SUMMARY
ID genes are required for breast cancer colonization of the lungs, but the mechanism remains poorly understood. Here, we show that Id1 expression induces a stem-like phenotype in breast cancer cells, while retaining epithelial properties, contrary to the notion that cancer stem-like properties are inextricably linked to the mesenchymal state. During metastatic colonization, Id1 induces a mesenchymal-to-epithelial transition (MET), specifically in cells whose mesenchymal state is dependent on the Id1 target protein Twist1 but not at the primary site, where this state is controlled by the zinc-finger protein Snail1. Knockdown of Id expression in metastasizing cells prevents MET and dramatically reduces lung colonization. Furthermore, Id1 is induced by TGFβ only in cells that have first undergone EMT, demonstrating that EMT is a pre-requisite for subsequent Id1-induced MET during lung colonization. Collectively, these studies underscore the importance of Id-mediated phenotypic switching during distinct stages of breast cancer metastasis.
INTRODUCTION
Metastasis accounts for 90% of carcinoma related deaths, making a detailed understanding of this complex phenomenon essential in reducing the lethality of this disease (Gupta and Massague, 2006). The multistep process of metastasis can be organized into two major phases: (1) physical dissemination of the cancer cell from its site of origin and (2) colonization of distant organs (Chaffer and Weinberg, 2011). The first phase is accompanied by re-activation of the developmental program called the “epithelial to mesenchymal transition” (EMT), which endows cancer cells with a highly invasive phenotype (Thiery et al., 2009). During EMT, immobile epithelial cells lose their epithelial traits and acquire mesenchymal properties and the ability to migrate. The importance of EMT in metastasis is supported by findings showing that cancer cells that have undergone EMT share key characteristics with tumor initiating cells (TICs) (Mani et al., 2008), which are functionally defined by their ability to seed new tumors and restore the heterogeneity of the original tumor (Dick, 2008). The generation of breast cancer TICs by the overexpression of EMT inducing transcription factors, such as Twist1 (Mani et al., 2008), has provided a direct molecular link between EMT driven metastatic dissemination and the generation of TICs. However, less is known about the biology of TICs during the second phase of metastasis, the colonization of distant organs.
The idea that breast cancer TICs which colonize the lung permanently retain their mesenchymal character has been challenged by clinical observations showing most metastases present a differentiated epithelial morphology (Tarin et al., 2005). This suggests that EMT is a transient process and that the re-differentiation of carcinoma cells by a “mesenchymal to epithelial transition” (MET) is a driving force of metastatic colonization at least in some cancers (Brabletz, 2012). While EMT governs different steps during cancer cell dissemination, including invasion of the local parenchyma, intravasation into the circulatory system, survival during migration, and finally extravasation into the secondary site, the loss of a mesenchymal phenotype may enhance the formation of macro-metastatic colonies, in part by overcoming the growth arrest associated with EMT (Brabletz et al., 2001; Vega et al., 2004). Evidence for the importance of MET in breast cancer metastasis has been provided by studies showing that after dissemination, engineered loss of the EMT transcription factor Twist1 (Tsai et al., 2012) and the expression of microRNAs inhibiting the EMT transcription factors Zeb1/2 (Korpal et al., 2011) enhance lung colonization of metastatic breast cancer cells. Furthermore, the transcription factor Prrx1, which induces EMT during dissemination but suppresses stemness traits necessary for lung colonization (Ocana et al., 2012), must be lost prior to colonization, uncoupling in this instance EMT from the TIC phenotype. However, the details of how a colonizing cancer cell sheds its mesenchymal phenotype while retaining the TIC properties that promote its ability to serve as a founder for a metastatic colony, remain unclear.
Comparison of gene expression data between cell lines and their derivatives with variable metastatic potential has led to the identification of candidate genes required for the metastatic cascade (Minn et al., 2005). Such analyses showed that the expression of the ID1 and ID3 genes was essential during lung colonization of breast cancer cells (Gupta et al., 2007). The Id (Inhibitor of DNA-binding) proteins are dominant negative regulators of basic helix-loop-helix (bHLH) transcription factors (Perk et al., 2005). During development, Id proteins play a key role in the maintenance of embryonic stem cell self-renewal (Romero-Lanman et al. 2012; Ying et al., 2003) and they continue to serve this function in many adult tissue stem cells, including neural (Nam and Benezra, 2009) and hematopoietic stem cells (Jankovic et al., 2007). ID genes have also been implicated as key regulators of the TIC phenotype in glioblastoma (Anido et al., 2010; Barrett et al., 2012) and colon cancer (O’Brien et al., 2012). The role of Id proteins in breast cancer metastasis and their importance in self-renewal makes them prime candidates for the study of stem cell character as it relates to metastatic colonization of breast cancer cells.
bHLH transcription factors act as obligate dimers, usually formed between ubiquitously expressed E proteins and tissue specific bHLH proteins, such as Twist1 (Perk et al., 2005; Spicer et al., 1996). Id proteins lack the basic DNA binding motif and thus negatively regulate bHLH transcription factors upon the formation of heterodimers (Ruzinova and Benezra, 2003). During cranial fusion in developing mouse embryos, Id1 regulates the differentiation state of cells found in osteogenic fronts by controlling the dimerization of Twist1 and E proteins (Connerney et al., 2006). However, the functional and regulatory role of Id proteins in the context of EMT-driven metastasis induced by bHLH transcription factors such as Twist1 remains to be elucidated.
In human breast cancer, Id1 is predominantly expressed in the more aggressive triple negative (negative for estrogen receptor, progesterone and HER2) and metaplastic subtype, with high expression correlating with poor clinical outcome (Gupta et al., 2007). The mechanism by which Id1 is induced in breast cancer cells is not understood. In glioblastoma, the cytokine TGFβ is required to maintain the TIC population through the upregulation of Id1 and Id3 (Anido et al., 2010). TGFβ plays a similar role as a component of an autocrine signaling program that controls self-renewal in normal and malignant breast cancer cells (Scheel et al., 2011). While TGFβ represses Id1 expression in normal epithelial cells (Kang et al., 2003), TGFβ induces Id1 in breast cancer cells derived from the pleural effusion fluid of patients with metastatic disease (Padua et al., 2008). This divergent effect of TGFβ parallels the dichotomous role of TGFβ as a tumor suppressor early in cancer formation versus a metastasis enhancer in late cancer progression (Massague, 2012).
In this report we examine the ability of Id1 to induce breast cancer TICs necessary for seeding new tumors during pulmonary colonization. We hypothesize that Id1 governs MET through antagonism of the bHLH transcription factor Twist without the loss of the TIC phenotype that is required for metastatic colonization. We also examine how Id1 is induced in disseminated breast cancer cells by investigating the regulation of Id1 by TGFβ in TICs.
RESULTS
Id1 Induces Tumor-Initiating Properties Independently of EMT
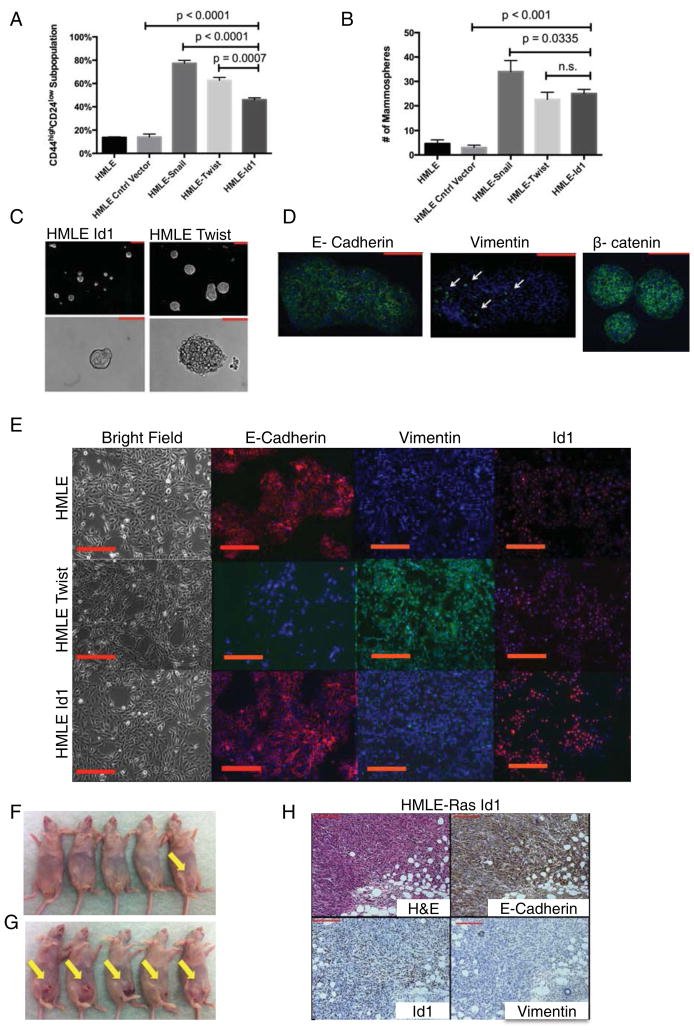
To determine whether Id1 can induce a TIC phenotype in human mammary cells, Id1 was overexpressed in immortalized human mammary epithelial cells (HMLEs) (Supplemental Figure 1A) (Elenbaas et al., 2001). Generation of TICs by expression of the transcription factors Snail1 and Twist1 has been previously described in HMLEs (Mani et al., 2008). HMLE cells transduced with an ID1 lentivirus vector (HMLE-Id1) were examined for TIC enrichment using flow cytometry analysis of CD44 and CD24 cell surface marker expression (Al-Hajj et al., 2003). HMLE-Id1 cells acquired a two-fold enrichment in the CD44high/CD24low expressing subpopulation compared to control cells (Figure 1A). An increase in the same antigenic phenotype was observed in cells expressing Snail and Twist (Figure 1A) as previously reported (Mani et al., 2008).
Figure 1. HMLE-Id1 Cells Exhibit Tumor-Initiating Properties Independently of EMT.
(A) Quantification of FACS analysis for cell surface markers CD44 and CD24 in HMLE cells expressing control vector, Snail, Twist1, and Id1.
(B) Quantification of mammospheres formed by cells described in (A).
(C) Mammospheres formed by HMLE-Id1 (left) and HMLE Twist cells. Scale bar = 200μm.
(D) Immunofluorescence (IF) images of mammospheres formed by HMLE-Id1 cells stained for E-Cadherin, β-catenin and Vimentin (white arrows). Scale bar = 200μm.
(E) (Top Row) Parental HMLE cells express E-Cadherin are negative for Vimentin. (Middle Row) HMLE cells expressing Twist1 undergo EMT as seen by the loss of E-Cadherin and gain of Vimentin. (Bottom Row) HMLE cells expressing Id1 express E-Cadherin and are negative for Vimentin. Scale bar = 100μm.
(F) Tumor incidence (yellow arrows) of 105 control HMLER cells injected into the mammary fat pads of nude mice compared to that of HMLER-Id1 cells (G).
(H) Immunohistochemistry (IHC) performed on tumors generated by HMLER-Id1 cells shows positive staining for Id1 and E-Cadherin, and absence of Vimentin. Scale bar = 200μm.
Self-renewing normal mammary and breast cancer cells form mammospheres in suspension culture (Dontu et al., 2003; Ponti et al., 2005). Similar to Twist or Snail, Id1 expression conferred HMLE cells with a higher capacity to form mammospheres compared to control HMLE cells (Figure 1B). However, HMLE-Id1 mammospheres had smaller diameters and more defined borders compared to HMLE-Twist mammospheres (Figure 1C), although no differences in proliferation were observed between them when grown under adherent conditions (Supplemental Figure 1B). This discrepancy could be due to the epithelial nature of HMLE-Id1 mammospheres as evidenced by expression of epithelial markers E-Cadherin and membrane bound β-catenin (Figure 1D) with a minority of cells staining positively for Vimentin (Figure 1D). In contrast, HMLE-Twist mammospheres were mostly positive for Vimentin with a minority of cells expressing E-Cadherin (Supplemental Figure 1C).
Recent studies have shown a correlation between the expression of several EMT-inducing embryonic transcription factors, such as Snail (Mani et al., 2008), Slug (Guo et al., 2012), Twist1 (Mani et al., 2008), Zeb1/2 (Eger et al., 2005) and the induction of TICs in breast cancer cells. Since Id1 induces mammospheres with an epithelial phenotype, we next examined if Id1 induces TICs independently of EMT. HMLE-Id1 cells did not change their epithelial morphology compared to control HMLE cells (Figure 1E), while HMLE-Twist cells underwent typical morphological changes accompanying EMT: they acquired a spindle-like shape and formed few contacts between neighboring cells (Figure 1E). HMLE-Id1 cells continued to express the epithelial marker E-Cadherin and were negative for the mesenchymal marker Vimentin (Figure 1E and Supplemental Figure 1A). The loss of E-Cadherin and the gain of Vimentin expression are hallmarks of EMT and are observed in HMLE-Twist cells (Figure 1E, Supplemental Figure 1D). Thus, a dominant negative transcriptional regulator can induce a TIC phenotype in mammary cells independently of the induction of EMT.
The standard in vivo assay for examining TIC properties of cancer cells is the ability to form tumors in xenotransplantation assays from limiting dilutions (Dick, 2008). Tumorigenicity of V12H-Ras transformed HMLE cells expressing Id1 (HMLER-Id1) (Elenbaas et al., 2001) was measured by injecting limiting dilutions of cells into mammary fat pads of immune-compromised mice. In vitro, HMLER-Id1 cells displayed the same antigenic phenotype, mammosphere formation properties, and morphology as HMLE-Id1 cells (data not shown). When 105 control HMLER cells were injected into mammary fat pads, only 1/5 mice developed palpable tumors one month after injection (Figure 1F and Supplemental Figure 1E). In contrast, 5/5 animals (p=0.02) injected with HMLER-Id1 (Figure 1G and Supplemental Figure 1E) cells had visible tumors one month after injection, identical to results obtained with HMLER-Twist and HMLER-Snail cells (data not shown) (Mani et al., 2008). HMLER-Id1 tumors continued to express the epithelial marker E-Cadherin and did not express the mesenchymal marker Vimentin (Figure 1H). Notably, no lung metastases were detected 3 months after tumor cell injection when mice were sacrificed due to heavy tumor burden at the primary site. We speculate that HMLER-Id1 cells do not metastasize efficiently because of a failure to acquire a fully invasive mesenchymal phenotype required for dissemination from the primary site. Indeed, Id1 expression does not have an effect on breast cancer cell intravasation or extravasation, early key steps in the metastatic cascade (Gupta et al., 2007). Taken together, our results demonstrate the ability of Id1 to endow breast cancer cells with TIC properties as defined by both in vitro and in vivo assays independent of EMT induction.
Id1 Expression Correlates with an Epithelial Phenotype in Pulmonary Metastases
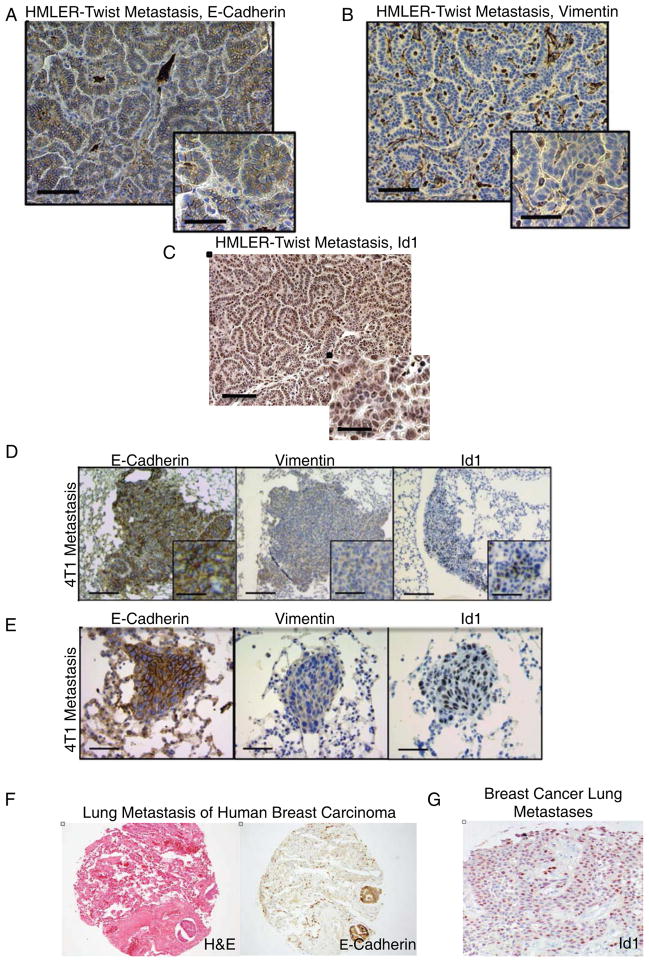
Loss of Id1/3 expression in human breast cancer cells does not affect extravasation from the circulation into the lung parenchyma but inhibits the generation of macroscopic colonies (Gupta, 2007). Recent reports have demonstrated that repression of EMT and the concomitant induction of MET is a necessary step for successful lung colonization of cancer cells (Korpal et al., 2011; Ocana et al., 2012; Siemens et al., 2011; Tsai et al., 2012). The observation that Id1 expression correlates with an epithelial TIC phenotype led us to hypothesize that Id1 plays a role in facilitating MET during lung colonization. To test this, we used the metastatic breast cancer cell line HMLER-Twist that has undergone EMT due to ectopic expression of Twist (Supplemental Figure 2A). Orthotopic injections of HMLER-Twist cells generated primary tumors that maintained a mesenchymal phenotype (Supplemental Figure 2B). Two months post injection, the tumor-bearing mice developed Twist positive lung metastases (on average 4±1SD metastatic lesions per lung) that had undergone MET as seen by the acquisition of E-Cadherin and loss of the Vimentin (Figure 2A&B and Supplemental Figure 2C, D and E). The metastatic colonies also showed strong nuclear staining for Id1 (Figure 2C), supporting our hypothesis that Id1 correlates with an epithelial phenotype during lung colonization.
Figure 2. Id1 Expression is Associated with an Epithelial Phenotype in Breast Cancer Lung Metastases.
(A) IHC of lung metastases from HMLER-Twist cells injected mice reveals tumor cells positive for E-Cadherin and negative for Vimentin (B). Scale bar = 200μm, inset scale bar = 100μm.
(C) Id1 staining in HMLER-Twist lung metastases. Scale bar = 200μm, inset scale bar = 100μm.
(D) 4T1 cells form lung metastases positive for Id1 and E-Cadherin, and negative for Vimentin as determined by IHC. Scale bar = 500μm, inset scale bar = 100μm
(E) IHC for Id1, E-Cadherin, and Vimentin in macrometastases 5 days after 105 4T1 cells were injected into the tail vein of Balb/c mice. Scale bar = 200μm.
(F) Hematoxylin-eosin and E-Cadherin staining of human lung parenchyma showing two tumor emboli of metastatic breast carcinoma.
(G) IHC for Id1 in lung metastasis of human breast arcinoma.
To confirm the generality of our results we examined Id1 expression in metastatic lesions derived from the murine mammary cancer cell line 4T1 (Supplemental Figure 2F). Mice injected orthotopically with 4T1 cells developed lung metastases positive for Id1, the epithelial marker E-Cadherin and negative for Vimentin (Figure 2D and Supplemental Figure 2G and 2H). To confirm Id1 accompanies colony initiation rather than solely proliferation in an established macrometastasis, we analyzed the presence of Id1 protein in early metastatic lesions (5 days post injections). 4T1 cells formed on average 12±2 SD micro-metastatic colonies exhibiting an epithelial morphology with uniform Id1 expression (Figure 2E). We were able to corroborate our findings in patients of the triple negative breast cancer subtype (Figure 2F). Analysis of tissue microarrays (TMAs) containing 25 triple negative lung metastatic lesions showed E-Cadherin positivity in 23/25 samples (Supplemental Table 1). Additionally, Id1 was detected in 7/25 samples, all of which were also positive for E-Cadherin (Figure 2G and Supplemental Table 1). Thus, Id1 expression correlates with epithelial marker expression during early tumor initiation steps of breast cancer colonization of the lungs.
Id Expression Induces MET in Twist Expressing Cells
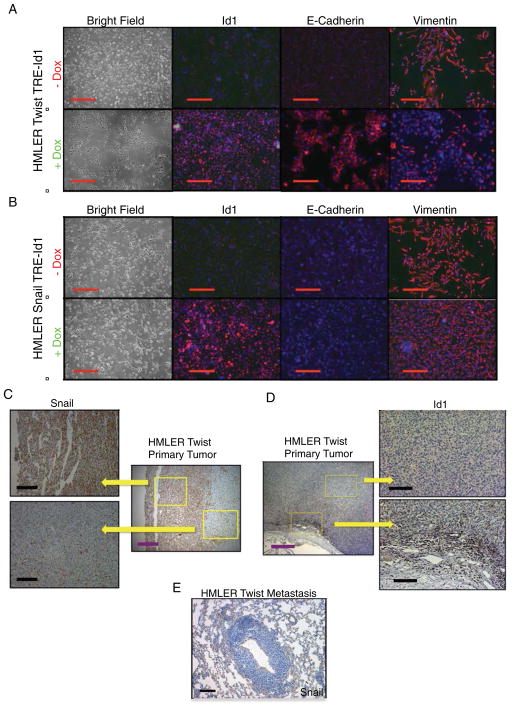
Since Id proteins act as dominant negative regulators of bHLH transcription factors, we hypothesized that Id1 antagonism of Twist controls the differentiation state of breast cancer cells that have undergone Twist-induced EMT. HMLER-Twist cells were transduced with a lentivirus expressing the tetracycline transactivator driven by the Ubiquitin C (UbC) promoter and the ID1 gene under control of a tetracycline response element (pTRIPZ TRE-Id1). In the presence of doxycycline, HMLER-Twist TRE-Id1 cells show robust Id1 expression (Supplemental Figure 3A and Supplemental Figure 4A) and undergo MET as evidenced by the loss of their mesenchymal morphology and re-acquisition of E-cadherin and reduced Vimentin expression (Figure 3A). We wanted to determine if Id1 induction of MET depends on its ability to antagonize the Twist1 bHLH transcription factor. Snail belongs to the zinc-finger family of transcription factors, lacks the helix-loop-helix protein-binding motif, and therefore is not antagonized directly by Id1. In contrast to HMLER-Twist cells, Id1 overexpression did not lead to MET in cells that had undergone Snail -induced EMT (Figure 3B). These findings suggest that Id1 does not induce MET in all mesenchymal cells, but rather likely does so by specifically antagonizing bHLH transcription factors, such as Twist.
Figure 3. Id1 Induces the Mesenchymal-to-Epithelial Transition in Cells that Express Twist but not in Cells Expressing Snail.
(A) Phase contrast images of HMLER-Twist cells infected with a lentivirus carrying the Id1 gene under a tetracycline regulatable promoter, without (top panel) and with (bottom panel) the addition of doxycycline to the growth media. IHC for Id1 (left column), E-Cadherin (middle column) and the Vimentin (right column). Scale bar = 200μm.
(B) Phase contrast images of HMLER-Snail cells carrying the Id1 gene under a tetracycline regulatable promoter, without (top panel) and with (bottom panel) the addition of doxycycline.
IHC for Id1 (left column), E-Cadherin (middle column) and Vimentin (right column). Scale bar = 200μm.
(C) IHC of HMLER-Twist tumors reveals Snail positive cells near the invasive edge, while no Snail protein is present in pulmonary metastases (E). Purple scale bar = 500μm, black scale bar = 200μm.
(D) IHC of HMLER-Twist tumors for Id1. Purple scale bar = 500μm, black scale bar = 200
A recent analysis of temporal and spatial expression of EMT transcription factors in human breast cancer revealed that Snail1 is required for EMT initiation at the invasive edge of the primary tumor, while Twist1 mainly acts to maintain EMT during later stages of metastatic dissemination and possibly in dormant micrometastases (Tran et al., 2011). We made a similar observation in HMLER-Twist tumors: Snail expression was confined to the primary tumor at the invasive edge (Figure 3C) but was absent from metastatic lesions in the lungs (Figure 3E). As expected, Twist was expressed in both primary tumor and metastases (Supplemental Figure 2C). Id1 was expressed in HMLER-Twist tumors at the primary site (Figure 3D) in addition to the lung metastatic site (Figure 2C). Interestingly, the Id1 staining pattern closely followed Snail expression in the primary site (Figure 3C and Figure 3D): absent in the tumor center and present at the invasive edge. Such spatial control may allow Id1 to exert its TIC and pro-proliferative properties while cells maintain their mesenchymal character required for extravasation.
Id Expression Promotes Pulmonary Metastasis via Induction of MET
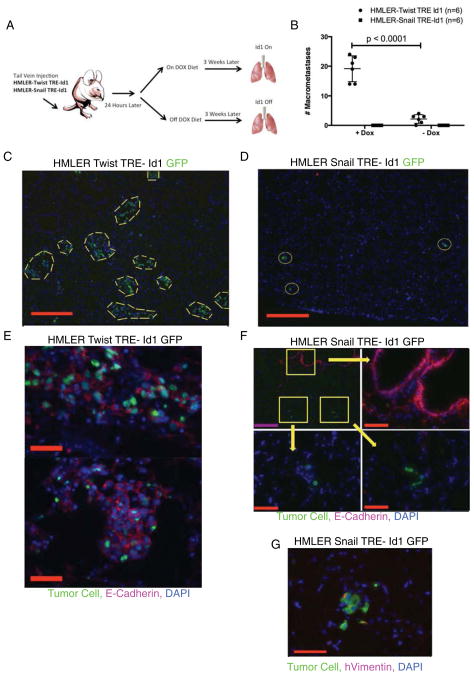
We next sought to determine if Id1 expression with concomitant MET induction is sufficient to increase metastatic colonization. HMLER-Snail TRE-Id1 and HMLER-Twist TRE-Id1 cells were injected into the tail veins of immune-compromised mice and after 24 hours when cells extravasated into the lung parenchyma, the animals were placed on a doxycycline feed diet in order to induce expression of Id1 (Figure 4A, Supplemental Figure 3A and 4A). This allowed us to assess the effect of Id1 specifically during the early colonization stages of the metastatic cascade. Similar levels of Id1 protein expression were detected in cells that extravasated successfully in both HMLER-Twist and HMLER-Snail injected animals (Supplemental Figure 4B). However, only the lungs of mice injected with HMLER-Twist Id1 overexpressing cells (Figure 4B&C and Supplemental Figure 4C) contained macrometastases (on average 20±4), while the lungs of mice injected with HMLER-Snail Id1 cells contained only single cancer cells and micrometastases (Figure 4B&D). Consistent with what we observed in vitro, the metastatic lesions formed by HMLER-Twist cells overexpressing Id1 underwent MET (Figure 4E), while HMLER-Snail TRE-Id1 micrometastases did not (Figure 4F). The failure of HMLER-Snail TRE-Id1 cells to undergo MET was further supported by the presence of Vimentin in the micrometastases (Figure 4G). We conclude that Id1 expression is sufficient to induce MET and promote colonization in Twist but not Snail expressing disseminated cells.
Figure 4. Id1 Enhances the Colonization of HMLER-Twist Cells via MET but not of HMLER-Snail Cells in Vivo.
(A) Schematic illustrating the tail vein injection of HMLER-Snail TRE-Id1 or HMLER-Twist TRE-Id1 cells into immune-compromised host mice.
(B) Quantification of the number of macrometastases in the lungs of mice 3 weeks after being injected with 105 HMLE-Ras Twist TRE-Id1 and HMLE-Ras Snail TRE-Id1 cells via tail vein (n = 5 for each group).
(C) Representative IF images of lungs from mice injected with GFP+ HMLE-Ras Twist TRE-Id1 and (D) GFP+ HMLE-Ras Snail TRE-Id1. Yellow dashed lines encircle macrometastases while yellow circles encircle micrometastases. Scale bar = 500μm.
(E) Two representative IF images of GFP+ HMLE-Ras Twist TRE-Id1 macrometastases expressing E-Cadherin (scale bar = 500μm), while (F) GFP+ HMLE-Ras Snail TRE-Id1 micrometastases (white arrows) do not express E-Cadherin. Purple scale bar = 500μm, red scale bar = 100μm.
(G) IF staining of GFP+ HMLE-Ras Snail TRE-Id1 micrometastases for Vimentin expression. Scale bar = 100μm.
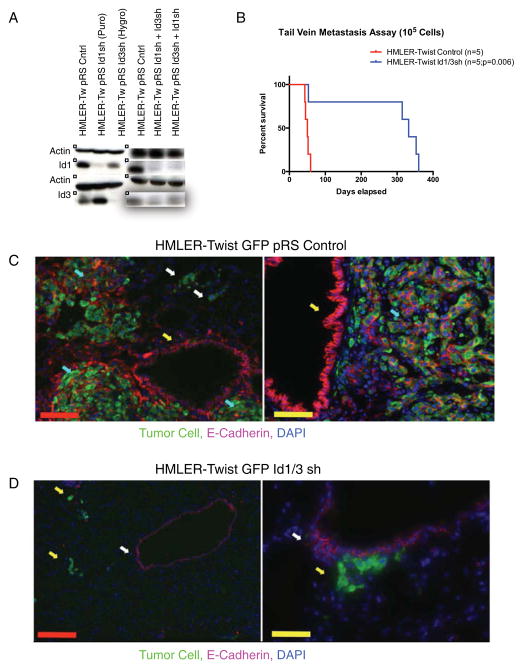
To determine if Id1 is necessary for MET during lung colonization, HMLER-Twist cells with reduced Id1/3 levels were generated using previously validated short-hairpin RNA sequences (Figure 5A and Supplemental Figure 5A) (Gupta et al., 2007). Knockdown of both Id1 and Id3 was required due to a redundancy in function between the two proteins (Gupta et al., 2007; Perk et al., 2005). The proliferation rate (Supplemental Figure 5A) and mesenchymal morphology (not shown) of these cells were unaffected. While all the animals injected with the control HMLER-Twist cells died within 60 days due to heavy lung metastatic burden (Figure 5B), only 1/5 mice injected with HMLER-Twist Id1/3 shRNA died due to pulmonary metastases (Figure 5B). Importantly, the lung metastatic lesions found in the one deceased mouse stained positive for Id1, indicating that metastases were derived from a subpopulation of cells escaping the Id1 knockdown (Supplemental Figure 5B). Metastases generated from control cells expressed E-Cadherin (Figure 5C). Interestingly, solitary cancer cells in the lungs that failed to transition from micro- to macro-metastases did not express E-Cadherin (Figure 5C, white arrows), indicating that HMLER-Twist cells likely reach the lungs while retaining their mesenchymal phenotype. The lungs of mice injected with HMLER-Twist Id1/3 shRNA cells contained micrometastatic colonies, supporting our previous finding that Id1/3 inhibition does not abrogate the extravasation ability of metastasizing breast cancer cells (Figure 5D) (Gupta et al., 2007). These micrometastatic colonies also failed to express E-Cadherin even when they reached colonies of approximately 10 cells (Figure 5D, right panel). We conclude that Id1/3 loss in metastasizing breast cancer cells results in a failure to initiate MET. Although HMLER-Twist and HMLER-Snail cells behave as bona fide TICs and are capable of generating mesenchymal-like tumors at the primary site from limited dilutions (data not shown) (Mani et al., 2008), our studies suggest that tumor initiation in the lungs places additional requirements on the metastasizing cell, which are in part fulfilled by the cell undergoing an MET. Thus, Id1 allows breast cancer cells to retain their TIC phenotype while re-acquiring their epithelial character necessary for lung colonization.
Figure 5. Id1 and Id3 Are Required for HMLER-Twist Colonization of the Lungs.
(A) Western blot showing knockdown of Id1 and Id3 in HMLER Twist cells using retroviruses expressing shRNA sequences targeting the two genes.
(B) Kaplan-Meier survival curves for mice injected via tail veins with 105 HMLE-Ras Twist control and HMLE-Ras Twist Id1/3 short-hairpin cells (n = 5 for each group).
(C) Two representative IF images from the lungs of a mouse injected with GFP+ HMLE-Ras Twist control cells. White arrows point to E-Cadherin negative micrometastases and cyan arrows point to E-Cadherin positive macro-metastases. The yellow arrow points to E-cadherin positive bronchiole cells. Red scale bar = 300μm, yellow scale bar = 100μm.
(D) Two representative IF images from the lungs of a mouse injected with GFP+ HMLE-Ras Twist Id1/3 short-hairpin cells. White arrows point to E-Cadherin negative micrometastases and the yellow arrow points to E-cadherin positive bronchiole cells. Red scale bar = 300μm, yellow scale bar = 100μm.
Id1 Expression is Regulated by TGFβ Signaling in Mesenchymal-Like Breast Cancer Cells
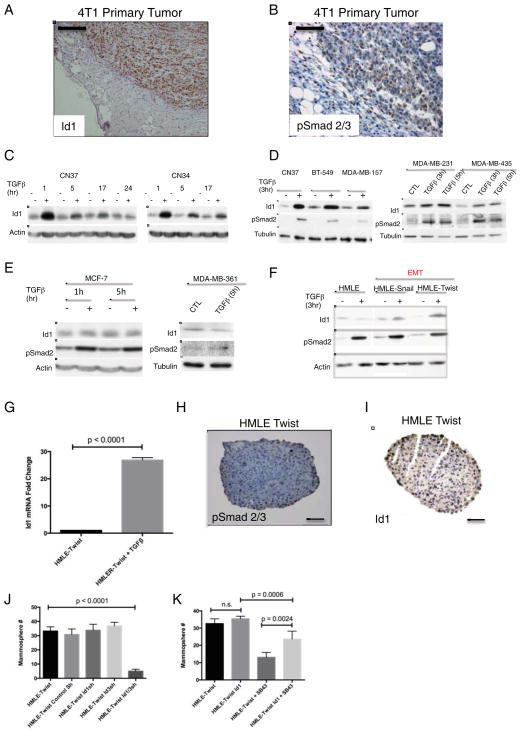
We next addressed the mechanism of Id1 regulation during metastatic progression. Our observation that Id1 is expressed at the invasive edge (Figure 3D & Figure 6A) of the primary tumor, led us to postulate that signals present at the tumor-stroma interface were responsible for Id1 induction. A factor implicated in switching cancer cells from a cohesive epithelial state into a mesenchymal migratory phenotype is the cytokine TGFβ (Massague, 2012). TGFβ pathway activation in primary breast tumors is associated with increased risk of lung metastases (Padua et al., 2008). The same study showed TGFβ upregulation of Id1 in cells isolated from pleural fluids of patients with metastatic breast cancer. These findings prompted us to explore the potential link between Id1 expression and TGFβ activity. Activation of the TGFβ pathway in 4T1 breast tumors was detected by nuclear accumulation of phosphorylated Smad2/3 and followed a similar although broader localization pattern as Id1 positive cells: staining was mostly at the invasive edge of the tumor and absent from the center of the tumor (Figure 6A&B). Importantly, the tumor invasive edge is where cells initially undergo EMT and acquire a migratory phenotype as evidenced by positive Vimentin staining (Supplemental Figure 6A).
Figure 6. Id1 is Induced by TGFβ in Breast Cancer Cell Belonging to the Basal Subtype and is Required for TGFβ Induction of TICs.
(A) IHC for Id1 shows Id1 positive cells present at the invasive edge of 4T1 tumor allografts from mammary fat pads of C57BL/6 mice. Scale bar = 100μm.
(B) IHC for phospho-Smad2/3 in 4T1 primary tumor allografts. Scale bar = 100μm.
(C) CN37 and CN34 cells were treated with 100pm TGFβ for the indicated time periods. Cells were serum starved for 3 h prior to TGFβ treatment. Protein levels of Id1, pSmad2 and actin were determined by immunoblot.
(D–E) Id1 response to TGFβ was evaluated in a panel of breast cancer cell lines classified either as basal (D) or luminal subtype (E). Total protein was subjected to Western Blot analysis.
(F) HMLE, HMLE-Snail, and HMLE-Twist cells were serum starved for 3h before being treated with 100 pM TGFβ for 3h. Protein levels of Id1, pSmad2 and actin were determined by immunoblot.
(G) Quantification of Id1 mRNA by qRT-PCR in HMLE-Twist mammospheres following the treatment with 100pM TGFβ for 3 h.
(H) IHC for p-Smad2/3 in HMLE-Twist mammosphere. Scale bar = 200μm.
(I) IHC for Id1 in HMLE-Twist mammosphere. Scale bar = 200μm.
(J) Quantification of mammospheres formed by HMLE-Twist cells and HMLE-Twist cells expressing RNA short-hairpins for Id1 and/or Id3.
(K) Quantification of mammospheres formed by HMLE-Twist cells, HMLE-Twist cells treated with the TGFβ inhibitor SB-431542 (2μM), and HMLE-Twist cells overexpressing Id1 with or without the TGFβ inhibitor.
Previous work demonstrated that TGFβ represses Id1 in non-tumorigenic human epithelial cell lines (Kang et al., 2003), while in some cancer cells TGFβ induces Id1 expression (Anido et al., 2010; Padua et al., 2008). Using cells isolated from the pleural effusions of patients with advanced breast cancer of the triple negative subtype (Gomis et al., 2006)(CN37 and CN34 cell lines), we found that TGFβ treatment leads to an increase in Id1 protein levels (Figure 6C). Continued treatment with TGFβ did not result in Id1 inhibition (Figure 6C) although a dampening of the effect was seen that could be reversed by the addition of fresh TGFβ (not shown). To further characterize Id1 responsiveness to TGFβ, a panel of breast cancer cell lines categorized as either of luminal or basal subtype were employed. Id1 induction in response to TGFβ was detected only in those belonging to the basal-like subtype (BT- 549, MDA-MB-157, MDA-MB-231, and MDA-MB- 435)(Figure 6D), while no Id1 induction was observed in MCF-7 and MDA-MB-361 luminal breast cancer cells (Figure 6E) (Finn et al., 2009). Since most basal-like cell lines are categorized as post-EMT due to expression of Vimentin, we hypothesized that the effect of TGFβ on Id1 expression may be dependent on the mesenchymal state of the cell. Indeed, while TGFβ did not induce Id1 in the epithelial parental cell line (HMLE), Id1 was upregulated in both mesenchymal derivatives (HMLE-Snail and HMLE-Twist)(Figure 6F and Supplemental Figure 6B). Collectively, these results suggest a novel link between the acquisition of mesenchymal characteristics and a switch in the Id1 response to TGFβ.
Given that both TGFβ and Id1 are inducers of TIC properties in breast cancer cells, we sought to determine if Id1 is a critical mediator of TGFβ activity. TGFβ treatment of mammospheres resulted in significant upregulation of Id1 mRNA (Figure 6G) demonstrating that Id1 remains under TGFβ control during mammosphere formation. Id1 and phosphorylated Smad2/3 were present in HMLE-Twist mammospheres (Figure 6H and 6I) consistent with the finding that endogenous TGFβ governs the TIC phenotype in breast cancer cells, which could be Id1/3 mediated (Scheel et al., 2011; Shipitsin et al., 2007). Consistent with this hypothesis, knockdown of Id1/3 led to fewer mammospheres formed by HMLE-Twist cells (Figure 6J). Inhibition of TGFβ by a small molecule inhibitor SB-431542 led to decreased pSmad2/3 levels in HMLE-Twist cells (Supplemental Figure 6B) and a reduced ability of HMLE-Twist cells to form mammospheres (Figure 6K). To determine if TGFβ governs the TIC phenotype via Id1 induction, HMLE-Twist cells overexpressing Id1 were treated with TGFβ inhibitor. Indeed, Id1 overexpression partially rescued the mammosphere formation potential suggesting that induction of Id1 by TGFβ is partly sufficient for this process (Figure 6K). Together, these results indicate that Id1 upregulation by TGFβ is necessary and sufficient for the acquisition of TIC phenotype in cultured breast cancer cells.
Id1 is Regulated by the Canonical Smad-dependent TGFβ Pathway and requires the Activity of CBP/p300 Co-Activators
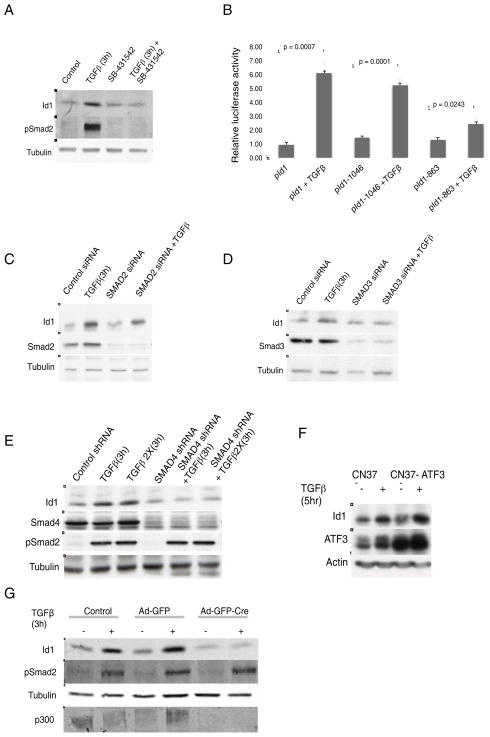
TGFβ driven induction of Id1 was abrogated by treatment of CN37 cells with either actinomycin D or cycloheximide, demonstrating that Id1 upregulation occurs at the level of transcription and requires de novo protein synthesis (Supplemental Figure 7A & B). TGFβ transduces signals through a heterodimeric receptor complex formed by the type I (TβRI) and the type II (TβRII) receptors. In the case of canonical TGFβ signaling, the activated receptor complex phosphorylates Smad2 and Smad3 transcription factors, which results in their binding to Smad4 and translocation to the nucleus where they regulate the transcription of target genes (Massague, 2012). To confirm that Id1 induction is mediated by TGFβ receptors, cells were treated with TGFβRI inhibitor, SB-431542, which resulted in a complete block of Id1 upregulation by TGFβ (Figure 7A and Supplemental Figure 6B). Deletion analysis of the Id1 promoter containing a luciferase reporter (Lopez-Rovira et al., 2002) identified the −1046 to −863 base-pair region as necessary for Id1 induction (Figure 7B, Supplemental Figure 7C). Interestingly, this sequence containing the Smad-binding element (SBE) is the same promoter region responsible for Id1 downregulation by TGFβ in normal epithelial cells (Kang et al., 2003). To examine the potential contribution of different Smads, individual knockdowns of Smad2, Smad3 and Smad4 were performed (Figure 7C–E). Smad2 inhibition did not block Id1 induction (Figure 7C), while the knockdown of either Smad3 or Smad4 prevented the Id1 response to TGFβ (Figure 7D & E).
Figure 7. TGFβ Induces Id1 Expression Through Smad Dependent Pathways and Requires the CBP/p300 Co-Activator.
(A) CN37 cells were treated with 100pM TGFβ for 3 h in the absence or presence of 2μM TβRI inhibitor SB431542. Levels of Id1, p-Smad2 and actin were determined by Western blot.
(B) Luciferase reporter activity characterizing the minimal ID1 gene promoter region necessary for Id1 induction by TGFβ. CN37 cells were transfected with indicated Id1 reporter constructs and treated with or without TGFβ for 20 h.
(C) Immunoblots using indicated antibodies were carried out using CN37 cells transfected with control, Smad2, Smad3 (D), or Smad4 shRNA (E) in the presence of absence of TGFβ for 3h.
(F) CN37 cells were transduced with vector control or lentivirus expressing ATF-3 and treated with or without TGFβ. ATF-3 protein levels were determined by Western Blot.
(G) Western blot of mouse embryonic fibroblasts (MEF) with the floxed CBP/p300 gene infected with control GFP+ adenovirus (Ad-GFP) or virus carrying Cre-Recombinase (Ad-GFP-Cre) and treated with TGFβ.
Id1 repression by TGFβ in normal epithelial cells is dependent on the transcriptional repressor ATF-3 (Kang et al., 2003). We observed that TGFβ still leads to the induction of ATF-3 protein in breast cancer cells (Figure 7F). Moreover, even after overexpression of ATF-3, TGFβ still maintains the ability to upregulate Id1, arguing that the effect of TGFβ is independent of low levels of ATF-3 protein (Figure 7F).
Smad complexes have the ability to recruit a wide variety of co-factors, which allow Smads to elicit a wide range of tissue specific responses. One of the known regulators of TGFβ signaling are co-activators CBP and p300 proteins, which in addition to acting as transcriptional activators also possess histone acetyltransferase activity. A recent report has implicated the importance of CBP/p300 in TGFβ mediated transcription of target genes in Vimentin positive mesangial cells (Yuan et al., 2013), prompting us to explore the role of these co-activators in TGFβ upregulation of Id1 in mesenchymal cells. Due to the redundant roles of CBP and p300 we used MEFs carrying homozygous conditional knockout alleles of both CBP and p300 (double knockout or dKO MEFs) (Kasper et al., 2010). Since MEFs express mesenchymal markers, we postulated that in this cellular context TGFβ induces Id1. As expected, TGFβ stimulation of non-transduced MEFs and those transduced with GFP control (Ad-GFP) led to strong upregulation of Id1. However, cells transduced with Cre-recombinase (dKO MEFs), which lost the expression of both CBP and p300, failed to induce Id1 following TGFβ treatment (Figure 7G). Thus, these results demonstrate that TGFβ mediated Id1 upregulation requires the activity of the CBP/p300 coactivators.
In summary, upregulation of Id1 by TGFβ in breast cancer requires a transition to a mesenchymal state, which manifests at the periphery of the primary tumor and primes the Id1 promoter for activation by canonical Smad3/4 signals in combination with CBP/p300. As described above, upregulation of Id1 is essential for the eventual colonization of the lung metastatic site.
DISCUSSION
Recently published studies support the notion that the epithelial to mesenchymal transition plays an important role in carcinoma metastasis (Thiery et al., 2009). While the bulk of the primary tumor is constrained by epithelial attachments, mesenchymal cells have an enhanced ability to migrate and traverse the barriers of endothelial cell junctions. This ability facilitates entry and exit from the circulation, critical steps in metastatic progression. The observation that induction of EMT in breast tumor cells by forced expression of the transcription factors Snail and Twist was associated with the enhancement of tumor sphere formation and the expression of stem-like markers, suggested the EMT transition also endows these cells with tumor initiating capacity (Mani et al., 2008). This concept connected the cell migration traits with the TIC programs such as the ability to colonize the metastatic site and potentially confer drug resistance. It appears now however that EMT and TIC properties are not always inextricably linked: loss of the homeobox protein Prrx1 was found to be required for metastatic colonization of BT-549 breast cancer cells and such loss is associated with a mesenchymal to epithelial transition (MET) and the acquisition of TIC capacity (Ocana et al., 2012). Additionally, forced repression of Twist protein at the metastatic site was shown to be associated with epithelial differentiation and enhanced colonization. Thus, the recent evidence not only uncouples “stemness” character from EMT, but also further fosters the notion that in order to form macrometastases, migrating cancer cells need to undergo MET. Therefore, understanding what contextual signals trigger MET is critical for understanding the underlying mechanism of metastatic progression and the development of improved clinical approaches.
Recent reports have elucidated some of the mechanisms that regulate MET, including the dynamic regulation of EMT inducing factors during the metastatic cascade. Snail1 is specifically required for EMT initiation at the invasive edge of primary tumors and high Snail1 expression in primary tumors relative to corresponding metastases correlates with distant relapses (Blanco et al., 2002; Peinado et al., 2004; Tran et al., 2011). Similarly, we observe Snail expression only at the invasive edge of the HMLER-Twist primary tumors. Conversely, Twist1 is dispensable for EMT initiation and Snail1 expression represses Twist1 transcription directly during Snail-induced EMT (Tran et al., 2011). Twist1 was shown to mainly act to maintain the EMT state in later stages of metastasis and while Twist1 expression in the primary tumors does not correlate with distant recurrence, high Twist1 expression in breast cancer bone marrow disseminated tumor cells is strongly associated with metastatic recurrences (Tran et al., 2011). A potential mechanism for how Snail expression is lost during metastatic colonization is provided by studies identifying micro-RNAs that target transcripts of various EMT factors for degradation and lead to induction of MET (Kim et al., 2011; Siemens et al., 2011). In particular, Snail and Zeb1/2 zinc finger transcription factors have been shown to be downregulated by miRNAs 34 and 200 respectively, which may contribute to their regulation in vivo (Burk et al., 2008; Korpal et al., 2008; Siemens et al., 2011). However, how other critical EMT transcriptional factors, such as bHLH factor Twist, are downregulated to allow for MET and colonization remains unclear. The studies presented here suggest that this is accomplished by the dominant negative transcription factor Id1, which opposes Twist activity likely by direct protein-protein interactions (Figure 8) (Spicer et al., 1996).
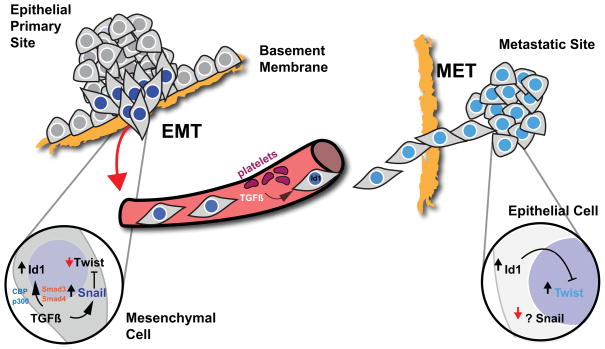
Figure 8. Proposed Model For Id1 Induced MET During Metastatic Colonization of Lungs by Breast Cancer Cells.
Integral for the metastatic dissemination of carcinoma cells is the acquisition of an invasive phenotype via induction of EMT. A key cytokine present at the tumor-stroma boundary, TGFβ, induces the EMT transcription factor Snail, which changes the Id1 response to TGFβ in disseminating cells. TGFβ mediated induction of Id1 is dependent on Smad3/4 and CBP/p300 cofactors. Id1 expression in the primary tumor does not revert Snail induced EMT, which allows Id1 expressing cells to complete the steps required for dissemination: intravasation, survival in circulation, and extravasation. In the circulation, platelet derived TGFβ continues to upregulate Id1. At the metastatic site, where Snail is absent, Id1 opposes the activity of EMT transcription factor Twist. Such inhibition leads to MET and the colonization of distant organs.
We also present evidence supporting a role for Id1 in endowing breast cancer cells with a TIC phenotype that is independent of EMT and show the TIC phenotype correlates with the ability to initiate tumor growth at the secondary site. Id1 expressing TICs retain an epithelial character and, importantly, are capable of initiating tumor growth both in mammary fat pads and during colonization of the lung parenchyma. We show here that while high levels of Id1 are present at the invasive edge of primary breast tumors, cells can retain the mesenchymal characteristics induced by Snail since Id1 does not have the ability to counter the activity of this factor. TGFβ mediated induction of Snail and EMT at the primary site makes the Id1 promoter receptive to TGFβ upregulation. In the bloodstream, platelets are the major source of TGFβ and platelet–specific loss of TGFβ is sufficient to prevent metastasis (Labelle et al., 2011). Thus, we speculate that in the circulation, TGFβ continues to induce Id1 and the mesenchymal state remains under the control of Snail1.
We show that TGFβ upregulates Id1 through the combined action of Smads3/4 and CBP/p300. In addition to being transcriptional co-activators, CBP/p300 possess intrinsic histone acetyltransferase activity (HAT) that modifies lysine residues of the core histone tails (Ogryzko et al., 1996). Thus, it is possible that CBP/p300 driven acetylation of Id1 promoter histones inhibits binding of ATF-3 to the Id1 promoter, while recruiting Smad3 for transcriptional activation of Id1. CBP/p300 have been demonstrated to acetylate non-histone substrates, such as Smad3, raising the possibility that similar to other context-dependent modifications of Smad proteins, acetylation may be an additional mechanism of regulatory control. Finally, a novel small-molecule inhibitor of p300 was shown recently to induce cytotoxicity and suppress tumor growth specifically in triple negative cell lines (Yang et al., 2013). These results are consistent with our data suggesting a role of CBP/p300 in mesenchymal breast cancer cells.
Upregulation of Id1 at the primary site may contribute to the enhanced primary tumor growth observed in breast cancer models due to its enhanced TIC properties and/or opposition to cell cycle inhibitors such as p16 and p21(O’Brien et al., 2012; Ohtani et al., 2001). In addition, upregulation of Id1 primes the cells for the eventual conversion back to the epithelial state via antagonism of Twist at the secondary site. Human samples of squamous cell carcinoma metastases often display strong Twist protein expression (Merikallio et al., 2011) and in human breast cancer Twist expression is present in disseminated tumor cells. In animal models used in our studies, Twist protein expression is present in metastatic colonies. However, we show that Twist activity is opposed by Id, leading to the induction of MET and the conversion of disseminated pre-metastatic tumor cells into macrometastatic colonies. Thus, our study provides insight as to how one of the critical EMT-inducers is inhibited to facilitate the formation of metastases.
We speculate that the maintenance of the TIC state after extravasation, at least in part mediated by Id1, is essential for early colonization since sphere formation assays (which define TICs) likely mimic the growth conditions for small numbers of cells in a non-adherent milieu prior to colonization. Once initiated, epithelialization is then required for the formation of macroscopic colonies, possibly by reversion of the growth inhibitory properties of the mesenchymal state (Brabletz et al., 2001). Such phenotypic plasticity between EMT and MET states has been proposed as an essential trait of malignant cells completing all the complex steps of the metastatic cascade in some cancers (Brabletz, 2012; Scheel and Weinberg, 2012). Recent work demonstrating tumor-initiating cell plasticity, as evidenced by conversions between a non-TIC and a TIC phenotype within basal breast cancer cells, further supports phenotypic plasticity during cancer metastasis (Chaffer et al., 2013)
Relevance to Human Disease
Id1 expression and the TGFβ -responsive gene signature correlate with poor prognosis in the triple negative subtype of breast cancer (Buck et al., 2004; Gupta et al., 2007). These tumors exhibit metaplastic morphology, mostly belong to the claudin-low subtype and are mesenchymal in character at the primary site (Gupta et al., 2007; Perk et al., 2006). Therefore, our results showing Id1 induction of MET in claudin-low cell lines (HMLER-Twist) (Morel et al., 2012)) during colonization and the requirement for this transition in animal models of metastasis suggests our results may be relevant to human breast cancer metastasis. Indeed, using human tissue microarrays of matched lung metastases obtained from patients with triple negative breast tumors, we show that while primary tumors are associated with mesenchymal features, their corresponding metastases display strong E-cadherin expression, suggesting the importance of epithelial traits in human metastatic colonization. Strong Id1 expression was detected in 7/25 human breast cancer metastases suggesting Id1 may be essential for the formation of some but not all metastases or that Id1 expression may be transient. We also note that the requirement for MET during metastatic colonization might not be universal as some tumor types retain their mesenchymal character at the secondary site (Acharyya et al., 2012).
The importance of targeting different populations of human tumor cells that play different roles in tumor relapse and responses to therapy has been established as an important future step in cancer drug development (Singh and Settleman, 2010). The compound salinomycin acts as a potent inhibitor of tumor growth in mouse tumor models by specifically targeting the mesenchymal, TIC sub-population (Gupta et al., 2009). However, our results point towards the existence of different populations even within the bounds of the tumor initiating cell phenotype that play different roles in the metastatic cascade and may be important to target as well. That is, colonizing epithelial-TICs might be important cells to target for effective metastasis intervention. Since distant organ colonization is the ultimate cause of death for most breast cancer patients, developing drugs that target this particular phase of metastasis is of paramount importance.
Experimental Procedures
Cell Culture and Reagents
MCF-7, MDA-MB-157, MDA-MB-231, MDA-MB-361, MDA-MB-435, BT-549 tumor cell lines and HMLE cell lines were cultured as described in Supplemental Data. CN37 and CN34 cells were isolated from the pleural effusion of patients with metastatic breast cancer (Gomis et al., 2006). HMLE (immortalized human mammary epithelial cells) and HMLER (V12H-Ras transformed HMLE cells) were a kind gift from Robert Weinberg (Elenbas et al., 2001). CBP/p300 dKO MEFs were a generous gift from Paul Brindle. TGF-β1 (R&D Systems), SB431542 (Sigma), cycloheximide (Sigma), and actinomycin D (Sigma) were used as indicated.
Additional Methods can be found in the Supplemental Data.
Supplementary Material
Highlights.
Id1 expression generates tumor-initiating cells independently of EMT
Id1 expression correlates with an epithelial phenotype in breast cancer metastasis
Id1 drives MET and pulmonary metastatic colonization by antagonizing Twist1
TGFβ mediated upregulation of Id1 is dependent on an EMT induced mesenchymal state
Significance.
The critical steps of the metastatic cascade include dissemination of cancer cells from the primary tumor and their subsequent colonization of the secondary site. Our findings identify Id1 as a potent inducer of tumor initiating properties in breast cancer cells. We show that high Id1 levels lead to the reversion of EMT to an epithelial phenotype, which is required for colonization of the metastatic site. Our work provides evidence for the importance of dynamic EMT-MET processes in metastatic progression, and identifies Id1 as a central regulator of such transitions.
Acknowledgments
We thank P. Brindle for CBP/p300 double null MEFs. We are grateful to L. Barrett, P. Cook, D. Marks, Eduard Nedea and J. Schvartzman for critical reading of the manuscript and their helpful suggestions. We would also like to thank the Laboratory of Comparative Pathology, Flow Cytometry Core Facility and Molecular Cytology Core Facility (MSKCC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Marko Stankic, Email: ms2826@cornell.edu.
Svetlana Pavlovic, Email: svp2003@med.cornell.edu.
Yvette Chin, Email: chiny@mskcc.org.
Edi Brogi, Email: brogie@mskcc.org.
David Padua, Email: dpadua@mednet.ucla.edu.
Larry Norton, Email: nortonl@mskcc.org.
Joan Massague, Email: massaguj@mskcc.org.
References
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D, Holland EC, Nam HS, Benezra R. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, Knuechel R, Kirchner T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MB, Fritz P, Dippon J, Zugmaier G, Knabbe C. Prognostic significance of transforming growth factor beta receptor II in estrogen receptor-negative breast cancer patients. Clin Cancer Res. 2004;10:491–498. doi: 10.1158/1078-0432.ccr-0320-03. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R, Massague J. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic V, Ciarrocchi A, Boccuni P, DeBlasio T, Benezra R, Nimer SD. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci U S A. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. Embo J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277:3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikallio H, Kaarteenaho R, Paakko P, Lehtonen S, Hirvikoski P, Makitaro R, Harju T, Soini Y. Zeb1 and twist are more commonly expressed in metastatic than primary lung tumours and show inverse associations with claudins. J Clin Pathol. 2011;64:136–140. doi: 10.1136/jcp.2010.086678. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, Devouassoux-Shisheboran M, Treilleux I, Tissier A, Gras B, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS genetics. 2012;8:e1002723. doi: 10.1371/journal.pgen.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, Tsatsanis A, Gallinger S, Dick JE. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Marin F, Cubillo E, Stark HJ, Fusenig N, Nieto MA, Cano A. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J Cell Sci. 2004;117:2827–2839. doi: 10.1242/jcs.01145. [DOI] [PubMed] [Google Scholar]
- Perk J, Gil-Bazo I, Chin Y, de Candia P, Chen JJ, Zhao Y, Chao S, Cheong W, Ke Y, Al-Ahmadie H, et al. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res. 2006;66:10870–10877. doi: 10.1158/0008-5472.CAN-06-2643. [DOI] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Romero-Lanman EE, Pavlovic S, Amlani B, Chin Y, Benezra R. Id1 maintains embryonic stem cell self-renewal by up-regulation of Nanog and repression of Brachyury expression. Stem Cells Dev. 21:384–393. doi: 10.1089/scd.2011.0428. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65:5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. discussion 6000-5991. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Tran DD, Corsa CA, Biswas H, Aft RL, Longmore GD. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Mol Cancer Res. 2011;9:1644–1657. doi: 10.1158/1541-7786.MCR-11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pinello CE, Luo J, Li D, Wang Y, Zhao LY, Jahn SC, Saldanha SA, Planck J, Geary KR, et al. Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents. Mol Cancer Ther. 2013;12:610–620. doi: 10.1158/1535-7163.MCT-12-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, Natarajan R. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol. 2013;304:F601–613. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.