Abstract
The phosphoglycerate kinase-2 (Pgk2) gene is regulated in a tissue-, cell type- and developmental stage-specific manner during spermatogenesis and is required for normal sperm motility and fertility in mammals. Activation of Pgk2 transcription is regulated by testis-specific demethylation of DNA and binding of testis-specific transcription factors to enhancer and core promoter elements. Here we show that chromatin remodeling including reconfiguration of nucleosomes and changes in histone modifications is also associated with transcriptional activation of the Pgk2 gene during spermatogenesis. Developmental studies indicate the order of events involved in transcriptional activation of the Pgk2 gene includes demethylation of DNA in T1- and T2-prospermatogonia, binding of a factor to the CAAT-box in type A and B spermatogonia, followed by recruitment of chromatin remodeling factors, displacement of a nucleosome from the Pgk2 promoter region, binding of factors to the Pgk2 core promoter and enhancer regions and, finally, initiation of transcription in primary spermatocytes. Transgene studies show that Pgk2 core promoter elements are required to direct demethylation of DNA and reconfiguration of nucleosomes, whereas both enhancer and core promoter elements are required to direct changes in histone modifications and initiation of transcription. These results provide novel insight into the developmental order of molecular events required to activate tissue-specific transcription of the Pgk2 gene, the distinct elements in the 5′-regulatory region of the Pgk2 gene that regulate each of these events, and the relationship among these events in that each step in this process appears to be a necessary prerequisite for the subsequent step.
Keywords: spermatogenesis, nucleosomes, histone modifications, CAAT-box
Introduction
Spermatogenesis is a highly ordered process by which the germ cell lineage gives rise to functional gametes in the male. As with other specific lineages in multicellular organisms, the characteristic differentiation of the male germ line proceeds as a result of expression of unique combinations of genes (Eddy 2002, Shima et al. 2004). Among the many germ cell type-specific genes expressed during spermatogenesis in mammals is a set encoding testis-specific glycolytic isozymes. These include the Ldhc, Gapdhs, Hk1s, Pgm2, Pdha2 and Pgk2 genes, each of which encode sperm-specific isozymes of glycolytic enzymes (Fitzgerald et al. 1992, Mori et al. 1993, Eddy 2002, Boussouar & Benahmed 2004, Tang et al. 2008).
Phosphoglycerate kinase (PGK) is a key enzyme involved in the metabolism of glucose via glycolysis. In most mammals, PGK is encoded by two genes, Pgk1 and Pgk2 (VandeBerg et al. 1973, VandeBerg 1985, McCarrey & Thomas 1987). Pgk1 is an X-linked gene that is ubiquitously expressed in all somatic cells, oogenic cells and premeiotic spermatogenic cells (McCarrey et al. 1992b). The autosomal Pgk2 gene is expressed in a tissue-, cell type- and developmental stage-specific manner during spermatogenesis, and is required for normal sperm motility and fertility (VandeBerg et al. 1973, McCarrey et al. 1992b, Danshina et al. 2010). Because the spermatogenic cell lineage is particularly well characterized and accessible in mammals, it provides an excellent system in which to chronicle events leading up to activation of tissue-specific gene expression (McCarrey 1998).
Pgk2 transcription is first seen in primary spermatocytes, with transcript levels increasing throughout first meiotic prophase and on into postmeiotic round spermatids (McCarrey et al. 1992).. Transcription of this gene continues until midway through the process of spermiogenesis when the spermatid nucleus becomes so condensed that all transcription ceases (Kumari et al 1996). We have previously demonstrated that a fragment carrying as little as 230 bp of the Pgk2 5′-regulatory region is sufficient to direct proper tissue-, cell-type and developmental-stage-specific transcription of a reporter gene in transgenic mice (Robinson et al. 1989, Zhang et al. 1999), suggesting this region contains sequence elements sufficient to direct all molecular changes required for properly regulated transcriptional activation of this gene. In addition, we characterized protein-DNA interactions associated with transcriptional activation of the Pgk2 gene and showed that the Sp3 factor is bound to a GC-box in the Pgk2 core promoter, and that PBX4, along with its co-activator, PREP1, are bound to the E3-E4 enhancer region immediately upstream of the Pgk2 core promoter in spermatogenic cells in which this gene is actively expressed (Yoshioka et al. 2007). Surprisingly however, we found that a CAAT-box, which is also present in the Pgk2 core promoter, is unbound in these same expressing cells although we did observe an in vivo footprint at this element in earlier spermatogenic cell types (spermatogonia) prior to the onset of Pgk2 transcription (Yoshioka et al. 2007).
We have also examined changes in DNA methylation at the Pgk2 locus and found that a domain of demethylation begins to develop over the Pgk2 promoter region in T1-prospermatogonia (McCarrey 2013) and becomes complete by the primitive type A spermatogonium stage (Geyer et al. 2004). This demethylation precedes transcription factor binding at the Pgk2 promoter/enhancer by several days (Geyer et al. 2004). Changes in DNA methylation are often associated with changes in chromatin structure (Robertson 2002), and at least two classes of chromatin modifications have been shown to contribute to regulation of transcription. One is ATP-dependent chromatin remodeling, which mobilizes nucleosomes by weakening the contacts between histone octamers and DNA, including transient unwrapping of the DNA from core histones or displacement of nucleosomes relative to specific landmarks in the promoter regions of genes, both of which enhance accessibility of key promoter elements to transcription factors (Becker & Horz 2002, Flaus & Owen-Hughes 2004, Smith & Peterson 2005, Cairns 2009).
A second predominant type of chromatin modification involves post-translational modifications of histones, which include a variety of different types of modifications occurring on different amino acids in various positions within the core histones (Sterner & Berger 2000, Jenuwein & Allis 2001). Such modifications are particularly prominent on the N-terminal tails of histones H3 and H4, and include lysine acetylation, lysine or arginine methylation, and serine or threonine phosphorylation among others (Berger 2002, Lachner et al. 2003, Kurdistani et al. 2004). These modifications can lead directly to altered structures of chromatin and/or can provide binding sites for regulatory proteins that further influence chromatin structure and transcription (Cairns 2009, Ho & Crabtree 2010). In general, chromatin structure and nucleosome positioning have been found to be distinctly different in a tissue-specific gene in cell types in which the gene is actively expressed or terminally repressed, respectively (Narlikar et al. 2002, Yuan et al. 2005, Mavrich et al. 2008, Schones et al. 2008), suggesting that these parameters are related to mechanisms regulating transcription.
The objectives of the study reported here were to: 1) examine histone modifications and nucleosome reconfiguration in the Pgk2 gene to identify features of chromatin structure that are unique to the actively expressed state of this gene, 2) determine the timing of appearance of these features relative to the timing of initiation of transcription of the Pgk2 gene during spermatogenesis and the timing of other molecular changes associated with activation of this gene, and 3) identify elements in the 5′-regulatory region of the Pgk2 gene that are involved in signaling or directing these chromatin changes. Our results indicate there is an ordered series of molecular events that lead to activation of transcription of the Pgk2 gene in spermatogenic cells, and that these events are signaled by distinct elements in the Pgk2 promoter/enhancer region. In addition, our data suggest that the occurrence of each of these events is largely dependent upon the occurrence of the preceding event.
Materials and Methods
Generation of transgenic mice
515 Pgk2-CAT/WT and 188 Pgk2-CAT transgenic mice (Mus musculus) were prepared for previous studies as described (Robinson et al. 1989). Three new lines of mice carrying a 515 Pgk2-CAT/CAAT-minus transgene were generated for this study using similar procedures. The transgene used to generate these new lines was identical to that used for the 515 Pgk2-CAT/WT lines except that the CAAT-box in the Pgk2 core promoter was mutated to ATCA by in vitro site-directed mutagenesis using recombinant PCR as described (Higuchi 1990, Barik 1996). The cesium chloride-purified, linearized transgene construct was introduced into early BDF1 (C57Bl6 × DBA2 F1 hybrid) embryos by pronuclear injection at the University of Texas Health Science Center at San Antonio transgenic mouse core as previously described (Hogan et al. 1986). Tail snips from pups were initially screened for transgene integration by PCR with specific primers shown in Supplementary Table 1 as previously described (Walter et al. 1989). PCR-positive mice were further confirmed as founders by Southern blot hybridization to genomic DNA from tail tissue (data not shown). Three transgenic founders (#9, #10 and #20) were identified and used to establish separate lines for the study described here.
Preparation of cells
Somatic spleen cells and spermatogenic germ cells for micrococcal nuclease (MNase) sensitivity and chromatin immunoprecipitation (ChIP) assays were recovered from either the transgenic mouse lines described above or from wild-type CD-1 mice purchased from Charles River Laboratories (Wilmington, MA). All procedures involving live animals were approved in advance by the University of Texas at San Antonio Institutional Animal Care and Use Committee.
For isolation of spleen cells, spleens were dissected from freshly euthanized mice and placed in phosphate-buffered saline (PBS), then mechanically teased open with hypodermic needles. The resulting cell suspension was filtered through 100-μm nylon mesh (Millipore, Billerica, MA), then treated with hypotonic buffer to lyse any contaminating erythrocytes and pelleted by centrifugation at 900 rpm for 10 min at 4°C.
For isolation of germ cells, testes were dissected from freshly euthanized prepuberal (6-8 days postpartum [dpp]), puberal (18 dpp) or adult (≥60 dpp) CD-1 wild type or BDF1 transgenic mice, decapsulated and transferred into KREBS buffer [120.1 mM NaCl, 4.8 mM KCl, 25.2 mM NaHCO3, 1.2 mM KH2PO4, 1.2 mM MgSO4·7H2O, 1.3 mM CaCl2, 11.1 mM glucose, 1 mM glutamine, 10 ml/L of essential amino acids, 10 ml/L of nonessential amino acids, 100 μg/ml streptomycin, and 100 U/ml penicillin G (K + salt)] containing 0.5 mg/ml collagenase Type 2 (Worthington) and incubated in a water-bath for 15 minutes at 33°C with vigorous shaking. Cells from prepuberal or puberal testes were then collected by centrifugation at 900 rpm for 10 min at 4°C, while seminiferous tubule segments were collected from adult testes by pouring off the collagenase-containing KREBS buffer as described (Bellve 1993). The recovered cells or tubule segments were then washed with fresh KREBS buffer and incubated for another 15 minutes at 33°C with vigorous swirling in KREBS buffer containing 0.5 mg/ml trypsin (Sigma). At the end of this incubation, 1μg/ml deoxyribonuclease I (DNase I) was added and the cells were pipetted up and down repeatedly to dissociate the cells, and then filtered through 80μ nylon mesh to yield a single cell suspension. This yielded a “mixed germ cell” population representative of the spermatogenic cell types present in testes at each age of male mice utilized.
To obtain purified populations of specific spermatogenic cell types, the mixed germ cell population derived from testes at each age was loaded onto a 2–4% bovine serum albumin “StaPut” gradient and the cells were allowed to sediment through the gradient at unit gravity for 4 hours at 4°C as previously described (McCarrey et al. 1992, Bellve 1993, Yoshioka et al. 2007). Primitive type A spermatogonia were isolated from testes of male mice at 6 dpp. Type A and type B spermatogonia were isolated from testes of 8 dpp male mice. Preleptotene spermatocytes, leptotene-plus-zygotene spermatocytes, and early (juvenile) pachytene spermatocytes were isolated from the testes of 18 dpp male mice. Pachytene spermatocytes, round spermatids, and elongated spermatids plus residual cytoplasmic bodies were isolated from the testes of adult (≥60 dpp) male mice. Testicular spermatozoa were isolated from the testes of adult mice by dissociation of tissue and sonication to lyse all other cells. The purity of each cell population was determined by morphological examination under phase optics. Purities of spermatogonial cell types and juvenile spermatocytes were ≥85%, while those of adult spermatocytes, spermatids, and spermatozoa were ≥95%.
Standard RT-PCR and real-time qRT-PCR
For standard RT-PCR and real-time qRT-PCR assays, total cellular RNA was extracted from each cell population using TRIzol reagent according to the manufacturer’s instructions (Invitrogen). The resulting RNA was quantified by Nanodrop (Thermo Scientific). Before reverse transcription, 1 μg of total RNA was treated with 1 unit of RQ1 RNase-free DNase (Fisher Scientific) for 30 minutes at 37°C to remove any DNA contamination. The RNA was then used as template to synthesize first-strand cDNA using the iScript cDNA synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. To further control for the possibility of genomic DNA contamination, an additional first-strand cDNA synthesis reaction was conducted without reverse transcriptase (RT-) and exposed to PCR amplification as a no-RT control.
RT-PCR was performed to detect the CAT transgene or the coding sequence of the constitutively expressed housekeeping gene, Gapdh. These target sequences were amplified using the RT-PCR primers shown in Supplementary Table 1. PCR products were resolved by electrophoresis through 1.5% agarose gels and visualized by ethidium bromide staining and detection on a VersaDoc imaging system (Bio-Rad).
For real-time qRT-PCR, first-strand cDNAs were amplified using SYBR Premix Ex Taq (Perfect Real Time) (Takara Bio INC) on a Chromo4 Continuous Fluorescence Detector (Bio-Rad) using the primers listed in Supplementary Table 1. Relative expression of target mRNAs and 18s rRNAs expressed as copy number, was determined using a standard curve generated from DNA samples of known concentration.
Bisulfite genomic sequencing of DNA
To assess the status of DNA methylation at the Pgk2 promoter, we used bisulfite conversion followed by sequencing as previously described (Frommer et al. 1992, Clark et al. 1994). Total genomic DNA was extracted from germ cells or spleen cells using TRIzol reagent according to the manufacturer’s instructions (Invitrogen). After quantification by Nanodrop (Thermo Scientific), 500 ng of genomic DNA were treated with 130 μl of CT conversion reagent from the EZ DNA methylation-Gold Kit according to the manufacturer’s instructions (Zymo Research Corporation). Converted DNA was then purified using the QIAquick Gel Extraction Kit (QIAGEN), and amplified using primers delineating the promoter or coding regions of the Pgk2 gene (Supplementary Table 1). Anticipated bands were excised from the 1.0 % agarose gel and DNA was recovered using the QIAquick Gel Extraction Kit (QIAGEN). The products were cloned into the TOPO TA cloning vector (Invitrogen) according to the manufacturer’s instructions. Positive (white) colonies were selected and prepared for sequencing by the Nucleic Acids Core Facility at the University of Texas at San Antonio Health Science Center. The methylation status of each CpG dinucleotide was determined by comparison of sequences from amplimers of bisulfite-converted and untreated DNA, respectively.
Micrococcal nuclease sensitivity assay
A micrococcal nuclease sensitivity assay was used to map nucleosome positioning as previously described with minor modifications (Sekinger et al. 2005, Shim et al. 2007, Lam et al. 2008). Briefly, spleen or germ cells were isolated from transgenic (BDF1) or wild type (CD-1) mice, washed three times with ice-cold PBS buffer and resuspended in Tris buffered saline (TBS) prior to permeabilization with Tween 20 plus a proteinase inhibitor cocktail and phenylmethanesulfonyl fluoride (PMSF) Cells were then homogenized in a Dounce homogenizer using several strokes to release the nuclei, which were then pelleted and resuspended in digestion buffer [0.32 M sucrose, 50 mM Tris-HCl (pH7.5), 4 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF and 5 mM Na-butyrate] and incubated with the appropriate concentration of micrococcal nuclease (Sigma, N5386) at 37°C. The reaction was stopped by adding SDS lysis buffer and proteinase K, and incubation at 55°C overnight, followed by extraction of the DNA with phenol/chloroform and precipitation in ethanol. Subsequently the DNA was resuspended in TE buffer and RNase A was added and incubated for 30 min at 37°C, followed by electrophoresis of the resulting mononucleosome DNA fragments through 1% agarose gel. The fragments were then recovered using a QIAquick Gel Extraction Kit (QIAGEN) and redissolved in PCR grade water in preparation for real time qPCR.
For analysis of MNase sensitivity in the endogenous Pgk2 gene and at Pgk2 transgenes, we used real-time qPCR with a series of primer pairs that delineated amplimers spanning the 5′-half of the Pgk2 gene, with each amplimer of 100 ± 8 bp representing regions of the gene located 40 ± 10 bp away from each other (Supplementary Table 1). The relative enrichment of target sequences was determined on the basis of copy numbers, which were calculated using a standard curve generated from DNA of known concentrations as described above.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using an immunoprecipitation assay kit (Abcam, #ab500) according to the manufacturer’s instructions, with minor modifications. After isolation from mice, spleen and germ cells were fixed in a final concentration of 1% formaldehyde for 10 min at 37°C, and then the reaction was stopped by adding 125 mM glycine. After cells were washed once with PBS, the cells were lysed, the lysate was sonicated using a Branson Digital Sonifier (Branson Ultrasonics) for 6 minutes with alternating 20 seconds on and 20 seconds off intervals at 45% amplitude, and with a longer pause every 2 minutes to cool down the samples. This sheared the chromatin to a fragment size range of 100-1,000 bp. The sample was centrifuged to remove cell debris and immunoprecipitations were performed with antibodies against acetyl-histone H3, acetyl-histone H4, and tri-methylation of lysine 4 of histone H3 (H3k4me3) using antibodies from Active Motif. Antibody to rabbit IgG (Abcam) was used as a non-specific, negative control. After samples were incubated with antibody overnight at 4°C, protein G beads were added to pull down immunoprecipitated complexes, which were then washed with ChIP buffer (provided in the Abcam KIT), followed by DNA purification using the DNA purifying slurry (provided in the Abcam KIT) plus 1μl of proteinase K (provided in the Abcam KIT) to digest proteins following the kit instructions.
The resulting DNA was then analyzed by real-time qPCR on a Chromo4 real-time PCR detection system (Bio-Rad) using SYBR Premix Ex Taq (Perfect Real time, Takara Bio INC.) Relative amounts of DNA in the input and bound fractions were determined on the basis of copy numbers, using a standard curve generated from DNA samples of known concentration. Values for enrichment were calculated as the average from three independent samples.
Results
Transgenic mice examined in this study
Three different transgenes were used in this study (Fig. 1B). These included a construct carrying 515 bp of 5′-flanking sequence from the human Pgk2 gene including the Pgk2 core promoter plus the E1/E2 and E3/E4 enhancer regions (515 Pgk2-CAT/WT). We have previously shown that this transgene is expressed in the same tissue-, cell type- and developmental stage-specific manner as the endogenous mouse Pgk2 gene, indicating all regulatory information required to direct this transcriptional pattern is included in this fragment of the Pgk2 gene (Robinson et al. 1989, Zhang et al. 1999). This reflects the fact that both the human and mouse Pgk2 gene promoters contain highly conserved regulatory elements including a GC-box, a CAAT-box and an enhancer region just upstream from the CAAT-box (McCarrey, 1987, 1990). A second transgene (188 Pgk2-CAT/WT) carried the human Pgk2 core promoter, but not the upstream E1/E2 + E3/E4 enhancer region. This transgene is not expressed in any tissue in transgenic mice, including spermatogenic cells (Robinson et al. 1989, Zhang et al. 1999). To investigate the function of the CAAT-box in the Pgk2 core promoter, we produced three new lines of transgenic mice carrying a transgene identical to the 515 Pgk2-CAT/WT transgene except that the CAAT-box was mutated from CAAT to ATCA (515 Pgk2-CAT/CAAT-minus). Because this transgene carries all other sequence information normally required to direct properly regulated transcription of the Pgk2 gene, this construct allowed us to focus on the function of the CAAT-box in this process. Importantly, as noted in our previous studies (Robinson et al. 1989, Zhang et al. 1999), expression of the promoter-reporter transgene, which encodes the prokaryotic chloramphenicol acetyl transferase enzyme, imposes no measurable phenotypic effects on testis or systemic functions or characteristics in the transgenic mice
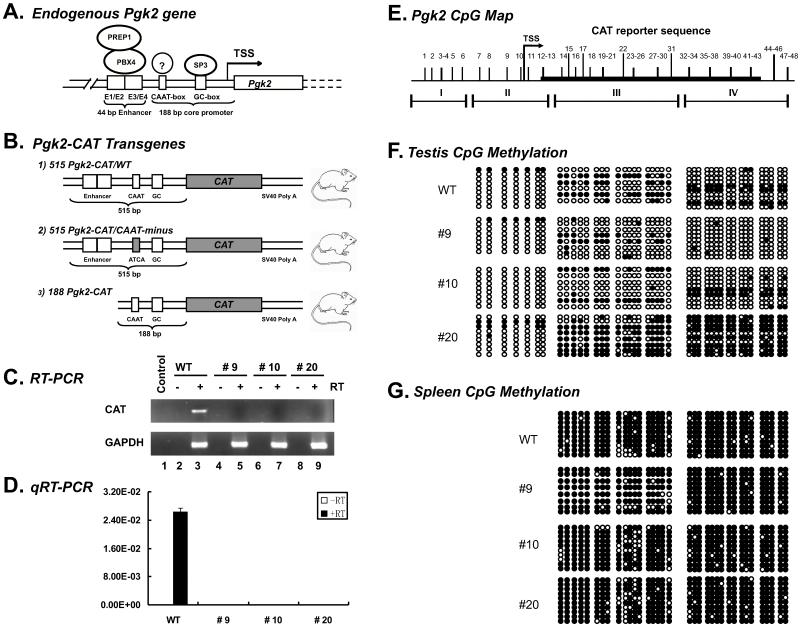
FIGURE 1. Effects of disruption of the Pgk2 CAAT-box on transcription and demethylation of the Pgk2 gene.
(A) A map of the 5′ regulatory region of the Pgk2 gene is shown. A 188 bp core promoter includes the transcription start site (TSS) at the +1 position, a GC-box at −30 to −22, and a CAAT-box at −105 to −101. A 44 bp enhancer region lies immediately upstream from the core promoter and includes two sub-regions, E1/E2 and E3/E4. In cells actively expressing the Pgk2 gene, the Sp3 factor is bound to the GC-box in the core promoter and the PBX4 transcriptional activator and its co-activator, PREP1, are bound to the E1/E2 portion of the enhancer region. Prior to initiation of transcription (but not during transcription), an unknown factor (?) is bound to the CAAT-box. (B) Three different transgenes were used for this study: 515 Pgk2-CAT/WT – a transgene which includes the wild type core promoter and enhancer region sequences, 515 Pgk2-CAT/CAAT-minus – a transgene identical to the 515 Pgk2-CAT/WT transgene except that the CAAT-box has been mutated to ATCA, and 188 Pgk2-CAT – a transgene which includes the wild type Pgk2 core promoter sequence but lacks the enhancer region. Mice carrying the 515 Pgk2-CAT/WT and 188 Pgk2-CAT transgenes were generated for a previous study (Robinson et al. 1989), whereas three lines carrying the 515 Pgk2-CAT/CAAT-minus transgene were generated for this study. All transgenes were composed of human Pgk2 promoter fragments ligated to the bacterial chloramphenicol acetyltransferase (CAT) reporter gene plus the simian virus 40 (SV40) 3′-UTR poly(A) addition sequences. The 515 Pgk2-CAT/WT transgene showed strong expression in mouse testis (WT), however, the 515 Pgk2-CAT/CAAT-minus showed no detectable expression in testis tissue from any of the three lines carrying this transgene (#9, #10, #20) as measured by either standard RT-PCR (C) or qRT-PCR (D). A map of the 5′ half of the Pgk2 gene (E) shows the locations of 42 CpG dinucleotides and the four amplicons, I, II, III and IV, used to analyze DNA methylation at these sites by bisulfite genomic sequencing analysis of testis tissue (F), or somatic spleen tissue (G) from each line. Unmethylated CpGs are represented as open circles, whereas methylated CpGs are represented as filled circles. Each row of circles represents analysis of a different original DNA strand. The 515 Pgk2-CAT/WT line and all three 515 Pgk2-CAT/CAAT-minus lines all showed complete or partial demethylation in testis tissue, but remained hypermethylated in spleen tissue. TSS = transcription start site, GC = GC-box, CAAT = CAAT-box, ATCA = mutant sequence substituted for the CAAT-box in the 515 Pgk2-CAT/CAAT-minus transgene.
The CAAT-box in the Pgk2 core promoter is required for initiation of transcription
We used RT-PCR to examine expression of the 515 Pgk2-CAT/WT and 515 Pgk2-CAT/CAAT-minus transgenes in testis RNA (Fig. 1C). As expected, we confirmed expression of the 515 Pgk2-CAT/WT transgene in testis tissue. However, the 515 Pgk2-CAT/CAAT-minus transgene was not expressed in testis tissue recovered from males of any of the three different lines we generated carrying this transgene (#9, #10, #20). This indicates that activation of transcription cannot occur in the absence of an intact CAAT-box in the Pgk2 core promoter. To confirm these results, we also performed real time qRT-PCR to detect the expression of these same transgenes, and this also showed that the 515 Pgk2-CAT/WT transgene was expressed in testis tissue whereas the 515 Pgk2-CAT/CAAT-minus transgene was not (Fig. 1D).
The CAAT-box in the Pgk2 core promoter is not required for tissue-specific demethylation of DNA in the Pgk2 gene
We used bisulfite genomic sequencing to determine if the absence of an intact CAAT-box would inhibit the demethylation process that normally occurs over the 5′-half of the Pgk2 gene prior to activation of Pgk2 transcription (Geyer et al. 2004). We previously showed that both the 515 Pgk2-CAT/WT and 188 Pgk2-CAT transgenes undergo tissue-, cell type-, and developmental stage-specific demethylation of DNA in early spermatogenic cells (T1- and T2-prospermatogonia and type A spermatogonia) in a manner analogous to that which the endogenous Pgk2 gene undergoes prior to initiation of transcription in spermatogenic cells in vivo (Ariel et al. 1994, Geyer et al. 2004). Importantly, both of those constructs carried an intact CAAT-box. Here we examined DNA methylation of the 515 Pgk2-CAT/CAAT-minus transgene in testicular cells from the three different lines of mice carrying this transgene (Fig. 1F).
While the 515Pgk2-CAT/CAAT-minus transgene in line #20 showed only partial demethylation of the 5′-portion of the transgene, lines #9 and #10 showed demethylation to a similar extent as that seen in the 515 Pgk2-CAT/WT transgene. Further, in each case, the observed demethylation event was testis-specific as indicated by the hypermethylated status of both the 515 Pgk2-CAT/WT and 515 Pgk2-CAT/CAAT-minus transgenes in spleen tissue from the same mice in which methylation of the transgenes in germ cells was assessed (Fig. 1G). This indicates that the CAAT-box alone is not the signal directing tissue-specific demethylation of the Pgk2 gene.
Testis-specific reconfiguration of nucleosomes in the Pgk2 promoter accompanies active Pgk2 tramscription
Most genomic DNA in eukaryotes is incorporated into regularly, albeit randomly spaced nucleosomes to facilitate packaging and various processes associated with the DNA (Rando & Chang 2009). However, regions of DNA closely associated with nucleosome core histones are typically inaccessible for binding by regulatory factors and/or the transcriptional pre-initiation complex (Jenuwein & Allis 2001). This conflict is normally resolved by reconfiguration of nucleosome presence and/or positioning relative to specific promoter sites required for protein-DNA interactions that facilitate activation of transcription (Cairns 2009). Thus, multiple studies have revealed differences in nucleosome positioning associated with the same gene in cells where the gene is actively expressed or terminally repressed, respectively (Sekinger et al. 2005, Lam et al. 2008, Leimgruber et al. 2009). In addition, genome-level methodologies such as ChIP-Chip or ChIP-Seq have revealed tissue- or cell type-specific patterns of nucleosomal reconfiguration in multiple species including Saccharomyces cerevisiae (Yuan et al. 2005, Lee et al. 2007), Drosophila melanogaster (Mavrich et al. 2008) and Caenorhabditis elegans (Valouev et al. 2008), as well as in promoter (Ozsolak et al. 2007, Schones et al. 2008) and enhancer (He et al. 2010) regions in the human genome. These studies reveal a characteristic pattern of nucleosome reconfiguration leading to unobstructed access facilitating binding of factors to enhancer, core promoter and transcription start site regions in tissue-specific genes.
We examined nucleosome positioning at the Pgk2 gene promoter and enhancer region to determine if similar tissue-specific differences in nucleosome positioning are associated with different transcriptional states of the Pgk2 gene. We isolated somatic spleen cells in which the Pgk2 gene is terminally repressed, and spermatogenic cells at stages prior to and during the time when the Pgk2 gene is actively transcribed, and used the micrococcal nuclease (MNase) sensitivity assay to map nucleosome positioning in the 5′-flanking region of the Pgk2 gene in each cell type. Consistent with similar studies of other tissue-specific genes, we found that nucleosome positioning in the Pgk2 gene is distinctly different in non-expressing spleen cells and expressing spermatogenic cells. Thus, in spleen cells, we found occupancy at the −1 nucleosome position which includes the Pgk2 promoter/enhancer region plus the nearby transcription start site (Fig. 2B), while in a mixed population of pachytene spermatocytes and round spermatids in which the Pgk2 gene is actively expressed, the −1 nucleosome position was unoccupied (Fig. 2C). At the same time, the +1 nucleosome position remained occupied in both non-expressing spleen cells (Fig. 2B) and actively expressing germ cells (Fig. 2C), indicating a location-specific reconfiguration of nucleosomes in the Pgk2 5′-regulatory region.
FIGURE 2. Nucleosome positioning in the endogenous Pgk2 gene in somatic and spermatogenic cells in the mouse.
The micrococcal nuclease (MNase) sensitivity assay was used to examine nucleosome positioning in the endogenous mouse Pgk2 gene in somatic cells in which the Pgk2 gene is terminally repressed, premeiotic spermatogonia in which the Pgk2 gene is undergoing potentiation prior to initiation of expression, and meiotic spermatocytes and postmeiotic spermatids in which the Pgk2 gene is actively expressed. Regions containing elevated amplimer levels represent the site of a nucleosome that inhibited access of the MNase to the underlying DNA, whereas regions of low amplimer levels represent more accessible sites devoid of nucleosomes. (A) Maps of the 5′ half of the Pgk2 gene provide context for each set of MNase assays. (B) Somatic spleen cells show the presence of nucleosomes (dark grey ovals) at the −1 region, which covers the Pgk2 enhancer and core promoter regions and at the +1 position within the Pgk2 coding sequence downstream of the transcription start site. (C) A mixed population of spermatogenic cells consisting primarily of Pgk2-expressing spermatocytes and spermatids shows the absence of a nucleosome at the −1 position in these cells (dashed oval), but the continued presence of a nucleosome at the +1 position. (D) A purified population of primitive type A spermatogonia in which the Pgk2 gene is not yet expressed shows the presence of nucleosomes at both the −1 and +1 positions, whereas in pachytene spermatocytes (E) the −1 nucleosome is completely missing, and in round spermatids (F), the −1 nucleosome presence is reduced (light grey dashed oval), while the +1 nucleosome persists in all of these cell types.
To determine the timing of the reconfiguration of nucleosomes normally associated with activation of the Pgk2 gene in spermatogenic cells, we examined nucleosome positioning in the 5′ portion of the Pgk2 gene in premeiotic spermatogonia – a stage at which transcription of the Pgk2 gene has not yet begun (Gebara & McCarrey 1992, McCarrey et al. 1992, McCarrey et al. 2005), and in pachytene spermatocytes and round spermatids – meiotic and postmeiotic spermatogenic cell types in which active expression of the Pgk2 gene is ongoing (McCarrey et al. 1992, 2005). We found that the −1 nucleosome position in the Pgk2 promoter was still occupied in premeiotic spermatogonia (Fig. 2D), but then became unoccupied in pachytene spermatocytes (Fig. 2E) and was still partially, though not as completely, unoccupied in round spermatids (Fig. 2F). Note that transcription of the Pgk2 gene ceases immediately after the round spermatid stage in elongating spermatids (McCarrey et al. 1992). Importantly, although we observed some variation among the different cell types investigated in the overall extent of post-MNase digestion signal intensity detected for each gene region, the differences were consistent within each gene region and were consistently distinct between those cell types in which the corresponding gene was or was not expressed. The post-MNase digestion signals do not reach 100% or 0% in any case because a) the purities of the enriched cell populations investigated were 85-95%, and b) reconfiguration of nucleosomes on any promoter is likely to reflect an equilibrium state that never reaches 100% or 0% simultaneously in all cells examined, even within a particular cell type.
The CAAT-box is required for reconfiguration of nucleosomes in the Pgk2 promoter
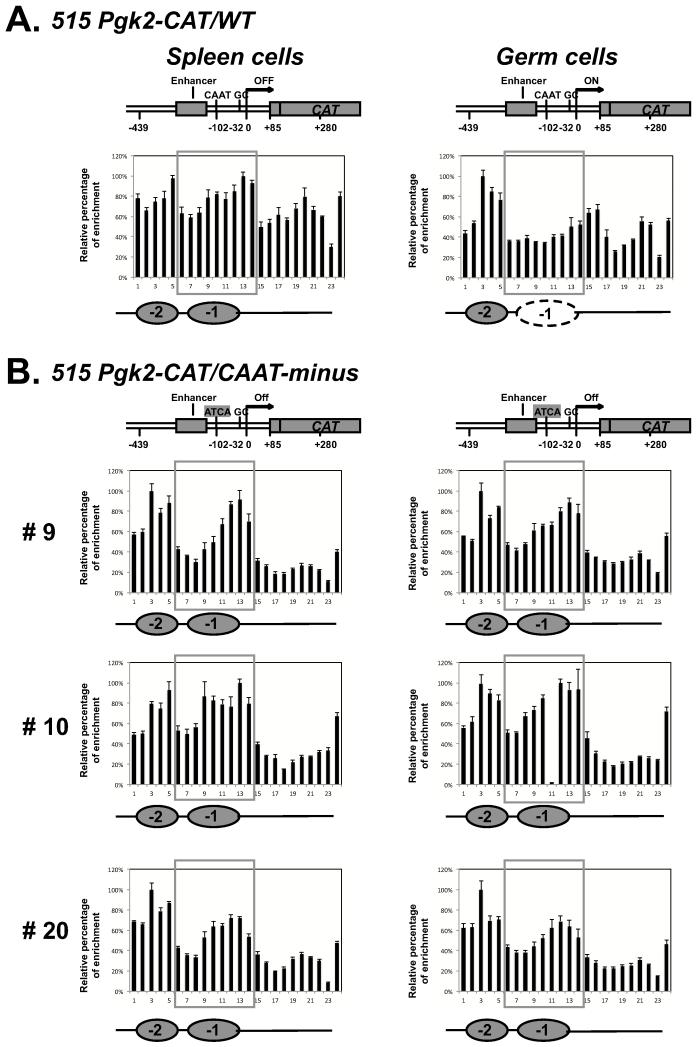
The tissue- and location-specific reconfiguration of nucleosomes at the −1 position in the 5′-region of the Pgk2 gene implies a highly regulated process. Because we had previously observed an in vivo footprint over the CAAT-box in spermatogenic cells prior to, but not during the time of active transcription of the Pgk2 gene, we next examined the extent to which normal nucleosome reconfiguration occurred in 515 Pgk2-CAT transgenes lacking an intact CAAT-box (i.e. in 515 Pgk2-CAT/CAAT-minus transgenes). We first confirmed that the 515 Pgk2-CAT/WT transgene carrying an intact CAAT-box undergoes a tissue-specific reconfiguration of nucleosomes at the −1 position in a manner similar to that we observed in the endogenous Pgk2 gene (Fig. 3A). Interestingly, in addition to nucleosome occupancy at the −1 position in this transgene, we also observed occupancy at a site upstream from the −1 position in both spleen and germ cells that we termed the −2 position (Fig. 3A). This was not observed in the endogenous Pgk2 gene, presumably because the sequence differs in the regions upstream of the endogenous Pgk2 gene and any integrated Pgk2 transgenes, respectively. We also did not observe consistent nucleosome occupancy at the +1 position in these transgenes, which we suspect also reflects sequence differences between the transgenes (carrying the CAT reporter gene downstream of the Pgk2 promoter) and the endogenous Pgk2 gene. However, the key observation was that when nucleosome mapping was performed on the 5′-portion of the 515 Pgk2-CAT/WT transgene in non-expressing spleen cells and in expressing spermatocytes or spermatids, respectively, only occupancy at the −1 position changed, becoming unoccupied in expressing germ cells consistent with the result we observed for the endogenous Pgk2 gene.
FIGURE 3. Nucleosome positioning in Pgk2 transgenes with or without intact CAAT-boxes.
(A) The MNase sensitivity assay was used to examine nucleosome positioning in the 515 Pgk2-CAT/WT transgene in spleen cells and in a mixed population of male germ cells (pachytene spermatocytes and round spermatids). A nucleosome was present at the −1 position over the Pgk2 enhancer and core promoter regions in non-expressing spleen cells, but was absent from this position in expressing germ cells. An additional nucleosome was persistently present further upstream in what has been termed the −2 position, but no persistent nucleosome was detected at the +1 position in these transgenes. Note that the DNA sequence in the −2 and +1 regions of these transgenes differs from that in the equivalent positions of the endogenous Pgk2 gene. (B) The nucleosome at the −1 position persisted in the 515 Pgk2-CAT/CAAT-minus transgene in both spleen cells and male germ cells in all three lines investigated.
We next examined nucleosome positioning in the 515 Pgk2-CAT/CAAT-minus transgenes in these same cell types. In all three lines carrying the CAAT-minus transgene, there was no difference in nucleosome occupancy in spleen and germ cells, respectively (Fig. 3B), such that the −1 position remained occupied even in spermatogenic cell types in which the equivalent position in the endogenous Pgk2 gene or in the 515 Pgk2-CAT/WT transgene became unoccupied. This correlated with the lack of expression of this transgene in testis tissue from any of these lines described above. Thus, it appears that an intact CAAT-box is required for both reconfiguration of nucleosomes over the Pgk2 promoter and tissue-specific initiation of transcription from this promoter.
Testis-specific modifications of histones accompany activation of Pgk2 transcription
A common molecular event that accompanies physical remodeling of chromatin and activation of tissue-specific transcription is modifications of core histones (Jenuwein & Allis 2001). Both histone tails and globular domains are subject to a vast array of posttranslational modifications sometimes referred to as the histone code (Berger 2002, Barth & Imhof 2010). These modifications include acetylation, methylation, ubiquitination, ADP-ribosylation, and sumolation of lysine residues, methylation of arginine residues, and other modifications (Kouzarides 2007). Commonly, acetylation of histone H3 and histone H4 (acetyl-H3 and acetyl-H4), and tri-methylation of lysine 4 of histone H3 (H3k4me3), are associated with a euchromatic chromatin structure and active transcription. Therefore we used the chromatin immunoprecipitation (ChIP) assay to investigate these three modifications in different regions of the Pgk2 gene in somatic spleen cells in which the Pgk2 gene is terminally repressed, premeiotic spermatogonia in which the Pgk2 gene is not yet expressed, and spermatocytes and spermatids in which the Pgk2 gene is actively expressed (Fig. 4). We also investigated similar modifications in two control genes in the same cell populations – the ß-actin gene which is constitutively expressed in spermatogenic and spleen cells and the Cryaa gene which is terminally repressed in these same cell types.
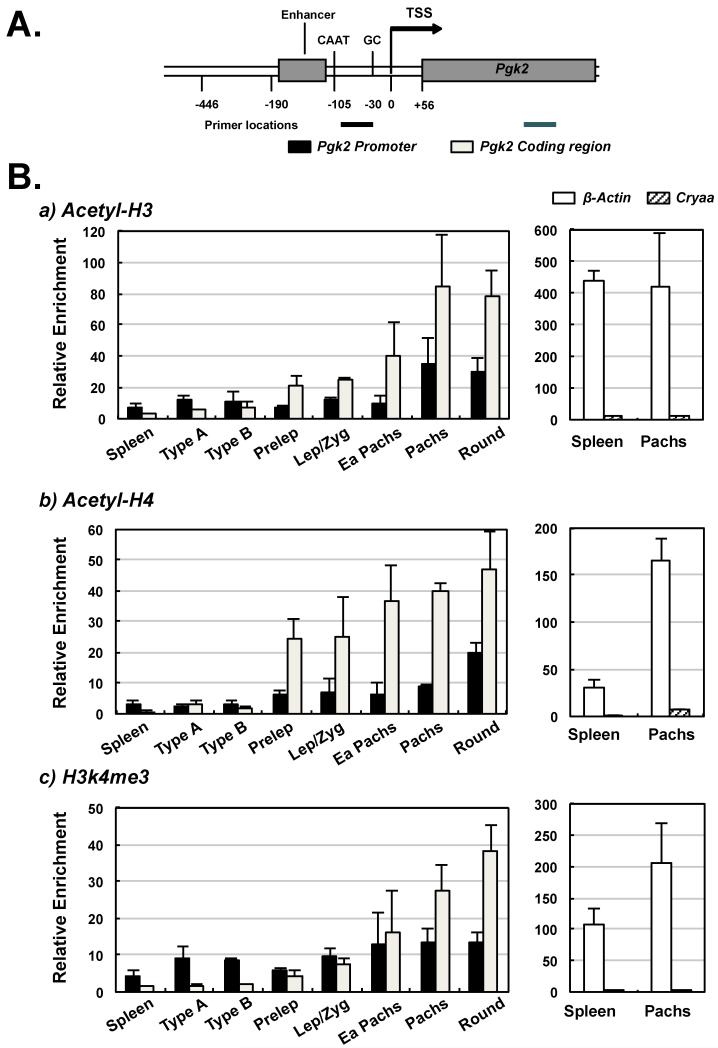
FIGURE 4. Chromatin immunoprecipitation (ChIP) analysis of activating histone modifications in the endogenous Pgk2 gene.
(A) A map of the intronless Pgk2 gene showing the positions of amplicons analyzed for histone modifications by ChIP. One amplicon (filled black bars in the graphs) was located within the Pgk2 promoter and a second amplicon (open bars in the graphs) was located in the Pgk2 coding sequence. (B) ChIP analysis of three different activating histone modifications, acetyl-H3, acetyl-H4, and H3k4me3, in the Pgk2 gene in different cell types, and in two control genes (ß-Actin or Cryaa) in spleen cells (Spleen) and pachytene spermatocytes (Pachs). The ß-Actin gene is ubiquitously active in all cell types and showed consistent enrichment for all three activating histone modifications in both spleen cells and spermatocytes, while the Cryaa gene is repressed in both spleen cells and spermatocytes, and showed an absence of activating histone modifications in both cell types. The Pgk2 gene also showed an absence of activating histone modifications in spleen cells where this gene is terminally repressed, and in premeiotic type A and type B spermatogonial cells in which the Pgk2 gene has not yet undergone initiation of transcription. However, all three activating histone modifications showed enrichment in the Pgk2 gene in cells actively expressing this gene, including successive types of primary spermatocytes – preleptotene spermatocytes (Prelep), leptotene plus zygotene spermatocytes (Lep/Zyg), early pachytene spermatocytes (Ea Pachs) and fully mature pachytene spermatocytes (Pachs), as well as in round spermatids (Round). Enrichment for these histone modifications was generally greater in the Pgk2 coding sequence than in the Pgk2 promoter in cells actively expressing this gene.
All three modifications were elevated in the ß-actin gene relative to the Cryaa gene in both somatic spleen cells and pachytene spermatocytes, confirming the association of these modifications with actively transcribed genes (Fig. 4B). Similarly, these modifications were generally low in the Pgk2 gene in spleen cells and in premeiotic spermatogonia, similar to levels seen for the repressed Cryaa gene in both somatic and germ cells. However, beginning in primary spermatocytes in which active transcription of the Pgk2 gene is initiated (McCarrey et al. 1992), we observed a gradual increase in all three modifications that reached a peak in either pachytene spermatocytes or round spermatids (Fig. 4B) in a pattern that directly mimicked the gradual increase in expression levels of this gene in the same cell types (McCarrey et al. 1992, Yoshioka et al. 2007). Levels of all three modifications associated with the Pgk2 gene in pachytene spermatocytes and round spermatids were similar to those seen for the same modifications in the constitutively expressed ß-actin gene, indicating these modifications are associated with a state of active expression of the Pgk2 gene. The gradually increasing pattern of these modifications in progressively later spermatogenic cell types was similar for all three modifications investigated (Fig. 4B), and correlates with previously published results describing ongoing transcription of the Pgk2 gene in primary spermatocytes and round spermatids (Kumari et al. 1996). In addition, the increase in these modifications was seen in both the promoter and coding regions of the Pgk2 gene, with the increase being greatest in the coding region.
The Pgk2 enhancer region plus the CAAT-box are required to direct activating histone modifications
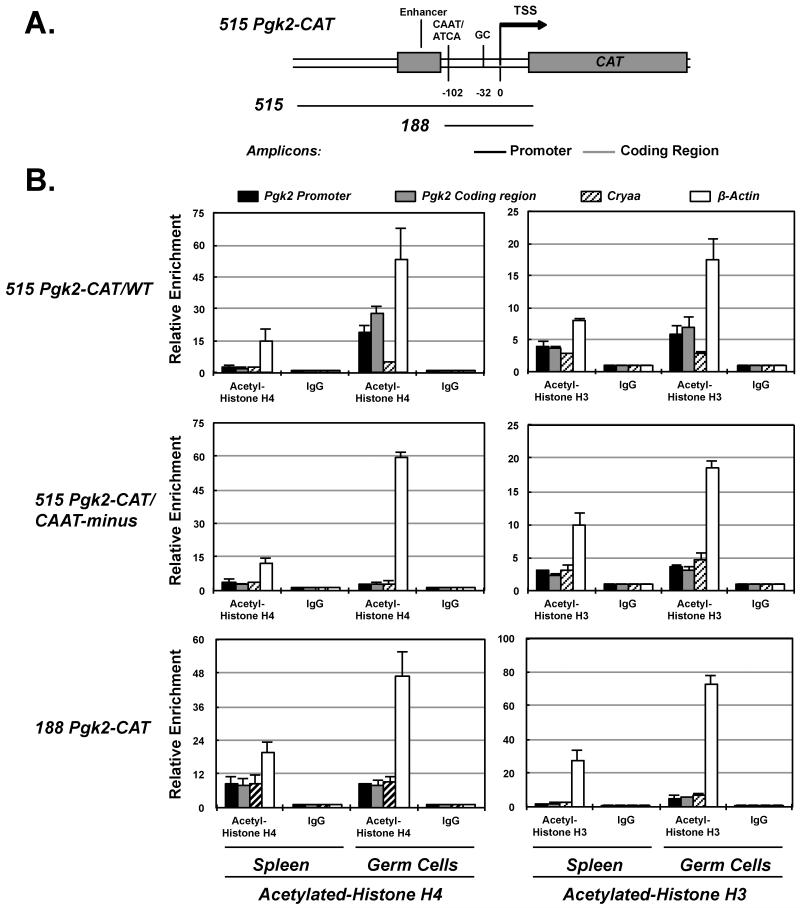
As with reconfiguration of nucleosomes, the tissue- and developmental stage-specificity of these histone modifications suggests a highly regulated process on a gene-specific level. To determine which part of the Pgk2 gene signals these modifications, we examined transgenes carrying the wild-type Pgk2 core promoter alone (= the 188 Pgk2-CAT transgene) or the wild-type Pgk2 core promoter plus enhancer regions (= the 515 Pgk2-CAT/WT transgene), as well as the transgene carrying the Pgk2 enhancer plus core promoter regions but with a mutated CAAT-box (= the 515 Pgk2-CAT/CAAT-minus transgene) (Fig. 5). The intact 515 Pgk2-CAT/WT transgene showed germ cell-specific enrichments of acetyl-H3 and acetyl-H4 (especially in spermatocytes and spermatids) that were diminished in the 188 Pgk2-CAT transgene (Fig. 5B). This indicated that at least part of the signal that directs tissue-specific acquisition of acetylation of histones H3 and H4 in the Pgk2 promoter appears to reside in the upstream enhancer region. However, the 515 Pgk2-CAT/CAAT-minus transgene also failed to acquire tissue-specific acetylation of H3 and H4, suggesting that the complete gene-specific signal that directs this modification may extend from the Pgk2 enhancer region through the 5′-portion of the Pgk2 core promoter where the CAAT-box resides.
FIGURE 5. Chromatin immunoprecipitation (ChIP) analysis of histone acetylation in Pgk2 transgenes.
(A). A map of the Pgk2-CAT transgenes shows the position of amplicons analyzed for acetyl-H3 or acetyl-H4 in Pgk2-CAT transgenes. (B) Relative enrichment of acetylated H4 (graphs on the left) or acetylated H3 (graphs on the right) is shown for each transgene as well as for the two control genes, ß-actin and Cryaa in spleen cells or mixed male germ (spermatogenic) cells. The 515 Pgk2-CAT/WT transgene, which is actively expressed in spermatogenic cells, shows significant enrichment for both acetyl-H3 and acetyl-H4 in male germ cells, but not in spleen cells where this transgene is not expressed. The 515 Pgk2-CAT/CAAT-minus transgene, which contains all regulatory sequences normally required to direct testis-specific expression except for an intact CAAT-box in the core promoter, and is not expressed in either spleen cells or spermatogenic cells, shows no enrichment of acetyl-H3 or acetyl-H4 in either cell type. Similarly, the 188 Pgk2-CAT transgene, which contains only the Pgk2 core promoter and no enhancer region and is not expressed in either spleen or spermatogenic cells, shows no enrichment for either histone modification in either cell type.
Summary of Pgk2 promoter regions required to signal each molecular event associated with transcriptional activation of the Pgk2 gene
Table 1 summarizes the occurrence of four different molecular events in the presence of different regions of the Pgk2 promoter + enhancer regions in transgenes. Thus, from a transgene carrying an intact Pgk2 enhancer plus core promoter region, transcription is initiated with normal tissue- and developmental stage-specificity (Robinson et al. 1989, Zhang et al. 1999). However, in the absence of the enhancer region, or of an intact CAAT-box in the core promoter region even when the enhancer region is present, transcription is not initiated from this promoter. Acquisition of activating histone modifications (acetyl H3, acetyl H4 and H3k4me3) requires an intact CAAT-box in the Pgk2 core promoter along with the presence of the Pgk2 enhancer region. Reconfiguration of nucleosomes also requires an intact CAAT-box in the core promoter. Whether or not the Pgk2 enhancer region is required for nucleosome reconfiguration could not be determined because the 188 Pgk2-CAT transgene that lacks the enhancer region also lacks a significant portion of the upstream sequence that normally makes up the −1 nucleosome position. Finally, the signal for cell type- and developmental stage-specific demethylation of DNA in the 5′ half of the Pgk2 gene appears to reside in the Pgk2 core promoter, and does not require the presence of the enhancer region. However, an intact CAAT-box in the core promoter does not appear to be a specific requirement for this signaling function.
Table 1. Regulation of molecular events associated with transcriptional activation of the Pgk2 gene.
| Molecular event | Specific Pgk2 promoter regions |
||
|---|---|---|---|
| Intact enhancer + intact core promoter |
Intact core promoter only |
Intact enhancer + core promoter lacking CAAT |
|
| Demethylation of DNA |
+ | + | + |
| Nucleosome reconfiguration |
+ | n/a* | − |
| Histone modifications | + | − | − |
| Initiation of transcription |
+ | − | − |
Note that in the absence of the enhancer region, the sequence over which the −1 position at which a nucleosome is normally found in non-expressing cells is missing.
Discussion
Spermatogenesis is a highly dynamic process of cellular differentiation that has been correlated with a highly dynamic series of changes in gene expression (Almstrup et al. 2004, Shima et al. 2004, Roy Choudhury et al. 2010). These changes include the activation of numerous testis-specific genes including several tissue-specific members of gene families that encode testis-specific isozymes (McCarrey & Thomas 1987, Mori et al. 1993b, Boussouar & Benahmed 2004, Tang et al. 2008). One example is the testis-specific Pgk2 gene, transcriptional activation of which occurs in primary spermatocytes coincident with the onset of meiotic prophase. Interestingly, this is also the time at which the X-linked Pgk1 gene, which encodes the form of the glycolytic enzyme phosphoglycerate kinase (PGK-A) that is expressed in all somatic or germ cell types carrying at least one transcriptionally active X chromosome, undergoes silencing due to the process of meiotic sex chromosome inactivation (MSCI) (McCarrey & Thomas 1987, McCarrey et al. 1992, Wang et al. 2005, Turner 2007). Thus, meiotic and postmeiotic spermatogenic cells are unique in the absence of at least one active X chromosome, and this presents a dilemma because these cells, especially spermatozoa, require phosphoglycerate kinase to carry out glycolysis. It appears that a gene duplication event that occurred early during mammalian evolution has solved this dilemma in mammals by providing an autosomal copy of the Pgk gene (Pgk2) which encodes a testis-specific isozyme of PGK-A (PGK-B) that is not subject to MSCI, and that undergoes tissue-specific transcriptional activation at the same time that the Pgk1 gene becomes silenced, such that a source of PGK protein is maintained throughout spermatogenesis (McCarrey et al. 1992, McCarrey et al. 1996).
Because the development and differentiation of the male germ line in mammals has been well characterized, and because many of the different cell types that make up this lineage can be recovered in relatively pure populations, spermatogenesis provides an excellent opportunity to study the developmental process of transcriptional activation of tissue-specific genes (Bellve 1993, McCarrey 1998). This has allowed us to chronicle molecular events that are associated with transcriptional activation of the spermatogenesis-specific Pgk2 gene (McCarrey et al. 1992, McCarrey et al. 1996, Geyer et al. 2004, Yoshioka et al. 2007). We previously showed that activation of the Pgk2 gene is associated with binding of ubiquitous transcription factors to the Pgk2 core promoter (Gebara & McCarrey 1992, Berg 1993, Yoshioka et al. 2007) and testis-specific factors to the Pgk2 enhancer region (Robinson et al. 1989, Gebara & McCarrey 1992, Yoshioka et al. 2007). We also showed that there is a general increase in nuclease (DNase I) sensitivity over the entire Pgk2 gene (Kramer et al. 1998), as well as the development of a DNase I hypersensitive site at the Pgk2 promoter (Kumari et al. 1996) in cells that express this gene. Finally, we showed that activation of the Pgk2 gene is preceded by demethylation of DNA over the 5′-portion of the gene (Ariel et al. 1994, Zhang et al. 1998, Geyer et al. 2004).
In the study described here, we have shown that two additional molecular events associated with chromatin remodeling – reconfiguration of nucleosomes and acquisition of activating histone modifications – also occur during the process of transcriptional activation of the Pgk2 gene. Thus, a nucleosome at the −1 position that corresponds to a region spanning the enhancer and core promoter region of the Pgk2 gene is present in non-expressing cells but is absent in cells in which the Pgk2 gene is actively transcribed. Interestingly, an adjacent nucleosome at the +1 position in the 5′ portion of the coding sequence remains present in both expressing and non-expressing cell types. This indicates that reconfiguration and/or displacement of the nucleosome at the −1 position is specifically correlated with events ongoing in the promoter region of the Pgk2 gene associated with transcriptional activation of this gene.
The acquisition of histone modifications including acetyl H3, acetyl H4 and H3k4me3 also shows a direct correlation with active expression of this gene. However these modifications occur in both the Pgk2 promoter and coding sequence regions. Indeed these modifications are most pronounced in the Pgk2 coding sequence in expressing cell types. This likely reflects the fact that nucleosome reconfiguration significantly reduces the overall representation of histones in the Pgk2 promoter region, and that these histone modifications facilitate the elongation as well as initiation phases of transcription (Berger 2002). Acetylation of lysine residues is known to contribute to changes in nucleosome/chromatin structure that facilitate recruitment of other transcription factors to promoter regions and/or more efficient processivity of RNA polymerase during the elongation phase of transcription (Brownell et al. 1996, Marmorstein & Roth 2001, Allis et al. 2007, Marmorstein & Trievel 2009). Our findings that reconfiguration of nucleosomes and modifcations of histones accompany activation of transcription of the Pgk2 gene are consistent with those previously reported for several other genes (Kouskouti & Talianidis 2005, Villagra et al. 2006, Plachetka et al. 2008), and are therefore not novel in that respect. However, our results do provide novel information regarding the order of these and other molecular events involved in transcriptional activation of the Pgk2 gene in particular and of tissue-specific genes in general.
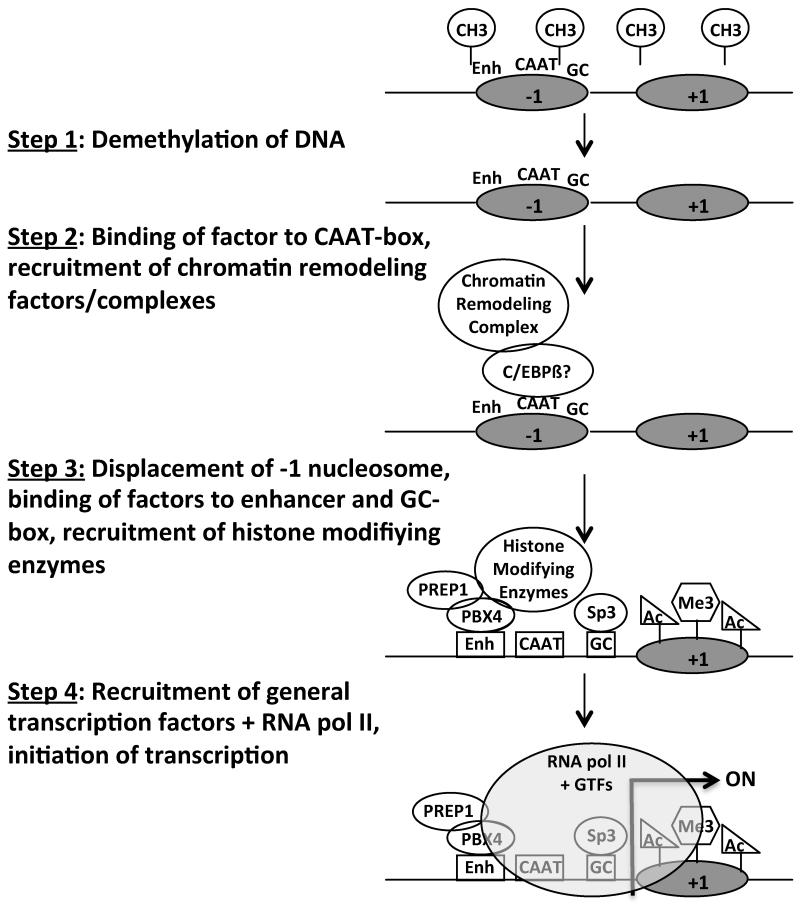
Thus, based on the data presented here, as well as that from our previously published studies including bisulfite sequencing analysis of demethylation of the Pgk2 gene (Zhang et al. 1998, Geyer et al. 2004), in vivo footprinting of the Pgk2 5′-regulatory region (Yoshioka et al. 2007), general nuclease sensitivity studies (Kramer et al. 1998), analysis of DNase I hypersensitive site formation (Kumari et al. 1996), and chromatin immunoprecipitation (ChIP) assays to detect binding of specific factors to the Pgk2 enhancer and core promoter regions in vivo (Yoshioka et al. 2007), we can now propose a developmental order of molecular regulatory events involved in activation of transcription of the Pgk2 gene (Fig. 6). These include two events that clearly precede initiation of transcription of the Pgk2 gene – step 1) demethylation of DNA in the 5′ half of the Pgk2 gene beginning in T1-prospermatogonia at 18 days postcoitum (dpc) and completed by the primitive type A spermatogonial stage at 6 days postpartum (dpp), and step 2) binding of one or more factors (possibly C/EBPß, see below) to a region in the Pgk2 core promoter that extends from the CAAT-box to just upstream of the GC-box in premeiotic spermatogonia (Geyer et al. 2004), plus recruitment of chromatin remodeling factors or complexes in early primary spermatocytes. These events are followed by a series of events that occur just prior to, or coincident with initiation of Pgk2 transcription in primary spermatocytes, beginning with step 3) reconfiguration of the −1 nucleosome covering the Pgk2 core promoter and enhancer regions as shown in this study, formation of a DNase I hypersensitive site as shown in a previous study (Kumari et al. 1996), binding of testis-specific factors (PBX4 and PREP1) to the E3/E4 portion of the Pgk2 enhancer region and Sp3 to the GC-box in the Pgk2 core promoter as shown in another previous study (Yoshioka et al. 2007), changes in histone modifications as shown in this study, and an increase in general nuclease sensitivity throughout the Pgk2 locus as shown in yet another previous study (Kramer et al. 1998). Finally, step 4) formation of a complete preinitiation complex is completed by recruitment of general transcription factors plus RNA polymerase II and transcription is initiated. This scheme is depicted in Figure 6.
FIGURE 6. Model of events leading up to transcriptional activation of the Pgk2 gene during spermatogenesis.
A series of molecular events involved in transcriptional activation of the Pgk2 gene during spermatogenesis is proposed based on observations reported in this and previous studies. Where possible, these events are shown in developmental chronological order and include Step 1 – demethylation of DNA in the 5′-half of the Pgk2 gene, which begins in T1-prospermatogonia and is completed by the primitive type A spermatogonia stage; Step 2 – binding of one or more factors (possibly C/EBPß) to the CAAT-box and surrounding sequences in the Pgk2 core promoter to attract a chromatin remodeling complex; Step 3 – displacement of the nucleosome at the −1 position, binding of factors to the enhancer and core promoter regions, and recruitment of histone-modifying enzymes; and Step 4 – formation of the complete preinitiation complex including recruitment of RNA polymerase II and initiation of transcription. CH3 = methyl groups on CpG dinucleotides, Enh = enhancer region, CAAT = CAAT-box, GC = GC-box, Ac = histone acetylation, Me3 = histone trimethylation.
In this study, we have also shown that an intact CAAT-box is required for both the acquisition of activating histone modifications and nucleosome reconfiguration. The CAAT-box is an element found in the core promoters or enhancers of many ubiquitously expressed and tissue-specific genes (Mantovani 1999, Villagra et al. 2006, Plachetka et al. 2008). It is known to serve as a binding site for multiple ubiquitous transcription factors including CTF/NF1 (CCAAT Transcription Factor/Nuclear Factor 1) (Nagata et al. 1982, Santoro et al. 1988), C/EBP (CCAAT/Enhancer Binding protein) (Chang et al. 1990, Roman et al. 1990) and NF-Y/Hap (Nuclear Factor Y/Hem activation protein) (Li et al. 1992, McNabb et al. 1995), among others (Nepveu 2001). Thus, while it is not surprising to find that the CAAT-box in the Pgk2 promoter is important for initiation of transcription, we were surprised to observe in our ChIP studies of protein-DNA interactions associated with the Pgk2 gene in spermatocytes or spermatids (Yoshioka et al. 2007) that none of the common ubiquitous factors known to bind to CAAT-boxes appear to be bound to this sequence in the actively expressed Pgk2 gene. This was confirmed by our in vivo footprinting studies (Yoshioka et al. 2007) that showed this site remains unoccupied in these same cells. Interestingly, we did observe in vivo footprints over the CAAT-box and nearby regions of the Pgk2 core promoter during the spermatogonial stage, prior to initiation of Pgk2 transcription in primary spermatocytes. This has led us to speculate that an intact CAAT-box is required for chromatin remodeling events that immediately precede initiation of Pgk2 transcription. Specifically, we suggested the CAAT-box and nearby sequences serve as signals to direct chromatin remodeling by binding factors or chromatin remodeling complexes in a gene-, cell type- and developmental stage-specific manner. Our observation that these events fail to occur in a transgene lacking an intact CAAT-box but possessing all other regulatory regions normally required for tissue-specific activation of transcription from the Pgk2 promoter suggests that the CAAT-box does indeed play a role in signaling chromatin remodeling, which is, in turn, a mandatory prerequisite for subsequent molecular events required for transcription from the Pgk2 promoter, including binding of some or all of the transcription factors that are normally required for initiation of Pgk2 transcription.
There have been previous reports of factors binding to CAAT-boxes to initiate chromatin remodeling in conjunction with tissue-specific activation of transcription. Tissue-specific activation of the bone-specific OC gene requires chromatin remodeling by the SWI/SNF complex recruited to the proximal promoter by C/EBPß, which binds to a CAAT-box-like sequence (Villagra et al. 2006). Similarly, C/EBPß has been shown to bind to an upstream enhancer site in the tissue-specific chicken mim-1 gene to induce decondensation of chromatin prior to activation of transcription (Plachetka et al. 2008). Both of these studies showed that the C/EBPß factor is required for chromatin remodeling associated with, and required for, activation of tissue-specific transcription. Our study shows that an intact CAAT-box is specifically required for this process in the testis-specific Pgk2 gene, and suggests this element may function by recruiting C/EBPß and chromatin-remodeling complexes to promote acquisition of activating histone modifications, decondensation of chromatin and reconfiguration of nucleosomes, although we have yet to directly demonstrate this.
A Pgk2 transgene lacking an intact CAAT-box but possessing all other 5′-regulatory sequences normally required for tissue-specific activation of Pgk2 transcription failed to acquire activating histone modifications or initiate transcription, indicating chromatin remodeling directed by the Pgk2 CAAT-box is a mandatory prerequisite for activation of transcription from the Pgk2 promoter. Our observation of in vivo footprints over the CAAT-box and nearby regions in spermatogonia, but not in spermatocytes or spermatids (Yoshioka et al. 2007) indicates that these chromatin remodeling steps normally occur prior to the initiation of transcription and do not need to be continually reinforced during active transcription. Thus the model we propose in Figure 6 suggests that a single chromatin remodeling event is required to facilitate access for most or all of the transcription factors that regulate ongoing expression of the Pgk2 gene to bind to their cognate sites in the Pgk2 core promoter and enhancer regions. We further observed that a transgene carrying the Pgk2 core promoter but not the upstream enhancer region failed to undergo activating histone modifications, suggesting that the enhancer region is also required to direct acquisition of these modifications.
We previously showed that the PBX4 transcription factor and its co-activator, PREP1, become bound to the Pgk2 E3/E4 enhancer region beginning in primary spermatocytes (Yoshioka et al. 2007), which is the same time at which we now show activating histone modifications appear. A previous report showed that the PBX4 factor can interact with histone lysine acetyltransferases to direct acetylation of chromatin (Choe et al. 2009). Taken together, our results are consistent with a model in which the CAAT-box in the Pgk2 core promoter is required to direct chromatin remodeling (potentially through binding of C/EBPß), and the E3/E4 enhancer region is required to direct acquisition of activating histone modifications through binding of the PBX4 activator and its co-activator, PREP1. Because a transgene carrying an intact Pgk2 enhancer region but a disrupted CAAT-box failed to undergo the histone modifications normally associated with transcriptional activation, we would further suggest that the chromatin remodeling step directed by the CAAT-box is a prerequisite for the histone modification step directed by the enhancer region in the Pgk2 gene.
Our previous studies have also shown that demethylation of DNA in the 5′ half of the Pgk2 gene precedes the chromatin remodeling events, as well as binding of transcription factors to the Pgk2 promoter and initiation of transcription. We previously showed that the Pgk2 enhancer region is not required to direct demethylation of the Pgk2 gene (Zhang et al. 1998, Geyer et al. 2004), and in this study, we showed that an intact CAAT-box is also not required for this demethylation event. Thus tissue-specific demethylation appears to be regulated independently from chromatin remodeling and activating histone modifications in the Pgk2 gene. How the demethylation step is signaled in a gene-, cell type- and developmental stage-specific manner, and whether or not that event is a mandatory prerequisite for the subsequent molecular events associated with activation of the Pgk2 transcription remain to be determined.
Supplementary Material
Acknowledgements
The authors thank Jacey Hornecker for technical assistance with preparations of purified spermatogenic cell types.
Funding - This work was supported by NIH grant RO1 HD46637 to JRM.
Footnotes
Declaration of interest
The authors delcare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Nielsen JE, Hansen MA, Tanaka M, Skakkebaek NE, Leffers H. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biology of Reproduction. 2004;70:1751–1761. doi: 10.1095/biolreprod.103.026575. [DOI] [PubMed] [Google Scholar]
- Ariel M, Cedar H, McCarrey J. Developmental changes in methylation of spermatogenesis-specific genes include reprogramming in the epididymis. Nature Genetics. 1994;7:59–63. doi: 10.1038/ng0594-59. [DOI] [PubMed] [Google Scholar]
- Barik S. Site-directed mutagenesis in vitro by megaprimer PCR. Methods in Molecular Biology. 1996;57:203–215. doi: 10.1385/0-89603-332-5:203. [DOI] [PubMed] [Google Scholar]
- Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends in Biochemical Sciences. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annual review of Biochemistry. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods in Enzymolology. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Berg WM, Gebara MM, McCarrey JR. Sp-1 is required for initiation and stimulation of transcription from the TATA-less PGK-2 promoter. Life Sciences Advances in Molecular Biology. 1993;12:85–91. [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Current Opinion in Genetics & Development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Boussouar F, Benahmed M. Lactate and energy metabolism in male germ cells. Trends in Endocrinology and Metabolism: TEM. 2004;15:345–350. doi: 10.1016/j.tem.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Chen TT, Lei HY, Chen DS, Lee SC. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Molecular and Cellular Biology. 1990;10:6642–6653. doi: 10.1128/mcb.10.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Developmental Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Research. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O’Brien DA. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biology of Reproduction. 2010;82:136–145. doi: 10.1095/biolreprod.109.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM. Male germ cell gene expression. Recent Progress in Hormone Research. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Hutchison WM, Dahl HH. Isolation and characterisation of the mouse pyruvate dehydrogenase E1 alpha genes. Biochimica et Biophysica Acta. 1992;1131:83–90. doi: 10.1016/0167-4781(92)90102-6. [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Current Opinion in Genetics & Development. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebara MM, McCarrey JR. Protein-DNA interactions associated with the onset of testis-specific expression of the mammalian Pgk-2 gene. Molecular and Cellular Biology. 1992;12:1422–1431. doi: 10.1128/mcb.12.4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Kiefer CM, Yang TP, McCarrey JR. Ontogeny of a demethylation domain and its relationship to activation of tissue-specific transcription. Biology of Reproduction. 2004;71:837–844. doi: 10.1095/biolreprod.104.028969. [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nature Genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. A Guide to Methods and Applications. Academic Press, Inc.; San Diego,CA: 1990. pp. 177–183. [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1986. [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. The EMBO Journal. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kramer JA, McCarrey JR, Djakiew D, Krawetz SA. Differentiation: The selective potentiation of chromatin domains. Development. 1998;125:4749–4755. doi: 10.1242/dev.125.23.4749. [DOI] [PubMed] [Google Scholar]
- Kumari M, Stroud JC, Anji A, McCarrey JR. Differential appearance of DNase I-hypersensitive sites correlates with differential transcription of Pgk genes during spermatogenesis in the mouse. The Journal of Biological Chemistry. 1996;271:14390–14397. doi: 10.1074/jbc.271.24.14390. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. Journal of Cell Science. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genetics. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Leimgruber E, Seguin-Estevez Q, Dunand-Sauthier I, Rybtsova N, Schmid CD, Ambrosini G, Bucher P, Reith W. Nucleosome eviction from MHC class II promoters controls positioning of the transcription start site. Nucleic Acids Research. 2009;37:2514–2528. doi: 10.1093/nar/gkp116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Mantovani R, Hooft van Huijsduijnen R, Andre I, Benoist C, Mathis D. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Research. 1992;20:1087–1091. doi: 10.1093/nar/20.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Current Opinion in Genetics & Development. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochimica et Biophysica Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR. Nucleotide sequence of the promoter region of a tissue-specific human retroposon: comparison with its housekeeping progenitor. Gene. 1987;61:291–298. doi: 10.1016/0378-1119(87)90192-2. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Molecular evolution of the human Pgk-2 retroposon. Nucleic Acids Research. 1990;18:949–955. doi: 10.1093/nar/18.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR. Spermatogenesis as a model system for developmental analysis of regulatory mechanisms associated with tissue-specific gene expression. Seminars in Cell & Developmental Biolology. 1998;9:459–466. doi: 10.1006/scdb.1998.0199. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biology of Reproduction. 2013 doi: 10.1095/biolreprod.113.110502. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ. Differential transcription of Pgk genes during spermatogenesis in the mouse. Developmental Biology. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Geyer CB, Yoshioka H. Epigenetic regulation of testis-specific gene expression. Annals of the New York Academy of Sciences. 2005;1061:226–242. doi: 10.1196/annals.1336.025. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Kumari M, Aivaliotis MJ, Wang Z, Zhang P, Marshall F, Vandeberg JL. Analysis of the cDNA and encoded protein of the human testis-specific PGK-2 gene. Developmental Genetics. 1996;19:321–332. doi: 10.1002/(SICI)1520-6408(1996)19:4<321::AID-DVG5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes & Development. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- Mori C, Welch JE, Fulcher KD, O’Brien DA, Eddy EM. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biology of Reproduction. 1993;49:191–203. doi: 10.1095/biolreprod49.2.191. [DOI] [PubMed] [Google Scholar]
- Nagata K, Guggenheimer RA, Enomoto T, Lichy JH, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nature Biotechnology. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Plachetka A, Chayka O, Wilczek C, Melnik S, Bonifer C, Klempnauer KH. C/EBPbeta induces chromatin opening at a cell-type-specific enhancer. Molecular and Cellular Biology. 2008;28:2102–2112. doi: 10.1128/MCB.01943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annual review of biochemistry. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- Robinson MO, McCarrey JR, Simon MI. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:8437–8441. doi: 10.1073/pnas.86.21.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman C, Platero JS, Shuman J, Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes & Development. 1990;4:1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- Roy Choudhury D, Small C, Wang Y, Mueller PR, Rebel VI, Griswold MD, McCarrey JR. Microarray-based analysis of cell-cycle gene expression during spermatogenesis in the mouse. Biology of Reproduction. 2010;83:663–675. doi: 10.1095/biolreprod.110.084889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C, Mermod N, Andrews PC, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Molecular Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Molecular and Cellular Biology. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biology of Reproduction. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Current Topics in Developmental Biology. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiolology & Molecular Biology Reviews. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Kung A, Goldberg E. Regulation of murine lactate dehydrogenase C (Ldhc) gene expression. Biology of Reproduction. 2008;78:455–461. doi: 10.1095/biolreprod.107.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Research. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeBerg JL. The phosphoglycerate kinase isozyme system in mammals: biochemical, genetic, developmental, and evolutionary aspects. Isozymes Current Topics in Biolology & Medical Research. 1985;12:133–187. [PubMed] [Google Scholar]
- VandeBerg JL, Cooper DW, Close PJ. Mammalian testis phosphoglycerate kinase. Nature New Biology. 1973;243:48–50. [PubMed] [Google Scholar]
- Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. The Journal of Biological Chemistry. 2006;281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- Walter CA, Nasr-Schirf D, Luna VJ. Identification of transgenic mice carrying the CAT gene with PCR amplification. Biotechniques. 1989;7:1065–1070. [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Human Molecular Genetics. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Geyer CB, Hornecker JL, Patel KT, McCarrey JR. In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Molecular and Cellular Biology. 2007;27:7871–7885. doi: 10.1128/MCB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang LP, Stroud J, Eddy CA, Walter CA, McCarrey JR. Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biology of Reproduction. 1999;60:1329–1337. doi: 10.1095/biolreprod60.6.1329. [DOI] [PubMed] [Google Scholar]
- Zhang LP, Stroud JC, Walter CA, Adrian GS, McCarrey JR. A gene-specific promoter in transgenic mice directs testis-specific demethylation prior to transcriptional activation In vivo. Biology of Reproduction. 1998;59:284–292. doi: 10.1095/biolreprod59.2.284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.