicated a complex time-variable regulation of metallothioneins to different metals that is homolog-dependent. We identified two groups of metallothioneins. The first cluster consisted of mt1 and mt3, homologous to D. magna mtB and mtA, and responsive to cadmium stress. The second cluster consisted of mt2, homologous to mtC in D. magna, and mt4, which are responsive primarily to copper stress. This variability in metallothioneins response to stressors over time underlines the critical need of considering the exposure period and choice of homolog in experiments and biomonitoring programs when interpreting mRNA tissue levels.
Highlights.
Transcription patterns of 4 metallothionein isoforms in Daphnia pulex.
Under cadmium and copper stress these patterns are time-dependent.
Under cadmium and copper stress these patterns are homolog-dependent.
The results stress the complex regulation of metallothioneins.
Acknowledgments
The authors thank Nancy De Saeyer, Emmy Pequeur, Gisèle Bockstaele, Dieter De Coninck and Zachary Smith for the technical assistance. Primers for Daphnia magna genes were developed by Dieter De Coninck. Jana Asselman is the recipient of a PhD grant provided by the Flemish Institute for the Promotion of Scientific and Technological Research in Industry (IWT, Belgium). This research benefits from, and contributes to the Daphnia Genomic Consortium. Funding was obtained from UGent Special Research Fund (BOF projects 01N01211 and 01SB1910U), from the Research Foundation Flanders (FWO projects 1.5.269.11, G.0614.11) and from the National Institute of Environmental Health Science (R01ES019324 awarded to JRS). Daphnia magna metallothionein sequence data were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu. This project is supported in part by NIH award 5R24GM078274 “Daphnia Functional Genomics Resources.” D. magna sequences were deposited in Genbank: Mta (KF561474), Mtb (KF561476), MtC (KF561475).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amiard JC, Amiard-Triquet C, Barka S, Pellerin J, Rainbow PS. Metallothioneins in aquatic invertebrates: Their role in metal detoxification and their use as biomarkers. Aquat Toxicol. 2006;76:160–202. doi: 10.1016/j.aquatox.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Andersen D, Weser U. Partial purification and translation in vitro of metallothionein messenger RNA. Experientia Suppl. 1979;34:303–310. doi: 10.1007/978-3-0348-6493-0_23. [DOI] [PubMed] [Google Scholar]
- Asselman J, Glaholt SP, Smith Z, Smagghe G, Janssen CR, Colbourne JK, Shaw DE, JR, Schamphelaere KAC. Functional characterization of four metallothionein genes in Daphnia pulex exposed to environmental stressors. Aquat Toxicol. 2012;110–111:54–65. doi: 10.1016/j.aquatox.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barka S, Pavillon J-F, Amiard J-C. Influence of different essential and non-essential metals on MTLP levels in the Copepod Tigriopus brevicornis. Comp Biochem Physiol C. 2001;128:479–493. doi: 10.1016/s1532-0456(00)00198-8. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. Genbank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Dallinger R, Gehrig P, Hunziker PE. Primary structure of a copper-binding metallothionein from mantle tissue of the terrestrial gastropod Helix pomatia L. Biochem J. 1997;328 (Pt 1):219–224. doi: 10.1042/bj3280219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodar CMW, Kluytmans JH, Van Montfort JCP, Voogt PA, Zandee DI. Cadmium resistance and the synthesis of metallothionein-like proteins in Daphnia magna. Proceedings of the 3rd International Conference of Environmental Contamination, CEP; Edingburgh. 1998. pp. 79–81. [Google Scholar]
- Bofill R, Orihuela R, Romagosa M, Domènech J, Atrian S, Capdevila M. Caenorhabditis elegans metallothionein isoform specificity – metal binding abilities and the role of histidine in CeMT1 and CeMT2. FEBS J. 2009;276:7040–7056. doi: 10.1111/j.1742-4658.2009.07417.x. [DOI] [PubMed] [Google Scholar]
- Capdevila M, Atrian S. Metallothionein protein evolution: a miniassay. J Biol Inorg Chem. 2011;16:977–989. doi: 10.1007/s00775-011-0798-3. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Cáceres CE, Carmel L, Casola C, Choi J, Detter JC, Dong Q, Dusheyko S, Eads BD, Fröhlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kültz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Singan VR, Gilbert D. wFleabase: the Daphnia genome database. BMC Bioinform. 2005;6:45. doi: 10.1186/1471-2105-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallinger R, Berger B, Hunziker P, Kägi JHR. Metallothioneins in Snail Cd and Cu metabolism. Nature. 1997;388:237–238. doi: 10.1038/40785. [DOI] [PubMed] [Google Scholar]
- Daphnia Genome Consortium. Daphnia magna genome sequence project. 2013 https://wiki.cgb.indiana.edu/display/DGC/Initiatives.
- Del Ramo J, Torreblanca A, Martinez M, Pastor A, Diaz-Mayans J. Quantification of cadmium-induced metallothionein in crustaceans by the silver-saturation method. Mar Environ Res. 1995;39:121–125. [Google Scholar]
- Ebert D. A genome for the environment. Science. 2011;331:539–540. doi: 10.1126/science.1202092. [DOI] [PubMed] [Google Scholar]
- Engel DW, Brouwer M. Metal regulation and molting in the blue crab, Callinectes sapidus: metallothionein function in metal metabolism. Biol Bull. 1987;173:239–251. doi: 10.2307/1541876. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fowler BAHC, Kojima Y, Webb M. Nomenclature of metallothionein. Experientia Suppl. 1987;52:19–22. doi: 10.1007/978-3-0348-6784-9_2. [DOI] [PubMed] [Google Scholar]
- Franzellitti S, Fabbri E. Differential HSP70 gene expression in the Mediterraneanmussel exposed to various stressors. Biochem Biophys Res Commun. 2005;336:1157–1163. doi: 10.1016/j.bbrc.2005.08.244. [DOI] [PubMed] [Google Scholar]
- Haq F, Mahoney M, Koropatnick J. Signalling events for metallothionein induction. Mutat Res. 2003;533:211–226. doi: 10.1016/j.mrfmmm.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet. 2010;11:97. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Jenny MJ, Warr GW, Ringwood AH, Baltzegar DA, Chapman RW. Regulation of metallothionein genes in the American oyster (Crassostrea virginica): Ontogeny and differential expression in response to different stressors. Gene. 2006;379:156–65. doi: 10.1016/j.gene.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Kato Y, Matsuura T, Watanabe H. Genomic integration and germline transmission of plasmid injected into crustacean Daphnia magna eggs. PLoS One. 7:e45318. doi: 10.1371/journal.pone.0045318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby C, Nordborg N, Kusuhara K, Kristensen KM, Neufer PD, Pilegaard H. Gene expression in human skeletal muscle: alternative normalization method and effect of repeated biopsies. Eur J Appl Physiol. 2005;95:351–360. doi: 10.1007/s00421-005-0022-7. [DOI] [PubMed] [Google Scholar]
- Mao H, Wang D-H, Yang W-X. The involvement of metallothionein in the development of aquatic invertebrate. Aquat Toxicol. 2012;110–111:208–213. doi: 10.1016/j.aquatox.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Martinez M, Del Ramo J, Torreblanca A, Diaz-Mayans J. Cadmium toxicity, accumulation and metallothionein induction in Echinogammarus echinosetosus. J Environ Sci Health. 1996;A31:1605–1617. [Google Scholar]
- Poyton HC, Varshavsky JR, Chang B, Cavigiolio G, Chan S, Holman PS, Loguinov AV, Bauer DJ, Komachi K, Theil EC, Perkins EJ, Hughes O, Vulpe CD. Daphnia magna ecotoxicogenomics provides mechanistic insights into metal toxicity. Environ Sci Technol. 2007;41:1044–1050. doi: 10.1021/es0615573. [DOI] [PubMed] [Google Scholar]
- Poynton HC, Loguinov AV, Varshavsky JR, Chan S, Perkins EJ, Vulpe CD. Gene expression profiling in Daphnia magna Part I: Concentration-dependent profiles provide support for the No Observed Transciptional Effect Level. Environ Sci Techol. 2008;42:6250–6256. doi: 10.1021/es8010783. [DOI] [PubMed] [Google Scholar]
- Riddell DJ, Culp JM, Baird DJ. Behavioral responses to sublethal cadmium exposure within an experimental aquatic food web. Environ Toxicol Chem. 2005;24:431–441. doi: 10.1897/04-026r.1. [DOI] [PubMed] [Google Scholar]
- Ritz C, Spiess A. qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics. 2008;24:1549–1551. doi: 10.1093/bioinformatics/btn227. [DOI] [PubMed] [Google Scholar]
- Roelofs D, Marien J, Van Straalen NM. Differential gene expression profiles associated with heavy metal tolerance in the soil insect Orchesella cincta. Insect Biochem Mol Biol. 2007;37:287–295. doi: 10.1016/j.ibmb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Roesijadi G. Metallothioneins in metal regulation and toxicity in aquatic animals. Aquat Toxicol. 1992;22:81–113. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saydam N, Adams TK, Steiner F, Schaffner W, Freedman JH. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J Biol Chem. 2002;277:20438–20445. doi: 10.1074/jbc.M110631200. [DOI] [PubMed] [Google Scholar]
- Shaw JR, Colbourne JK, Davey JC, Glaholt SP, Hampton TH, Chen CY, Folt CL, Hamilton JW. Gene response profiles for Daphnia pulex exposed to the environmental stressor cadmium reveals novel crustacean metallothioneins. BMC Genomics. 2007;8:477. doi: 10.1186/1471-2164-8-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soazig L, Marc L. Potential use of the levels of the mRNA of a specific metallothionein isoform (MT-20) in mussel (Mytilus edulis) as a biomarker of cadmium contamination. Mar Pollut Bull. 2003;46:1450–1455. doi: 10.1016/S0025-326X(03)00283-2. [DOI] [PubMed] [Google Scholar]
- Spanier KI, Leese F, Mayer C, Colbourne JK, Gilbert D, Pfrender ME, Tollrian R. Predator-induced defences in Daphnia pulex: selection and evaluation of internal reference genes for gene expression studies with real-time PCR. BMC Mol Biol. 2010;11:50. doi: 10.1186/1471-2199-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squibb KS, Cousin RJ. Synthesis of metallothionein in a polysomal cell-free system. Biochem Biophys Res Commun. 1977;75:806–812. doi: 10.1016/0006-291x(77)91544-3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom M, Jakubov E, Rinkevich B, Herut B. Monitoring of hepatic metallothionein mRNA levels in the Fish Lithognathus mormurus- evaluation of transition metal pollution in a Mediterranean coast. Mar Pollut Bull. 1999;38:503–508. [Google Scholar]
- Valls M, Bofill R, Gonzalez-Duarte R, Gonzalez-Duarte P, Capdevila M, Atrian S. A new insight into metallothionein (MT) classification and evolution. The in vivo and in vitro metal binding features of Homarus americanus recombinant MT. J Biol Chem. 2001;276:32835–32843. doi: 10.1074/jbc.M102151200. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viarengo A, Burlando B, Dondero F, Marro A, Fabbri R. Metallothionein as a tool in biomonitoring programmes. Biomarkers. 1999;4:455–466. doi: 10.1080/135475099230615. [DOI] [PubMed] [Google Scholar]
- Woo S, Yum S, Jung JH, Shim WJ, Lee CH, Lee TK. Heavy metal-induced differential gene expression of metallothionein in Javanes Medaka, Oryzias javanicus. Mar Biotechnol (NY) 2006;8:645–662. doi: 10.1007/s10126-006-6046-0. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]