Abstract
Recent years have seen a rise in publications demonstrating coupling between transcription and mRNA decay. This coupling most often accompanies cellular processes that involve transitions in gene expression patterns, for example during mitotic division and cellular differentiation and in response to cellular stress.
Transcription an affect the mRNA fate by multiple mechanisms. The most novel finding is the process of co-transcriptional imprinting of mRNAs with proteins, which in turn regulate cytoplasmic mRNA stability. Transcription therefore is not only a catalyst of mRNA synthesis but also provides a platform that enables imprinting, which coordinates between transcription and mRNA decay. Here we present an overview of the literature, which provides the evidence of coupling between transcription and decay, review the mechanisms and regulators by which the two processes are coupled, discuss why such coupling is beneficial and present a new model for regulation of gene expression. This article is part of a Special Issue entitled: RNA Decay mechanisms.
1. Introduction
In many ways, transcription can be regarded as the most important part in the mRNA life cycle. It is not only responsible for the synthesis of a transcript itself, but via 5′ capping, splicing and 3′ end formation it also converts a pre-mRNA into an export, translation and decay competent mRNA. These three processes occur co-transcriptionally while a pre-mRNA is still associated with a transcribing RNA II (RNAPII). As transcription proceeds, an RNAPII recruits pre-processing regulators thus temporally dictating the conversion of each pre-mRNA into mature mRNA (reviewed in [1]). Transcription also controls the length of 5′ and 3′ untranslated regions (UTRs) through alternative transcription start site (TSS) choice [2] and alternative polyadenylation (APA) [3]. Since longer UTRs normally contain more cis regulatory sequences, which can be targeted by RNA-binding proteins (RBPs) or microRNAs (miRNAs), alternative TSS and polyadenylation thus affect mRNA stability and/or translatability. RNAPII and associated transcription factors can also recruit various post-transcriptional regulators that are co-transcriptionally deposited or imprinted onto a nascent mRNA (reviewed in [4] and [5], Table 1). By modulating this recruitment process a cell could vary the way a single mRNA species is regulated in the cytoplasm. Such RNAPII-dependent post-transcriptional mRNA regulation could play an important role during growth, differentiation, development and in response to environmental signals.
Table 1.
Mechanisms that couple transcription and mRNA decay.
| Mechanism | Factors/elements | Description | References |
|---|---|---|---|
| mRNA imprinting (demonstrated) | Rpb4/7p | Subunits of RNAPII, which are imprinted on the mRNA in a transcription dependent manner. They regulate export, translation and cytoplasmic decay. Coordinator prototype. | [21], [22], [36], [37], [38] and [39] |
| Pab1p/PABPC1 | Poly-A binding protein. Regulates 3′-end processing, export, translation and decay. | [44], [45], [46], [50], [51], [52], [53], [54] and [55] | |
| Dbf2p | A mitotic kinase that is co-transcriptionally imprinted on SWI5 and CLB2 mRNAs and regulates their timely decay. | [9] | |
| mRNA imprinting (speculateda) | CCR4/NOT | Major cytoplasmic deadenylase found to have many roles in transcription, including a direct role in elongation. | [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [69], [70], [71] and [72] |
| Dbp5p/Rat8p | A DEAD-box helicase implicated in transcription, export, translation and P-body formation. | [73], [74], [75], [76] and [77] | |
| Ssd1p | RNA binding protein which directs its target mRNAs to P-bodies. Nuclear import is required for association with its target mRNAs. | [78] and [79] | |
| Sus1p | A co-factor of SAGA and TREX-2 complexes. Physically interacts with decay factors. | [81] | |
| αCP | RNA binding protein that regulates h α-globin mRNA 3′-end processing and stability during erythroid maturation. | [82] and [83] | |
| Cth2p | ARE binding protein, regulates 3′-end processing and Dhh1-dependent decay of iron deficiency-responsive mRNAs in a transcription-dependent manner. | [84], [85] and [86] | |
| TTP | ARE binding protein, regulates decay of immune response, cell cycle and carcinogenesis related mRNAs. Implicated in NF- κ B induced transcription. | [87], [88] and [89] | |
| KSRP | ARE binding protein implicated in transcription initiation, splicing, RNA editing, mRNA localization and mRNA decay | [91] | |
| ELAV/Hu family | ARE binding proteins that co-assemble with target mRNAs in the nucleus. Regulate 3′ end processing, export and decay, including HuR mRNA itself. | [92], [93], [94], [95] and [96] | |
| AUF1 | ARE binding protein that destabilizes cytokine mRNAs. Associates with pre-mRNAs. | [97], [98] and [99] | |
| TOB/BTG family | Anti-proliferative proteins that modulate transcription and deadenylation by association with transcription factors and PABPC1 and CCR4-NOT, respectively. | [100] | |
| Alternative transcription start site (TSS) | cis elements in 5′ UTR | Certain elements are required for human Dcp2 binding and subsequent decapping and decay. | [104] |
| Alternative splicing | cis elements in the ORF | The ORF of several mRNAs (e.g. APP, c-fos, Mn-SOD) determines their stability. | [110] |
| Alternative poly-adenylation (APA) | cis elements in 3′ UTR | 3′ UTRs contain many mRNA stability cis elements (e.g. ARE, PUF, GU-rich elements, miRNA binding sites); longer UTRs may contain more regulatory elements. | [3], [115] and [116] |
| Promoter-regulated decay | cis elements in promoters | Promoter elements in yeast and mammalian cells were shown to regulate mRNA decay. | [9], [24] and [25] |
Co-transcriptional mRNA imprinting and/or a post-transcriptional role has not been demonstrated.
An essential and well-controlled component of the gene expression system is the cytoplasmic mRNA decay pathway, considered to represent the end-point of the mRNA life. Following shortening of the mRNA poly(A) tail by deadenylases, the eukaryotic mRNA can then be degraded via two pathways: from 3′ to 5′ by the cytoplasmic exosome or from 5′ to 3′ by the Xrn1p exonuclease. The latter pathway involves prior removal of the 5′ -cap by the decapping complex. In yeast, it is composed of two proteins: Dcp2p, the decapping enzyme and Dcp1p, a regulatory subunit. In Drosophila and mammals a third protein, Ge-1/Hedls, is also a part of this complex. In mammalian cells there are multiple decapping enzymes, compared to a single enzyme in yeast. The decapping process is assisted and regulated by a multitude of proteins including Pat1p, Dhh1p, Edc3p and the Lsm1–7 heptamer (see reviews in this issue).
Commitment of an organism to a new physiological state involves transitions from one gene expression pattern to another. These transitions entail altered transcriptional profiles, which are often accompanied by changes in mRNA stability thus allowing an organism to quickly respond to cellular and environmental changes. For example, in budding yeast quiescence causes stabilization of newly transcribed G0 mRNAs [6] while cell cycle-dependent changes in transcriptional activity can be coupled with changes in mRNA stability [7], [8] and [9]. Similarly, fission yeast control meiotic gene expression via global coordination between transcriptional control and mRNA decay [10]. Furthermore, environmental stimuli, such as temperature and osmotic shock, oxidative stress, amino acid starvation and nitrogen source depletion all cause changes in transcriptional program often accompanied by changes in mRNA stability [11], [12], [13], [14], [15], [16], [17], [18] and [19]. These gene expression transitions involve groups of transcripts (RNA regulons), which can be post-transcriptionally regulated by one or more RBPs. Such co-regulation in turn facilitates synchronous cellular response to a particular stimulus [20]. For example, the Rpb4/7p heterodimer, an RNAPII subunit and a regulator of cytoplasmic mRNA stability, co-transcriptionally binds its target mRNAs [21] and [22]. These transcripts (and genes) thus comprise an Rpb4/7p-regulon. Coupling between transcription and decay via Rpb4/7p complex ensures two consequences: Rpb4/7p controls the cellular mRNA abundance by reducing the rate of decay thus preventing unnecessary mRNA synthesis (see 2.1.1 and 3.2), while precise titration of transcript levels involved in protein synthesis regulates cellular growth rate by globally fine-tuning the rate of translation in response to the environment and nutrient availability.
Coupling is also an evolutionary conserved phenomenon and is a strategy adopted by a variety of budding and fission yeast genes [23]. Mechanistically, the coupling is achieved via specific cis sequences or trans regulators and mutations in either of the two affect transcription and decay concurrently. Dori-Bachash et al. demonstrated that for most genes identified, the coupling occurs via Rpb4/7p and CCR4-NOT, two complexes involved in the regulation of both mRNA synthesis and mRNA decay, although several other regulators have also been identified (see Section 2.1). Interestingly, coupling can also involve specific promoters as well as transcription factors [23], raising a possibility that a promoter and a transcription factor recruit decay regulators, which are then imprinted onto an mRNA thus directly coupling transcription with decay. In support of this possibility are three publications that demonstrate promoter-regulated mRNA turnover in mammalian cells and in yeast [9], [24] and [25]. Coupling transcription and decay via a promoter is a unique regulatory mechanism because the specificity of mRNA turnover is encoded entirely in the promoter sequence itself. mRNAs that share their promoter elements will therefore share not only their transcription patterns but also their decay patterns without the need for common, yet specific sequence motives. These groups of mRNAs constitute promoter-specified mRNA regulons.
Transcription and decay are not only mechanistically coupled through shared cis and trans regulatory sequences or factors, but can influence each other kinetica In budding yeast and in higher eukaryotes, attenuated rate of transcription decreases the rate of mRNA turnover while an increase in the rate of transcription also increases the rate of mRNA decay. Such mutual feedback maintains the steady-state mRNA levels and either globally affects the cellular mRNA abundance [26] and [27] or acts in a gene-specific manner (see Section 3.3) [8]. These findings also imply that a single mRNA can exhibit several stabilities in its lifetime simply by responding to changes in transcription rates, presumably independently of specific cis mRNA sequences or trans regulators.
This review highlights the recent findings of coupling between transcription and decay. In many cases, it is this communication and mutual dependence between the two processes that finally shapes a gene expression response. Here, we propose a new model for gene expression regulation: coupling between transcription and decay lies at the core of eukaryotic gene expression regulation, a mechanism likely employed by the majority of genes and in a variety of organisms.
2. Mechanisms for coupling transcription and decay
The mechanism by which transcription affects mRNA decay in the cytoplasm is currently under intense investigation (summarized in Table 1, Fig. 1). Already, several findings suggest one possible mechanism, which involves direct imprinting of the mRNA with trans activating factors. These factors are recruited onto the mRNA during transcription, and affect post-transcriptional events, including decay. cis-Acting elements appear to be required for some imprinting mechanisms. In other cases, cis-acting elements directly regulate the stability of the mRNA, either by attracting cytoplasmic RNA binding factors that regulate decay, or by interacting with the decay factors themselves.
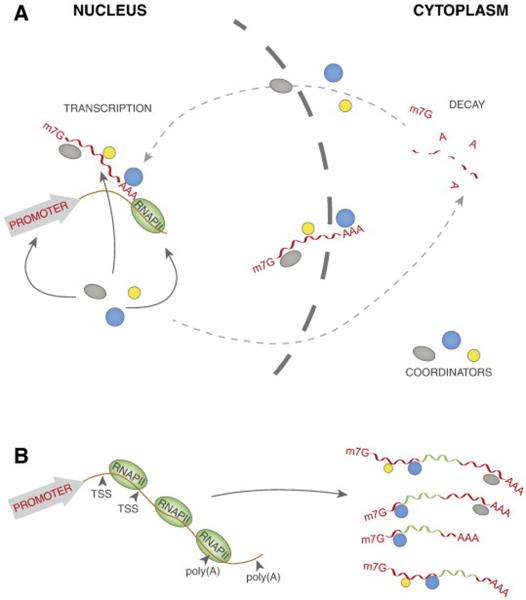
Fig. 1.
A. Transcription and decay are coupled processes through imprinting of coordinators onto a nascent mRNA. This imprinting can be mediated via cis or trans factors and it can be promoter or RNAPII dependent (solid lines). There are general coordinators, which are imprinted to a variety of mRNAs (Rpb4/7p, Pab1p (PABPC1), CCR4-NOT, EJC, Dbp5/Rat8p) or class-specific coordinators, which seem to bind to a subset of transcripts (Sus1p, Ssd1p, Dbf2p, αCP, Cth1p, TTP, KSRP, ELAV/Hu, AUF1, TOB/BTG). Transcription and decay can also regulate each other kinetically (dashed lines) to maintain the steady-state levels of cellular mRNAs. B. Alternative TSS and polyadenylation affect 5′ and 3′ UTR lengths. Shorter or longer UTRs allow less or more RBPs to associate with the mRNA and regulate its stability.
2.1. trans-Acting proteins: mRNA coordinators and mRNA imprinting
2.1.1. Rpb4/7p–the mRNA coordinator prototype
The best characterized imprinting process, and most direct evidence for coupling transcription and decay, is that of the yeast RNAPII subunits Rpb4p and Rpb7p. Rpb4p and Rpb7p were first identified as the fourth (Rpb4) and seventh (Rpb7) largest subunits of RNAPII and normally associate with the core polymerase as a heterodimer. Nevertheless, Rpb7p is an essential protein, whereas Rpb4p is dispensable under optimal environmental conditions but essential under some adverse conditions (reviewed in [4]). An early observation that singled Rpb4/7p out as unusual among RNAPII subunits was its sub-stoichiometric association with the RNAPII complex [28] (though free Rpb4p is found in excess of RNPII in yeast cells [29]) and its propensity to dissociate from the core polymerase [30]. The ability of Rpb4/7p to dissociate from RNAPII in a reversible manner has been exploited to demonstrate that this complex is required for promoter-directed initiation of transcription in vitro [30] and [31]. It is further required for recruitment of 3′ -end processing factors and proper usage of polyadenylation sites [32] (see also Section 2.2.3). Recently, a mechanism for Rpb4/7p-induced dissociation was suggested, in which ubiquitylation of Rpb1p, phosphorylated at serine 5 on the C-terminal domain, excludes Rpb4/7p from RNAPII [33]. However, since this event occurs during early elongation, and renders RNAPII inactive (at mechanism.
Two studies have provided evidence that during elongation, the extended RNA that exits the polymerase encounters Rpb7p and forms contacts with it [34] and [35]. It is probably through this strategy that Rpb4/7p becomes imprinted onto the mRNA. Although it is yet unknown exactly when during transcription Rpb4/7p leaves RNAPII and binds to the mRNA, evidence demonstrates that association of Rpb4/7p with the mRNA depends on the association with the core polymerase, presumably during transcription [21].
Finally, post-transcriptional roles for Rpb4/7p in regulating mRNA export, translation and mRNA decay have been described and depend on prior association of the complex with RNAPII [21], [22], [36], [37] and [38]. Rpb4/7p shuttles between the nucleus and the cytoplasm, in a transcription dependent manner [39] and the nuclear localization signal required for import of Rpb4p to the nucleus was identified to be important for post-transcriptional regulatory functions of Rpb4/7p [37]. Importantly, Rpb4/7p must associate with RNAPII in order for it to exert its post-transcriptional roles [21] and [37]. Rpb4/7p was therefore proposed to function as an “mRNA coordinator”, since it seems to coordinate all four major stages of gene expression (see Ref. [37] for further discussion).
It is unknown how exactly Rpb4/7p regulates mRNA decay. However, some findings suggest possible mechanisms. Rpb4/7p can interact directly with the mRNA decay sub-complex of Pat1–Lsm1–7 [22] and [38]. It is therefore possible that Rpb4/7p recruits Pat1p to the mRNA. Pat1p seems to be a hub for the decay complex, since it interacts with multiple decay factors and it is required for recruiting Lsm1–7p, Dcp1/2p and Xrn1p to the mRNA [40] and [41]. Consequently, Rpb4/7p may affect mRNA decapping and subsequent degradation through its interaction with Pat1p. Deadenylation, an early event in mRNA decay, is also regulated by Rpb4/7p [21], [22] and [38]. However, no direct interaction of Rpb4/7p with the deadenylation complexes, CCR4-NOT and Pan2/3p, has been detected [22] and [38]. Furthermore, lack of Pat1p does not affect deadenylation rates [42] rendering it unlikely that Rpb4/7p affects deadenylation through Pat1–Lsm1–7. It is possible that the effect of Rpb4/7p on deadenylation is mediated by its interaction with eukaryotic translation initiation factor 3 (eIF3) [37], which participates in both translation initiation and post-termination ribosome disassembly [43]. Although translation termination has been linked to deadenylation (see Section 2.1.2), the connection between eIF3-mediated post-termination events and deadenylation is unclear.
2.1.2. Poly(A) binding protein is a key coordinator
PABPC1 (Pab1p in yeast) is required for both maintaining the poly(A) tail and for inducing deadenylation. Pab1p is involved in the 3′ -end processing (cleavage and polyadenylation) of the mRNA [44] and is deposited on the poly(A) tail following adenylation; it is then exported with the mRNA into the cytoplasm [45], where it regulates translation initiation through its interaction with eIF4F [46], and translation termination through its interaction with eukaryotic release factor 3 (eRF3). eRF3 has been shown previously to mediate poly(A) shortening and mRNA decay in a manner coupled to translation termination [47]. Furthermore, yeast eRF3 directly interacts with the decapping protein Dcp1p [48], thus linking translation termination to decapping. Significantly, eRF3 and human PAN2/3 deadenylase compete for their binding to the poly(A) binding protein, PABPC1. The capacity of eRF3 to compete is dependent on translation. Thus binding of eRF3 to PABPC1 blocks access of the deadenylase enzymes to PABPC1-containing poly(A) tails [47]. Binding of PABPC1 and eRF1 to eRF3 might be mutually exclusive [49], suggesting that eRF3 can bind either to the poly(A) tail or to the terminating ribosome. It is assumed that following deadenylation, Pab1p is imported back into the nucleus. Significantly, impairing the import of Pab1p into the nucleus inhibits mRNA export [45] out of the nucleus, suggesting that the cytoplasmic and nuclear roles of Pab1p are coupled.
Pab1p has been shown to play a key role in deadenylation. For example, cells expressing Pab1–53p that lacks the RRM2 and RRM3 RNA binding domains, exhibit slow deadenylation of several mRNAs, suggesting that Pab1p is required for efficient deadenylation. Likewise, cells carrying pab1ΔRRM1 or pab1ΔP-domain (lacking residues 405–500) exhibit slow deadenylation, which is dependent on Ccr4p, but not Pan2p [50]. In vitro studies showed that Ccr4p/Caf1p can degrade naked poly(A) efficiently. Addition of Pab1p slowed down this enzymatic activity, yet, addition of Pab1ΔC slowed it down even further [51]. These observations suggest that Pab1p protects the poly(A) tail from degradation on one hand, yet it regulates deadenylation in a more complex manner. Pab1p seems to regulate Ccr4p/Caf1p activity either by acting on the enzyme or on the substrate. Pab1p interacts with the Pan2/3p deadenylase complex [52]. Likewise, PABPC1 interacts with hPan2/3 and hCaf1/Ccr4, the mammalian deadenylases homologous to the yeast Pan2/3p and Ccr4p/Caf1p respectively. Significantly, this interaction stimulates the deadenylation activities of these enzymes [53] and [54]. Thus, PABPC1 both recruits the deadenylation enzymes and stimulates their activity.
Pab1p can also affect decapping. Artificial tethering of Pab1p to the 3′ UTR of a reporter mRNA can stabilize mRNAs and uncouple decapping from deadenylation [55], suggesting that Pab1p represses decapping. Hence, removal of Pab1p from the mRNA after complete deadenylation might be responsible for decapping [55], as much as it affects translation initiation [46].
Thus, like Rpb4/7p, Pab1p (and conceivably PABPC1) seems to be an mRNA coordinator that affects the 3′ -end processing and is then imprinted on the mRNA during pre-mRNA processing and then further regulates export, translation initiation, translation termination and consequently deadenylation and decapping. However, the interplay between Rpb4/7p and Pab1p has not been investigated yet, and may yield interesting insight as to the role of both factors in translation termination and deadenylation.
2.1.3. The case of CCR4–NOT4
The CCR4–NOT complex is a large 9-subunit complex that is highly conserved from yeast to human. These subunits include the transcription activator/deadenylase Ccr4p, a second deadenylase (Pop2p/Caf1p) whose function is yet unclear, five Not proteins (Not1–5p) with various functions and two additional proteins, Caf130p and Caf40p (reviewed in [56]). Ccr4p was initially identified as a transcription activator [57] and [58]. Similarly, the five Not proteins were initially identified as transcription repressors of TATA-less promoters [59], [60] and [61]. Subsequent analysis in yeast and higher eukaryotes identified genetic, physical and functional interactions of CCR4–NOT with a plethora of promoters and transcription related proteins and protein complexes (recently reviewed in [62], [63] and [64]). Furthermore, early genetic interaction studies suggested that CCR4–NOT functions in transcription elongation [65] and [66]. Indeed, recently it was shown that CCR4–NOT directly promotes transcription elongation by preventing or somewhat modulating backtracking of RNAPII, in a mechanism that is independent of TFIIS [67]. In backtracked RNAPII, the 3′ OH end of the elongating transcript is displaced from the active site to the secondary channel. TFIIS reactivates backtracked RNAPII by stimulating the intrinsic endonuclease activity of RNAPII [68]. CCR4–NOT does not stimulate this activity. Instead, it was suggested that CCR4–NOT associates with the mRNA, and limits the ability of the 3′ end to be displaced, or helps pulling it out of the secondary channel. This activity requires direct interaction with the elongation complex, but does not involve phosphorylation of the C-terminal domain (CTD) of Rpb1p. However, CCR4–NOT requires a minimal RNA length, to which it binds, to exert its effect [67].
Over the past decade, the CCR4–NOT complex was also implicated in nuclear RNA processing [69], mRNA export [70], translation quality control and protein degradation (reviewed in [63]) and, relevant to this review, as one of the two major deadenylases, important for cytoplasmic mRNA decay [71] and [72].
Many mRNA binding proteins are assumed to recruit the CCR4-NOT complex to the mRNA and/or to stimulate its deadenylation activity. Such factors include Pab1p/PABPC1 (see Section 2.1.2), ARE-binding proteins, Dbf2p, PUF family proteins, TOB/BTG family proteins, Unr, Smaug, Bicaudal-C, NANOS2 and the miRNA induced silencing complex (RISC) (reviewed [63]; (also see 2.1.5 and 2.2.3)). To date however, the two roles of CCR4-NOT, as a transcriptional and mRNA decay regulator, have been investigated separately and thus no mechanistic connection between the two has been established. However, recent work suggests an indirect functional connection between these roles (see Section 3).
2.1.4. Dbp5p/Rat8p
Another possible general coordinator is the yeast Dbp5/Rat8, a DEAD-box RNA helicase that has been implicated in transcription initiation [73], mRNA export [74] and [75], translation termination [76] and mRNA decay [77]. The roles of Dbp5p/Rat8p in transcription and in decay are based mostly on genetic and physical interactions with TFIIH component Ssl1p and transcription regulator Bur6p and on co-localization with decay factors in P-bodies. It is still not clear whether the distinct functions of Dbp5p are mechanistically linked. Thus, involvement of this protein in the regulation of gene expression as a general coordinator of transcription and decay remains suggestive.
2.1.5. Possible class specific coordinators
Some proteins are implicated in coupling transcription and decay of specific mRNAs or mRNA families. Whereas Rpb7p seems to be coordinating transcription and decay of all gene families [38], its partner Rpb4p affects the decay rate of only subsets of mRNAs, such as those encoding protein biogenesis factors [22].
Ssd1p is an RNA binding protein implicated in aging, stress response and polarized growth in yeast. Many of its target mRNAs are involved in bud morphogenesis. Ssd1p requires its NLS in order to associate with its target mRNAs [78], suggesting co-transcriptional assembly with the mRNAs in the nucleus. In the cytoplasm, Ssd1 regulates mRNA localization to the bud tip [79], translation [80] and localizing mRNAs to P-bodies [78] and [79], possibly destined for degradation.
Sus1p, a co-factor of the transcription activator SAGA and the export complex TREX2, genetically and physically interacts with decay factors [81]. Thus, Sus1p may specifically coordinate degradation of SAGA-transcribed, TREX2-exported mRNAs.
Dbf2p, a yeast mitotic kinase, associates with SWI5 and CLB2 mRNAs co-transcriptionally and regulates their decay [9] (see also Section 2.2.4).
In human erythroid cells, α CP is an RNA binding protein which associates with the 3′ UTR of h α-globin mRNA, and regulates its 3′ -end processing [82] and its stability during erythroid maturation [83].
Cth2p, an AU-rich element (ARE) RNA binding protein, was found to affect both 3′-end processing [84] and Dhh1p-dependent mRNA decay [85] of specific iron deficiency-responsive genes. Furthermore, Cth2p is a shuttling protein whose export out of the nucleus, and mRNA decay activity, depend on active transcription [86]. Thus Cth2p possesses several characteristics of a class-specific coordinator. Importantly, Cth2p belongs to the conserved Tis11 protein family, which includes the human tristetraprolin (TTP) protein. TTP has a central role in targeting immune and inflammatory responses-related mRNAs for degradation, as well as some mRNAs related to cell cycle, carcinogenesis and development (reviewed in [87]). Recently, TTP was shown to regulate NF-κB induced transcription. This role has been suggested to involve both p65/NF-κB import to the nucleus [88], as well as p65/NF-κB acetylation through its interactions with histone-deacetylases (HDAC) 1, −3 and −7 [89]. TTP was also implicated in insulin-dependent transcription of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase in rats [90].
More examples of ARE-binding proteins which may act as class-specific coordinators are i) KSRP, a protein that has been implicated in transcription initiation, splicing, RNA editing, mRNA localization and mRNA decay (reviewed in [91]); ii) the ELAV/Hu proteins can co-assemble with their target mRNAs in the nucleus and facilitate their export [92] and later regulate mRNA stability [93]. Recently, it was found that Hu proteins can also affect alternative splicing (reviewed in [94]) and alternative polyadenylation [95] (see Section 2.2.3), including that of HuR mRNA itself [96]; and iii), AUF1 (heterogeneous nuclear ribonuclear protein D), is a nucleus-cytoplasm shuttling protein, and mediates cytoplasmic destabilization of cytokine mRNAs by binding to ARE sequences in the 3′ UTR during inflammatory response [97] and [98]. Although coupling between the transcription and decay has not been established for AUF1, the protein has been found to bind to pre-mRNA [99], suggesting that AUF1, like other coordinators, could become imprinted onto an mRNA co-transcriptionally.
Finally, the TOB/BTG family includes anti-proliferative proteins that are found in both cytoplasm and nucleus of mammalian cells. While these proteins have not been shown to coordinate transcription with decay, data suggest a possible involvement since these proteins modulate transcription by associating with a variety of transcription factors, and deadenylation by associating with PABPC1 and recruiting the CCR4-NOT complex (reviewed in [100]).
2.2. cis-acting elements
So far we have discussed the imprinting or association of different mRNAs with general or class-specific trans-activating factors. However, there are several types of cis-acting elements that affect the fate of the mRNA. Amazingly, in some cases the cis-elements are not found on the mRNA itself.
2.2.1. 5′ UTRs
5′ UTRs are mRNA sequences found upstream of the open reading frame (ORF). Some 5′ UTRs contain regulatory cis-elements such as upstream ORFs (uORFs), Internal Ribosome Entry Site (IRES) or Iron Responsive Elements (IRE) – all of which were shown to be involved in translation regulation (reviewed in [101], [102] and [103]). Thus, by regulating the translatability of an mRNA, these elements may affect the mRNA stability. However, the relative contribution of such elements to mRNA stability was not tested.
Recently, cis elements in the 5′ UTR of several hundred mRNAs transcribed from human genes were shown to be required for mRNA binding by human decapping enzyme, hDcp2, and ultimately for mRNA degradation [104]. Some of these genes (e.g. HMOX2) have alternative transcription start sites (TSS), which do not change the coding sequence [105]. Thus, decisions on TSS (or alternative splicing of 5′ UTR regions) during initiation of transcription can affect mRNA fate, including its stability, and therefore the expression and localization of the encoded protein, without affecting protein sequence.
2.2.2. Coding sequence
In mammalian cells, two RNA binding proteins associate with the coding region of the alpha-amyloid protein (APP) mRNA. Fragile X-mental retardation protein (FMRP) seems to assemble with mRNAs co-transcriptionally in the nucleus [106]. It inhibits APP mRNA translation and localizes the mRNA to P-bodies (presumably targeted for decay), whereas hnRNP C competes with FMRP on binding to the coding region and enables APP mRNA translation [107]. Unr, a cold shock domain RNA binding protein, associates with the coding region of c-fos mRNA and promotes its degradation via interaction with PABPC1 [108]. The coding region in the mRNA for manganese superoxide dismutage contains a cis element that determines the mRNA stability [109]. Several mRNAs also contain miRNA binding sites in their coding region. Binding of the miRNAs reduces translatability and stability of these mRNAs (reviewed [110]). Therefore, alternative splicing and other regulatory mechanisms that modulate the coding region can affect not only on the protein product but can also affect the translatability and stability of the alternatively-spliced mRNA. Introns, or more accurately exon-exon junctions (EEJ), can also be perceived as cis elements that guide coupling between transcription (i.e. splicing) and degradation via the quality control process of nonsense mediated decay (NMD). A large protein complex called the exon junction complex (EJC) is co-transcriptionally deposited 20–24 nucleotides upstream of an EEJ. Along with the NMD regulators, the EJC then co-translationally regulates the NMD process and in some instances, coordinately affects transcription and decay of mRNAs undergoing NMD (see Section 3.5) (reviewed in [111] and [112]).
2.2.3. 3′ UTR and downstream sequence elements
For some time, the 3′ UTRs have been known as central regulatory sequences that affect mRNA localization, translatability and stability. 3′ UTRs contain cis elements such as zipcode sequences, AU-rich elements (ARE), GU-rich elements, PUF response elements, miRNA binding sites as well as the poly(A) tail. The length of the 3′ UTR is determined by the maturation process of the 3′ end of the pre-mRNA, which results in cleavage and polyadenylation of the mRNA. The assembly of the 3′ processing complex, composed of multiple components, is regulated by many factors, including the CTD of the largest subunit of RNAPII, as well as cis elements such as the canonical polyadenylation signal AAUAAA and downstream U/GU-rich region. In recent years it is becoming apparent that many genes express transcripts with alternative polyadenylation (APA) (i.e. different 3′ UTR lengths) (reviewed in [3]). It is estimated that 72% of yeast genes [113] and ~ 54% of human genes [114] (or possibly more [113]) exhibit APA. Mechanisms that regulate APA include differential expression level of 3′ processing factors, RNA binding proteins (e.g. Pab1p (see Section 2.1.2), Nova2, polypyrimidine tract binding protein (PTB), hnRNP H, ELAV/Hu proteins (see Section 2.1.5) and U2AF65), cell signaling pathways, transcription elongation rate, chromatin and epigenetic modifications and several RNA cis elements that are found up or downstream of canonical AAUAAA sequences (reviewed in [3], [115] and [116]). Importantly, APA can affect the mRNA stability since longer 3′ UTRs may contain more cis regulatory elements compared to short 3′ UTRs (see above reviews for specific examples).
2.2.4. Promoters
Promoters are DNA cis-acting regulatory elements which direct transcription initiation and can affect post initiation events, including capping, splicing and 3′-end processing (reviewed in [117]). Three papers show that promoter elements also affect mRNA decay. Trcek et al. demonstrated that replacing the promoter of SWI5 and CLB2 genes with the promoter of ACT1, drastically affected the decay kinetics of these mRNAs [9]. They further showed that Dbf2p, a mitotic kinase, associates with SWI5 and CLB2 promoters and mRNAs and that the absence of Dbf2p affects decay of these mRNAs. It is unclear how this kinase becomes recruited to SWI5 and CLB2 promoters nor how it interacts with the nascent mRNAs. Dbf2p physically interacts with Cdc5p, a SWI5 and CLB2 transcription factor [118], which suggests a possible mechanism of recruitment. Since specificity of SWI5 and CLB2 mRNA decay is independent of cis mRNA sequences, it is likely that Dbf2p becomes imprinted onto a general mRNA feature, such as the cap structure, the poly(A) tail or its associated proteins. Interestingly, Dbf2p interacts with CCR4-NOT [119] and thus this kinase might regulate stability of SWI5 and CLB2 through regulation of CCR4-NOT activity [9].
In the second paper, Bregman et al. found that Rap1p binding sites (RapBS), which are short upstream activating sequences (UAS) found in ~ 5% of yeast promoters, affect the stability of the mRNA that is transcribed via this promoter [24]. Thus, eliminating RapBS from the promoter of RPL30 stabilized the RPL30 mRNA, an effect similar to when RPL30 was transcribed via an ACT1 promoter, which normally does not contain RapBS. When RPL30 mRNA synthesis was controlled by ACT1-RapBS promoter, this mRNA was destabilized again. Furthermore, binding of Rap1p to RapBS is required to regulate the stability of RPL30 mRNA, as well as mRNAs of other genes whose transcription is dependent on Rap1p. However, it seems that in this case Rap1p itself is not imprinted on the mRNA. Therefore, Rap1p may induce imprinting of (a) protein(s) that later affects decay of an mRNA. The third paper, a report from 1993, showed that in HeLa cells replacement of the β-globin promoter with that of the Herpes simplex virus 1 thymidine kinase (HSV1- TK) stabilizes a nonsense mutation containing β-globin mRNA, whereas this effect was not seen when the promoter was replaced with the CMV immediate early promoter [25]. This report implies that mammalian and viral promoters can also affect the stability of the mRNA (in this case probably via the NMD pathway – see 2.2.4 and 3.5), similar to yeast promoters, yet likely in a much more complex manner.
3. The advantages of coupling transcription and decay
3.1. Coupling shapes gene expression patterns
When gene expression reacts to cellular and environmental signals by parallel changes in transcription and mRNA decay, such coupling provides an enduring gene expression response. In Saccharomyces cerevisiae, mild osmotic stress leads to rapid up-regulation of transcriptional activity with the concurrent stabilization of “osmo-mRNAs” [120], while moderate heat shock and DNA damage response cause stabilization of transcriptionally induced mRNAs whereas repressed genes quickly degrade [12] and [18]. It has been proposed that these “high endurance” genes, where induced genes are further stabilized and repressed genes are destabilized, likely use the regulation of mRNA stability to enhance the changes in transcription and help maintain a new physiological state [12] and [121].
However, stress can also cause opposing effects on mRNA synthesis and decay. This counterintuitive phenomenon, which has been observed in budding and fission yeast as well as in mammalian cells, characterizes many mRNAs whose levels change rapidly and transiently (a “peaked ” behavior) in response to stress. Counteraction might seem counter-productive, but it enables an organism to quickly react to a signal, and importantly, it further allows an organism to quickly attain new steady state levels [10], [12], [121], [122] and [123].
The capacity of Rpb4/7p and CCR4-NOT complex to stimulate both mRNA synthesis and decay might underlie this counter action. Recently, Rap1p was found to stimulate mRNA decay in addition to its well-documented role as a transcription activator. Likewise, Aft1p is capable of stimulating both mRNA synthesis and decay of specific genes involved in the Ftr1/Fet3p-mediated reductive pathway. These kind of factors that both stimulate (or repress) mRNA synthesis and decay seems to be prevalent [121]. We named them “synthegradases” [24]. Synthegradases might serve as a mechanistic basis for the characteristic “peaked” behavior of many genes whose expression responds to environmental changes in a manner that stimulates (or represses) both mRNA synthesis and decay. We suspect that the twoarmed mechanism of synthegradases is more responsive to regulatory signals. Specifically, signaling pathways can modulate either the synthetic or the decay function of the synthegradases, thereby affecting the balance between mRNA synthesis and decay and fine-tuning the desired steady-state levels, as well as the kinetics with which they are achieved.
3.2. Coupling may facilitate evolution
The rate by which an organism evolves can be restricte be acquired in order to evolve a new trait, such as a new regulatory mechanism (reviewed in [121]). Coupling transcription with decay facilitates evolvability as it reduces the number of mutations needed to achieve coordination between the two processes. In the absence of coupling, each process would demand a separate set of mutations, resulting in a slower rate of fixation of a trait [124].
A unique example of coupling occurs when coupling proceeds via the promoter sequence. Two genes belonging to the CLB2 cluster, SWI5 and CLB2, couple their transcription with decay via a promoter sequence [9] (see Section 2.2.4). Thus SWI5 and CLB2 coordinate not only their transcription but also their decay. Since promoter provides specificity of decay and is independent of unique cis mRNA sequences, this strategy might be very efficient from an evolutionary point of view. To preserve regulation of mRNA decay and to maintain temporal coordination with transcription, only the promoter sequence needs to be subjected to evolutionary pressure while the mRNA sequence can vary independently without disrupting regulation of decay or coupling between the two processes.
It is likely that other genes in the CLB2 cluster are similarly regulated as SWI5 and CLB2, since they share promoter sequences with SWI5 and CLB2. Importantly, there needs to be no sequence similarity among mRNAs in an individual cluster to achieve temporal co-regulation of decay of multiple mRNAs. Such a gene cluster/mRNA regulon would ensure that during the cell cycle, gene expression profile of all mRNAs would oscillate as a group ensuring precise transitions from one cell cycle phase into the other. This strategy therefore would reduce the number of necessary mutations to create a novel regulatory mechanism and would thus likely prove to be evolutionary advantageous.
Co-transcriptional RNA-protein interaction has two advantageous features. First, the interacting partners are placed near each other in a manner that can stimulate (or in some cases represses) binding. Second, the protein is exposed to the emerging RNA in a temporal manner. Thus as a nascent mRNA emerges from an elongating polymerase, it provides an RNA-binding protein a temporal window for binding to acis sequence or a secondary mRNA structure, which could otherwise be occupied by other RBPs or would not exist in a mature, cytoplasmic transcript. Once in the cytoplasm, this RNP complex can be subject to competition with cytoplasmic regulators which can displace the imprinted protein. Co-transcriptional imprinting would thus affect specificity in a temporal fashion [121].
Finally, when activation of transcription is coordinated with mRNA stabilization, superfluous mRNA synthesis is avoided simply by not binding the limited stabilizer [9]. Thus, coupling also enables a more resourceful titration of cellular mRNA levels (also see below), ensuring a more energetically efficient gene expression system that could enhance evolvability. Coordination between the two processes via a cis sequence or a trans factor is a conserved mechanism employed by a variety of budding and fission yeast genes [23] indicating that at least in yeast, this strategy proved to be an evolutionary favorable mechanism of gene expression regulation.
3.3. Coupling ensures gene dosage compensation
Slowly but steadily, cumulative observations suggest a cross talk between mRNA synthesis and decay. Genome-wide attenuation of transcription can globally modulate the rate of mRNA turnover, which seems to maintain the steady-state levels of cytoplasmic mRNAs. In mouse fibroblasts, Mat1p, a TFIIH kinase subunit, is involved in both mRNA synthesis and decay; it is required for serine5 phosphorylation of the RNAPII C-terminal domain as well as mRNA turnover. Decreased RNAPII Ser5 phosphorylation compromised the transcript capping and severely attenuated transcription elongation. Nevertheless, the steady-state mRNA levels of the majority of genes in Mat1p−/−M cells are unaltered suggesting at a genome-wide mRNA stabilization [26]. This phenomenon was also observed in budding yeast. When a point mutation in RNAPII reduces the rate of transcription, cellular mRNA levels are maintained by global reduction in mRNA turnover rates [27]. Surprising however is the finding that deletion of CCR4-NOT complex impairs mRNA turnover, as anticipated, but importantly also causes a decrease in the rate of transcription [27]. However, it is yet unclear whether the reduced transcription rate is due to the increase in mRNA stability or due to the absence of the transcriptional activity of the CCR4-NOT complex.
Gene-dosage compensation can also be gene specific. In budding yeast, modulation of mRNA decay rate compensates for increased transcription to control the concentration of core histone mRNAs during the cell cycle [8]. Stability of canonical histone mRNAs is temporally co-regulated with their transcription: entry into S phase triggers transcription of stable mRNAs while exit from S phase represses transcription of canonical histone mRNAs while their transcripts rapidly degrade [7] and [125].
Yeast canonical histones are each encoded by two genes: H2A by HTA1 and HTA2, H2B by HTB1 and HTB2, H3 by HHT1 and HHT2 and H4 by HHF1 and HHF2[126]. In haploid cells, the presence of an additional copy of HTA1-HTB1 gene pair increases the rate of HTA1 and HTB1 production, but it does not affect the steady-state mRNA levels. Rather, increased mRNA degradation rate compensates for increased transcription thus maintaining the cytoplasmic steady-state levels [8]. Interestingly, the HTA2-HTB2 locus does not exhibit dosage compensation and when deleted, the lack of HTA2-HTB2 is compensated by an increased HTA1-HTB1 transcription [127] indicating that yeast maintain a precise concentration of core histone mRNAs through transcriptional and post-transcriptional coordination (reviewed in [128]). The Lsm1–7 heptamer is required for maintaining proper histone mRNA dosage in yeast [129]. Therefore, a plausible mechanism that couples histone mRNA transcription and decay would be through Rpb4p, which physically interacts with the Lsm1–7 heptamer [22].
Eukaryotes seem to evolve a buffering system that maintains steady-state mRNA levels by compensating transcription with degradation and vice versa when one of the processes is perturbed. mRNA stability however regulates not only the concentration of mRNAs already produced but may also indirectly impact transcription [27]. The mechanisms that enable mutual feedback between transcription and decay are unknown. However, as communication between transcription and cytoplasmic degradation is an evolutionary conserved process [23], it is likely that such mutual feedback between transcription and decay may be a global mechanism by which eukaryotes regulate their cytoplasmic mRNA levels.
3.4. Coupling provides a unified gene expression response
During the mitotic cycle of the budding yeast, transcription of histone mRNAs is temporally coordinated with their stability [7] and [125]. In addition, entry into mitosis creates a signal that triggers cessation of SWI5 and CLB2 transcription concomitantly with destabilization of otherwise stable SWI5 and CLB2 transcripts [9]. Thus, for at least some genes, transitions between cell cycle phases are accompanied by coupled changes in transcription and decay, alternating between high and low gene expression activity. When put in a sequence, a tight periodicity in gene expression patterns is created, which could aid in precisely timed execution of cell cycle phase-dependent events. Likewise, coupling between mRNA synthesis and decay plays an important role in shaping the proper levels of stress-responsive mRNAs in response to stresses [12] and [13] (see also Section 3.1).
More than 10% of the protein-coding genes are cell cycle regulated in S. cerevisiae[130]. Many of these genes are regulators that coordinate mitotic events such as DNA synthesis, buddi g and cytokinesis and temporal order of these cell cycle events is precisely controlled [130] and [131]. For instance, DNA synthesis must always precede cytokinesis to avoid mitotic division catastrophe [132]. Not surprisingly, many of the cell cycle genes reduce cellular viability or cause cell cycle defects when constitutively expressed [133] and [134]. As demonstrated for core histone, SWI5 and CLB2 mRNAs as well as for G0 transcripts, coordination between transcription and mRNA decay would prove important since it enables a cell to restrict expression of a gene only to the period of the cell cycle, when its activity is needed.
3.5. Coupling as an mRNA surveillance strategy
Coupling between transcription and mRNA turnover could act as a surveillance mechanism to ensure coordinate removal of erroneously synthesized mRNAs which, if not removed, could cause production of toxic proteins. An example of such coupling during mRNA surveillance is Nonsense-Mediated Decay (NMD). The NMD decay pathway targets a variety of mRNA substrates but the pathway is mostly known for degrading mRNAs that contain a premature translation termination codon (PTC). The recognition of the PTC is translation-dependent and involves the UPF1–3 proteins, in higher eukaryotes SMG1 and SMG5–9 proteins, and typically the presence of an exon-junction complex (EJC) located more than 30 nucleotides downstream of the PTC [111] and [112].
During pre-mRNA splicing, EJCs are co-transcriptionally deposited 20 to 24 nucleotides upstream of exon-exon junctions. While in the nucleus, the EJC components recruit UPF3/UPF3X proteins, which then further recruit UPF2 once the mRNA is exported into the cytoplasm. Here, an mRNA undergoes translation during which the ribosome displaces EJC complexes within the ORF from the mRNA [111] and [135]. However, if the ribosome stalls at a PTC and one or more EJCs further downstream remain bound to the mRNA, ribosome-associated UPF1 can interact with the EJC-bound UPF2, leading to UPF1 phosphorylation and subsequent induction of mRNA decay [112], [136] and [137]. NMD however does not only regulate decay of the PTC-containing mRNAs but also their transcription. In mammalian cells, unspliced variants of PTC-containing mRNAs (and not their PTCfree counterparts) are retained at their site of transcription and this retention cases, a PTC can also induce a reversible transcriptional silencing of its cognate gene [140]. How precise coupling between the two processes is achieved during NMD is not understood. However, coordination between transcription (retention of aberrant mRNAs at the site of transcription, recruitment of UPF1 and SMG6 to the site of transcription and transcriptional silencing) and mRNA decay (induction of mRNA turnover upon translational recognition of a PTC followed by UPF1 phosphorylation) ensures that at any given time the synthesis and life-span of an aberrant mRNA is minimal thus reducing the probability of translation of truncated and potentially toxic proteins.
4. Perspective
Over or under expression of factors can be deleterious to living cells. Therefore, it is essential to maintain steady state mRNA levels in a robust manner. In other cases, it is essential to rapidly increase and/or rapidly decrease mRNA levels in response to external or internal cues. These levels are determined by the rate of mRNA synthesis In order to maintain the required mRNA levels, both synthesis and decay must be coordinated in the cell. Indeed, experimental data from yeast and higher eukaryotes demonstrate the existence of mRNA regulons, which coordinate between their transcription and degradation, despite the physical barrier of the nuclear envelope between the two processes. However, we suggest that these regulons are not exceptional. Instead, coupling between transcription and decay is likely an inherent characteristic of the gene expression process. The cellular level of mRNAs is maintained by direct coupling between the transcription and decay machineries.
The mechanism of coordinating transcription and decay is only now beginning to unravel. There are currently more than 800 known mammalian mRNA binding proteins [141] and [142]; some of them may be coordinators (see Section 2.1, Table 1). A major hurdle in the field is the lack of direct measurements of transcription and decay rates simultaneously, though recently effort has been made to advance the field in this direction [9], [24], [27] and [143].
One important mechanism involves imprinting of the mRNA with general and/or class specific coordinator, yet, not much is known about how imprinting occurs. At which step during transcription does a certain factor associate with the mRNA? Which signals are required to create the imprinting? Are there physiological conditions that modulate the imprinting process? Do imprinted factors have to participate in (any) step of the transcription process in order to be imprinted, or is the mRNA imprinted also by factors which have no role in transcription? Does a factor have to be imprinted on all transcripts from a certain gene or only a fraction of them? In the latter case, does this fraction change under different conditions?
Furthermore, the current field of mRNA imprinting concentrates on imprinting the mRNA with proteins. However, mRNAs may also be imprinted structurally (by acquisition of a certain structure), chemically (e.g. methylation), or even with other RNAs, creating RNA-RNA hybrids at specific sequences.
Cis elements seem to play a major role in determining the mRNA fate. Though cis elements in 3′ and 5′ UTRs have been known for many years, it seems that regulated inclusion of these elements, by APA and alternative TSS can affect the cytoplasmic fate of the mRNA. Unexpectedly, cis elements in non-transcribed regions - promoter regions - can also affect the mRNA fate. As discussed in Section 2.2.4, these elements probably affect the imprinting of the mRNA. However, promoter elements can also affect TSS choice [144], [145] and [146] and APA [147], which in turn can affect mRNA decay. It is also becoming evident that most mammalian genes have multiple alternative promoters [2]. Thus, depending on the choice of promoter, transcripts from the same gene may have entirely different post-transcriptional properties.
The realization that promoter cis-elements affect mRNA decay also implies that some transcription factors, which bind these elements, regulate not only transcription initiation but also mRNA decay. It could be that, like Dbf2p, some transcription factors act post-transcriptionally – i.e. they are directly imprinted on the mRNA, whereas others are not imprinted on the mRNA themselves, but promote the imprinting of another protein.
An often overlooked way to couple transcription and decay is by signal transduction. Signal transduction pathways transmit an exogenous or endogenous signal through a series of steps to affect many processes, including transcription, cellcycle, apoptosis and more. Recently, some known signal transduction pathways were shown to affect mRNA decay factors. Thus, in yeast, the stress activated MAPKKKK kinase Ste20p directly regulates P-body and stress granule formation, as well as affect the decay of some mRNAs by direct phosphorylation of the decapping enzyme, Dcp2p. Another signal transduction pathway in yeast that affects P-body, but not stress granule assembly is the PKA pathway. PKA subunits were found associated with P-bodies in yeast and Pat1p phosphorylation by PKA was found to be required for large P-body assembly [148], [149] and [150]. Similarly, in mammals, the JNK pathway may regulate the dynamic transfer of mRNAs from polysomes to stress granules and P bodies during stress, via a novel JNK binding protein, WDR62 [151] or by direct phosphorylation of the decapping regulator DCP1a [152]. The insulin signal transduction pathway regulates the activity of the decapping enhancer and P-body core component EDC3, via phosphorylation by AKT and binding to 14-3-3 protein. Thus, phosphorylation of EDC3 is required for proper P-body formation, miRNA induced translation repression and the relative interaction of EDC3 with other RNA binding proteins [153]. Since signal transduction pathways also regulate transcription, it is tempting to see if there are cases in which both the synthesis and decay of some classes of mRNAs are simultaneously affected by these pathways. In these cases, synthegradases are good targets because they affect both mRNA synthesis and decay (see Section 3.1).
In recent years, a new model of gene expression is beginning to emerge (Fig. 1). According to the old view, after its export from the nucleus to the cytoplasm, the mRNA begins a new life, where it meets new partners that regulate its fate. The new model proposes that the fate of the mRNA is already determined during transcription, even before its synthesis is completed. Thus, mRNA localization, translatability, and stability are already pre-determined in the nucleus. We propose that DNA cis-elements, chromatin structure, location of the gene in the nuclear space, transcription factors, ncRNAs and any other features of nuclear molecules can affect the structure and composition of the mRNP, thereby affecting its functions in the cytoplasm. This model therefore views mRNA synthesis as the key event in the expression of protein encoding genes.
Acknowledgements
We would like to thank Dr. Oliver Mühlemann for his help with the writing of the NMD-dependent mRNA surveillance section. This review was supported by the Gruss Lipper family postdoctoral fellowship awarded to GH and the HHMI fellowship of The Jane Coffin Childs Memorial Fund for Medical Research awarded to TTP. Work in MC laboratory is supported by grants from “Israel Science Foundation”, “US-Israel Binational Foundation” and by the Israeli Ministry of Health. Work in RHS lab is supported by NIHGM57071.
References
- [1].Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- [2].Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- [3].Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression, Wiley Interdiscip. Rev. RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- [4].Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- [5].Choder M. mRNA imprinting: additional level in the regulation of gene expression. Cell. Logist. 2011;1:37–40. doi: 10.4161/cl.1.1.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Talarek N, Cameroni E, Jaquenoud M, Luo X, Bontron S, Lippman S, Devgan G, Snyder M, Broach JR, De Virgilio C. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′–3′ mRNA decay pathway. Mol. Cell. 2010;8:345–355. doi: 10.1016/j.molcel.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Osley MA, Hereford LM. Yeast histone genes show dosage compensation. Cell. 1981;24:377–384. doi: 10.1016/0092-8674(81)90327-5. [DOI] [PubMed] [Google Scholar]
- [9].Trcek T, Larson DR, Moldon A, Query CC, Singer RH. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Amorim MJ, Cotobal C, Duncan C, Mata J. Global coordination of transcriptional control and mRNA decay during cellular differentiation. Mol. Syst. Biol. 2010;6:380. doi: 10.1038/msb.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, Martin DE, Tresch A, Cramer P. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 2008;4:223. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 2011;7:e1002273. doi: 10.1371/journal.pgen.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- [17].Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Castells-Roca L, Garcia-Martinez J, Moreno J, Herrero E, Belli G, Perez-Ortin JE. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One. 2011;6:e17272. doi: 10.1371/journal.pone.0017272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Castells-Roca L, Muhlenhoff U, Lill R, Herrero E, Belli G. The oxidative stress response in yeast cells involves changes in the stability of Aft1 regulon mRNAs. Mol. Microbiol. 2011;81:232–248. doi: 10.1111/j.1365-2958.2011.07689.x. [DOI] [PubMed] [Google Scholar]
- [20].Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- [21].Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 2005;19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:e1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- [25].Enssle J, Kugler W, Hentze MW, Kulozik AE. Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10091–10095. doi: 10.1073/pnas.90.21.10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Helenius K, Yang Y, Tselykh TV, Pessa HK, Frilander MJ, Makela TP. Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover. Nucleic Acids Res. 2011;39:5025–5035. doi: 10.1093/nar/gkr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Lariviere L, Maier KC, Seizl M, Tresch A, Cramer P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kolodziej PA, Woychik N, Liao SM, Young RA. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell. Biol. 1990;10:1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rosenheck S, Choder M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol. 1998;180:6187–6192. doi: 10.1128/jb.180.23.6187-6192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Edwards AM, Kane CM, Young RA, Kornberg RD. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- [31].Orlicky SM, Tran PT, Sayre MH, Edwards AM. Dissociable Rpb4–Rpb7 subassembly of RNA polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 2001;276:10097–10102. doi: 10.1074/jbc.M003165200. [DOI] [PubMed] [Google Scholar]
- [32].Runner VM, Podolny V, Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell. Biol. 2008;28:1883–1891. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19649–19654. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen CY, Chang CC, Yen CF, Chiu MT, Chang WH. Mapping RNA exit channel on transcribing RNA polymerase II by FRET analysis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:127–132. doi: 10.1073/pnas.0811689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- [36].Farago M, Nahari T, Hammel C, Cole CN, Choder M. Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol. Biol. Cell. 2003;14:2744–2755. doi: 10.1091/mbc.E02-11-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2011;143:552–563. doi: 10.1016/j.cell.2010.10.033. [DOI] [PubMed] [Google Scholar]
- [38].Lotan R, Goler-Baron V, Duek L, Haimovich G, Choder M. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 2007;178:1133–1143. doi: 10.1083/jcb.200701165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Selitrennik M, Duek L, Lotan R, Choder M. Nucleo-cytoplasmic shuttling of the Rpb4p and Rpb7p subunits of yeast RNA polymerase II by two pathways. Eukaryot. Cell. 2006;5:2092–2103. doi: 10.1128/EC.00288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- [42].Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- [48].Kofuji S, Sakuno T, Takahashi S, Araki Y, Doi Y, Hoshino S, Katada T. The decapping enzyme Dcp1 participates in translation termination through its interaction with the release factor eRF3 in budding yeast. Biochem. Biophys. Res. Commun. 2006;344:547–553. doi: 10.1016/j.bbrc.2006.03.174. [DOI] [PubMed] [Google Scholar]
- [49].Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [50].Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol. Cell. Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Simon E, Seraphin B. A specific role for the C-terminal region of the Poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 2007;35:6017–6028. doi: 10.1093/nar/gkm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- [53].Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Uchida N, Hoshino S, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- [55].Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem. Soc. Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Draper MP, Liu HY, Nelsbach AH, Mosley SP, Denis CL. CCR4 is a glucose-regulated transcription factor whose leucinerich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- [60].Collart MA. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Oberholzer U, Collart MA. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene. 1998;207:61–69. doi: 10.1016/s0378-1119(97)00605-7. [DOI] [PubMed] [Google Scholar]
- [62].Miller JE, Reese JC. Ccr4–Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 2012;47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Collart MA, Panasenko OO. The Ccr4–not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- [64].Reese JC. The control of elongation by the yeast Ccr4–Not complex. Biochim. Biophys. Acta. 2013;1829:127–133. doi: 10.1016/j.bbagrm.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Denis CL, Chiang YC, Cui Y, Chen J. Genetic evidence supports a role for the yeast CCR4–NOT complex in transcriptional elongation. Genetics. 2001;158:627–634. doi: 10.1093/genetics/158.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Biswas D, Yu Y, Mitra D, Stillman DJ. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4–Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics. 2006;172:837–849. doi: 10.1534/genetics.105.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- [69].Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4–NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One. 2009;4:e6760. doi: 10.1371/journal.pone.0006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kerr SC, Azzouz N, Fuchs SM, Collart MA, Strahl BD, Corbett AH, Laribee RN. The ccr4–not complex interacts with the mRNA export machinery. PLoS One. 2011;6:e18302. doi: 10.1371/journal.pone.0018302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- [73].Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol. Biol. Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- [77].Scarcelli JJ, Viggiano S, Hodge CA, Heath CV, Amberg DC, Cole CN. Synthetic genetic array analysis in Saccharomyces cerevisiae provides evidence for an interaction between RAT8/DBP5 and genes encoding P-body components. Genetics. 2008;179:1945–1955. doi: 10.1534/genetics.108.091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kurischko C, Kuravi VK, Herbert CJ, Luca FC. Nucleocytoplasmic shuttling of Ssd1 defines the destiny of its bound mRNAs. Mol. Microbiol. 2011;81:831–849. doi: 10.1111/j.1365-2958.2011.07731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kurischko C, Kim HK, Kuravi VK, Pratzka J, Luca FC. The yeast Cbk1 kinase regulates mRNA localization via the mRNA-binding protein Ssd1. J. Cell Biol. 2011;192:583–598. doi: 10.1083/jcb.201011061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jansen JM, Wanless AG, Seidel CW, Weiss EL. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr. Biol. 2009;19:2114–2120. doi: 10.1016/j.cub.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cuenca-Bono B, Garcia-Molinero V, Pascual-Garcia P, Garcia-Oliver E, Llopis A, Rodriguez-Navarro S. A novel link between Sus1 and the cytoplasmic mRNA decay machinery suggests a broad role in mRNA metabolism. BMC Cell Biol. 2010;11:19. doi: 10.1186/1471-2121-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol. Cell. Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Morales J, Russell JE, Liebhaber SA. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- [84].Prouteau M, Daugeron MC, Seraphin B. Regulation of ARE transcript 3′ end processing by the yeast Cth2 mRNA decay factor. EMBO J. 2008;27:2966–2976. doi: 10.1038/emboj.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol.Chem. 2008;283:28527–28535. doi: 10.1074/jbc.M804910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Vergara SV, Puig S, Thiele DJ. Early recruitment of AU-rich element-containing mRNAs determines their cytosolic fate during iron deficiency. Mol. Cell. Biol. 2011;31:417–429. doi: 10.1128/MCB.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay, Wiley Interdiscip. Rev. RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J. Biol. Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J. Biol. Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ness GC, Edelman JL, Brooks PA. Involvement of tristetraprolin in transcriptional activation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase by insulin, Biochem. Biophys. Res. Commun. 2012;420:178–182. doi: 10.1016/j.bbrc.2012.02.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation, Wiley Interdiscip. Rev. RNA. 2010;1:230–239. doi: 10.1002/wrna.2. [DOI] [PubMed] [Google Scholar]
- [92].Pieper D, Schirmer S, Prechtel AT, Kehlenbach RH, Hauber J, Chemnitz J. Functional characterization of the HuR:CD83 mRNA interaction. PLoS One. 2011;6:e23290. doi: 10.1371/journal.pone.0023290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- [96].Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- [99].Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 2010;222:66–72. doi: 10.1002/jcp.21919. [DOI] [PubMed] [Google Scholar]
- [101].Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, transacting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- [102].Wethmar K, Smink JJ, Leutz A. Upstream open reading frames: molecular switches in (patho)physiology. Bioessays. 2010;32:885–893. doi: 10.1002/bies.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Leipuviene R, Theil EC. The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cell. Mol. Life Sci. 2007;64:2945–2955. doi: 10.1007/s00018-007-7198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li Y, Song MG, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol. Cell. Biol. 2008;28:939–948. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. T. Mammalian Gene Collection Program, Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol. Cell. Biol. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, Worley PF, Mattson MP, Gorospe M. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010;17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu AB. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Davis CA, Monnier JM, Nick HS. A coding region determinant of instability regulates levels of manganese superoxide dismutase mRNA. J. Biol. Chem. 2001;276:37317–37326. doi: 10.1074/jbc.M104378200. [DOI] [PubMed] [Google Scholar]
- [110].Lee EK, Gorospe M. Coding region: the neglected post-transcriptional code. RNA Biol. 2011;8:44–48. doi: 10.4161/rna.8.1.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the doublelife of NMD factors. Cell. Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond, Wiley Interdiscip. Rev. RNA. 2010;1:494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- [118].Darieva Z, Bulmer R, Pic-Taylor A, Doris KS, Geymonat M, Sedgwick SG, Morgan BA, Sharrocks AD. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature. 2006;444:494–498. doi: 10.1038/nature05339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu HY, Toyn JH, Chiang YC, Draper MP, Johnston LH, Denis CL. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5289–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Romero-Santacreu L, Moreno J, Perez-Ortin JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. doi: 10.1261/rna.1435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Tirosh I. Transcriptional priming of cytoplasmic post-transcriptional regulation. Transcription. 2011;2:258–262. doi: 10.4161/trns.2.6.18608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Molin C, Jauhiainen A, Warringer J, Nerman O, Sunnerhagen P. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA. 2009;15:600–614. doi: 10.1261/rna.1403509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Dahan O, Gingold H, Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 2011;27:316–322. doi: 10.1016/j.tig.2011.05.008. [DOI] [PubMed] [Google Scholar]
- [125].Hereford LM, Osley MA, Ludwig TR, II, McLaughlin CS. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- [126].Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Norris D, Osley MA. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 1987;7:3473–3481. doi: 10.1128/mcb.7.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Eriksson PR, Ganguli D, Nagarajavel V, Clark DJ. Regulation of histone gene expression in budding yeast. Genetics. 2012;191:7–20. doi: 10.1534/genetics.112.140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Herrero AB, Moreno S. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 2011;30:2008–2018. doi: 10.1038/emboj.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- [132].Lew DJ. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 2000;10:47–53. doi: 10.1016/s0959-437x(99)00051-9. [DOI] [PubMed] [Google Scholar]
- [133].Niu W, Li Z, Zhan W, Iyer VR, Marcotte EM. Mechanisms of cell cycle control revealed by a systematic and quantitative overexpression screen in S. cerevisiae. PLoS Genet. 2008;4:e1000120. doi: 10.1371/journal.pgen.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [135].Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- [136].Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phos- phorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].de Turris V, Nicholson P, Orozco RZ, Singer RH, Muhlemann O. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA. 2011;17:2094–2107. doi: 10.1261/rna.02918111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Muhlemann O, Mock-Casagrande CS, Wang J, Li S, Custodio N, Carmo- Fonseca M, Wilkinson MF, Moore MJ. Precursor RNAs harboring nonsense co-dons accumulate near the site of transcription. Mol. Cell. 2001;8:33–43. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- [140].Buhler M, Mohn F, Stalder L, Muhlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol. Cell. 2005;18:307–317. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- [141].Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. In-sights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- [142].Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- [143].Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kawaji H, Frith MC, Katayama S, Sandelin A, Kai C, Kawai J, Carninci P, Hayashizaki Y. Dynamic usage of transcription start sites within core promoters. Genome Biol. 2006;7:R118. doi: 10.1186/gb-2006-7-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kasahara K, Ki S, Aoyama K, Takahashi H, Kokubo T. Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA poly- merase II. Nucleic Acids Res. 2008;36:1343–1357. doi: 10.1093/nar/gkm1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, Tian B. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Shah KH, Zhang B, Ramachandran V, Herman PK. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics. 2013;193:109–123. doi: 10.1534/genetics.112.146993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Ramachandran V, Shah KH, Herman PK. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell. 2011;43:973–981. doi: 10.1016/j.molcel.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tudisca V, Recouvreux V, Moreno S, Boy-Marcotte E, Jacquet M, Portela P. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur. J. Cell Biol. 2010;89:339–348. doi: 10.1016/j.ejcb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- [151].Wasserman T, Katsenelson K, Daniliuc S, Hasin T, Choder M, Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell. 2010;21:117–130. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Rzeczkowski K, Beuerlein K, Muller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, Holtmann H, Kracht M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 2011;194:581–596. doi: 10.1083/jcb.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Larance M, Rowland AF, Hoehn KL, Humphreys DT, Preiss T, Guilhaus M, James DE. Global phosphoproteomics identifies a major role for AKT and 14-3-3 in regulating EDC3. Mol. Cell. Proteomics. 2010;9:682–694. doi: 10.1074/mcp.M900435-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]