Abstract
Bioremediation is one of the most promising methods to clean up polluted environments using highly efficient potent microbes. Microbes with specific enzymes and biochemical pathways are capable of degrading the tobacco alkaloids including highly toxic heterocyclic compound, nicotine. After the metabolic conversion, these nicotinophilic microbes use nicotine as the sole carbon, nitrogen, and energy source for their growth. Various nicotine degradation pathways such as demethylation pathway in fungi, pyridine pathway in Gram-positive bacteria, pyrrolidine pathway, and variant of pyridine and pyrrolidine pathways in Gram-negative bacteria have been reported. In this review, we discussed the nicotine-degrading pathways of microbes and their enzymes and biotechnological applications of nicotine intermediate metabolites.
1. Introduction
Tobacco (Nicotiana, Solanaceae family) is mainly cultivated in Brazil, China, Cuba, India, and USA. An annual production of 6.7 million tonnes of tobacco has been reported. China is the largest tobacco producer (39.6%) followed by India (8.3%), Brazil (7.0%), and the USA (4.6%) [1]. India is the third largest tobacco consumer (275 million) in the world followed by China and USA [2]. It is anticipated that the tobacco industry would produce 3,00,274 tonnes of nicotine wastes every year [3]. Nicotine usually accounts for more than 90% of the whole plant alkaloid fraction in commercial tobacco, Nicotiana tabacum [4]. The entire or part of the tobacco leaf was used as raw material for tobacco products such as cigarette, cigars, chewing tobacco, and snuff.
In the year 2000, it was estimated that 4.9 million deaths occur due to smoking [5]. By the year 2020, it is also expected to exceed 9 million deaths annually [5]. Due to the increased usage of tobacco products, the industry generated solid and liquid tobacco wastes containing high concentrations of nicotine [6]. The tobacco industries produce tobacco waste with an average content of nicotine of 18 g per kg of dry weight [7]. The Environmental Protection Agency (EPA) has classified these nonrecyclable powdery tobacco wastes as Toxic Release Inventory (TRI) chemicals [8]. When the concentration of nicotine content exceeds more than 0.05% (w/w), it is designated as “toxic and hazardous” by the European Union Regulations (EUR) [7]. These tobacco wastes are dumped on the ground without proper storage and processing [7, 8]. Nicotine dissolves easily in water leading to the contamination of the ground water [7, 8]. Hence, the nicotine-contaminated water disturbs the ecological balance of soil [7, 8]. Therefore, it is important to remove nicotine from tobacco polluted soil and water [7, 8].
2. Nicotine
Nicotine 3-(1-methyl-2-pyrrolidinyl) pyridine is a heterocyclic compound with a pyridine and a pyrrolidine ring moiety. Nicotine is a pale yellow to dark brown liquid with slight fishy odor when warm. It is water-soluble. The chemical formula of nicotine is C10H14N2. Its molecular weight is 162.234 with melting point –79°C and boiling point 247°C. Nicotine is present up to 2 to 8% of the dry mass of the tobacco leaves [9].
2.1. Effect of Nicotine on Human Beings
Nicotine is a hazardous compound that causes tobacco related lung cancer and peripheral arterial disease [10]. Although more than 4000 substances are present in the tobacco cigarette smoke, nicotine is the major substance [11, 12]. Nicotine has a blood half-life period of approximately 2 h and causes severe vascular diseases [11, 12]. Nicotine can cause cancer, gene mutation, and malformation [13]. An array of toxic nicotine intermediate metabolites such as N′-nitrosonornicotine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, cotinine, and N-nitrosamine causes tobacco lung cancer [14, 15]. Neurotoxin developmental effects of nicotine can naturally affect a variety of cellular processes such as generation of oxidative radicals, apoptosis, and hyperplasia of cell, enhancing gene expression to secrete hormones and regulation of enzymatic activity [13, 16, 17]. The lungs rapidly absorb 90% of the nicotine that is present in the cigarette smoke inhaled by human beings [18]. Nicotine in the human body can easily pass the biological membranes and blood-brain barrier and increases heart rate, mean arterial blood pressure and mimics the venous endothelial dysfunction [15, 19, 20].
Nicotine is an additive substance that can lead to nicotine dependence and addictive behavior in human populations [43]. Nicotine dependence is complex, multidimensional trait that involves psychological, physiological behavioral, and social factors [44]. Nicotine, the primary psychoactive ingredient of tobacco, contributes to physical dependence, by acting on nicotinic acetylcholine receptors in the central nervous system and leads to the release of neurotransmitters (e.g., dopamine and serotonin), which produce reinforcing effects by activating the mesocorticolimbic dopamine system [45].
3. Microbial Degradation of Nicotine
Physical and chemical methods are available to degrade nicotine in the tobacco. These methods are often time-consuming, expensive and involve solvent extraction procedures [46]. Bioremediation is one of the promising methods to clean up polluted environments using microbes [47–49]. Briski et al. [50] reported that aerobic composting is an effective method to reduce 80% of nicotine and 50% of the volume and mass of tobacco solid wastes in 16 d. Meher et al. [51] used a technique called biomethanation that removed 60% of nicotine, 75.6% of chemical oxygen demand, and 80% of biological oxygen demand from the tobacco wastes. Biological method employs a variety of nicotine-degrading bacteria and fungi [41, 46, 51]. These ecofriendly biological methods are extensively used in wastewater treatment due to its high efficiency and low cost. Microbes that degrade nicotine are reported to adapt to polluted environment easily [52, 53]. Native strains of bacteria and fungi that live in the tobacco environment have the ability to degrade nicotine [54]. These microbes utilize nicotine as sole carbon, nitrogen, and energy source for their growth [6]. Batham [55] reported that the nitrate content of the soil increased due to the microbial degradation of nicotine.
3.1. Optimal Conditions of Nicotine-Degrading Bacteria
Nicotine degradation experiments are carried out with different culture media such as distilled water, minimal salt medium, inorganic salt medium, and basal salt medium (BSM). The culture media influence the nicotine degradation efficiency. Wang et al. [28] reported that 3 g/L nicotine was fully degraded in 5 h when the degradation experiment was carried out with 0.05 M sodium phosphate buffer (pH 7.0). However, it took more than 8 h to complete degradation when carried out with distilled water. The optimal culture conditions and nicotine degradation efficiency of nicotinophilic bacteria vary from each other. Most of the bacteria grow at 30°C, pH range of 6.4 to 7.5, and degrade maximum concentration of nicotine up to 6 g/L [2, 5]. Huang et al. [56] reported that the rate of nicotine degradation efficiency was high at 37°C when compared to 30°C. The optimal conditions and nicotine degradation efficiency of various nicotine-degrading bacteria are presented in Table 1.
Table 1.
The optimal conditions, nicotine degradation efficiency, and degradation pathways of nicotine-degrading bacteria.
| Microorganism | Medium | Optimal conditions (pH and Temperature) |
Nicotine degradation efficiency (g/L) | Pathway | Reference |
|---|---|---|---|---|---|
| Fungi | |||||
| Pellicularia filamentosa JTS-208 | Synthetic | ND | 0.04/20 d | Demethylation | [21] |
| Cunninghamella echinulata IFO-4444 | Synthetic | ND | 0.54/13 d | Demethylation | [21] |
| Aspergillus oryzae 112822 | Synthetic | 6.5, 28°C | 2.12/40 h | A variant of pyridine and pyrrolidine | [22] |
| Gram-positive bacteria | |||||
| Arthrobacter nicotinophagum N. sp. | Synthetic | 6.8 to 7.0, 20 to 25°C | 1.62/56 h | Pyridine | [23] |
| Arthrobacter sp. HF-2 | Synthetic | 7.0, 30°C | 0.70/43 h | ND | [24] |
| Gram-negative bacteria | |||||
| Pseudomonas sp. 41 | Synthetic | 6.4, 30°C | 1.30/24 h | Pyrrolidine | [25] |
| Pseudomonas sp. HF-1 | Synthetic | 6.5 to 7.5, 30°C | 1.30/25 h | Pyrrolidine | [26] |
| Ochrobactrum intermedium DN2 | Synthetic | 30°C | 0.50/36 h | ND | [27] |
| P. putida S16 | Synthetic | 7.0, 30°C | 3.00/10 h | Pyrrolidine | [28] |
| Pseudomonas sp. Nic 22 | Synthetic | 6.5, 30 to 34°C | 2.89/52 h | ND | [29] |
| P. putida J5 | Synthetic | 30°C | 3.97/48 h | ND | [30] |
| Rhodococcus sp. Y22 | Synthetic | 7.0, 28°C | 1.00/52 h | ND | [31] |
| Agrobacterium tumefaciens S33 | Synthetic | 7.0, 30°C | 5.00/18 h | A variant of pyridine and pyrrolidine | [32] |
| Gram-negative bacteria | |||||
| Ensifer sp. N7 | Synthetic | 7.0, 28°C | 3.35/48 h | ND | [33] |
| Pseudomonas sp. ZUTSKD | Synthetic | 7.5, 30°C | 1.55/12 h | ND | [34] |
| Pseudomonas sp. HZN6 | Synthetic | 7.0, 30°C | 0.50/12 h | Pyrrolidine | [35] |
| Shinella sp. HZN1 | Synthetic | 7.0, 30°C | 0.50/9 h | ND | [36] |
| Acinetobacter sp. TW | Synthetic | 7.0, 30°C | 1.00/12 h | ND | [37] |
| Sphingomonas sp. TY | Synthetic | 7.0, 30°C | 1.00/18 h | ND | [37] |
| Pseudoxanthomonas sp. 5–52 | Synthetic | 7.0, 28°C | 1.00/28 h | ND | [38] |
| P. stutzeri | Synthetic | 7.5, 37°C | 2.50/36 h | ND | [39] |
| Sinorhizobium sp. 5–28 | Synthetic | 7.0, 28°C | 1.50/28 h | ND | [38] |
| Ochrobactrum sp. 4–40 | Synthetic | 7.0, 28°C | 0.50/28 h | ND | [38] |
| P. plecoglossicida TND35 | Synthetic | 7.0, 30°C | 3.0/12 h | A variant of pyrrolidine | [2] |
ND: not detected.
3.2. Effect of Trace Elements, Carbon, and Nitrogen Sources on Nicotine Degradation
Trace elements and other carbon and nitrogen sources play a significant role in the biodegradation of nicotine [26]. The presence of (NH4)2SO4 in the nicotine containing medium could decrease the nicotine degradation efficiency [26]. The nicotine degradation rate was increased in the presence of yeast extract, glucose, and Tween 80 in tobacco waste extract (TWE) containing media [57]. However, the nicotine degradation rate was dependent on the concentration of yeast extract and Tween 80 in the TWE medium [27]. In contrast, the medium containing glucose and (NH4)2SO4 reduced the efficiency of nicotine degradation [32]. ZnSO4·7H2O and NiCl2·6H2O had no influence on nicotine degradation, whereas Na2MoO4 and CuCl2·4H2O inhibited the rate of nicotine degradation [39]. Glucose is an important carbon source that promotes bacterial cell growth and improves the rate of nicotine degradation [24, 34, 39]. The concentration of glucose above 10 g/L inhibited the nicotine degradation by Pseudomonas sp. ZUTSKD [34]. Raman et al. [2] reported that 1 g/L dextrose increased nicotine degradation rate. In contrast, glucose is not influenced in the nicotine degradation rate when the experiment was carried out in solid-state fermentation process [39, 56]. Jiang et al. [36] reported that the nicotine degradation rate was slow in the presence of glucose. Conversely, P. stutzeri ZCJ could not utilize carbon sources such as sucrose and maltose and nitrogen source, namely, NaNO2, for their growth which in turn inhibits nicotine degradation [39].
3.3. Effect of Nicotine-Degrading Bacteria on Tobacco Leaves and Tobacco Wastes
Nicotine-degrading bacteria played an important role in the improvement of quality of tobacco leaves during aging (fermentation) process. These bacteria accumulated more in younger tobacco leaves when compared to aged leaves [56]. The desirable flavor, taste, and smoking properties of tobacco remained unaltered when treated with nicotinophilic Pseudomonas [6, 29]. Li et al. [42] reported that Arthrobacter sp. successfully reduced 28 to 39% of nicotine during flue curing of tobacco leaves and enhanced the quality of tobacco leaf. The nicotine degradation efficiency of different nicotine-degrading bacteria on tobacco leaves and tobacco wastes is listed in Table 2.
Table 2.
The nicotine degradation efficiency of different nicotine-degrading bacteria on tobacco leaves and tobacco wastes.
| Bacterium | Source | Duration (time/temperature) |
Nicotine (mg/g) |
Nicotine degradation (%) |
Reference |
|---|---|---|---|---|---|
| Micrococcus nicotianae | Tobacco leaves | NM | NM | 0.83 | [40] |
| Debaryomyces nicotianae | Tobacco leaves | NM | NM | 0.45 | [40] |
| Cellulomonas sp. | Tobacco leaves | NM | NM | 15.00 | [41] |
| A. oxidans α-2 | Tobacco waste | 212 h | 700 | NM | [7] |
| A. oxidans pAO1 | Tobacco waste | 125 h | 3400 | NM | [7] |
| P. putida | Tobacco waste | 40 h | 3300 | NM | [7] |
| P. putida J5 | Tobacco leaves | 7 d | 2.8 | 11.72 | [30] |
| Pseudomonas sp. Nic 22 | Tobacco leaves | 30°C | NM | 33.10 | [29] |
| Ensifer sp. N7 | Tobacco leaves | NM | 4.1 | 16.00 | [42] |
| Arthrobacter sp. | Tobacco leaves | NM | NM | 28.00 to 39.00 | [33] |
| Pseudoxanthomonas sp. 5–52 | Tobacco leaves | NM | NM | 47.20 | [38] |
| Sinorhizobium sp. 5–28 | Tobacco leaves | NM | NM | 72.50 | [38] |
| Ochrobactrum sp. 4–40 | Tobacco leaves | NM | NM | 51.50 | [38] |
| P. stutzeri ZCJ | Tobacco leaves | 7 d/37°C | NM | 32.24 | [39] |
| P. plecoglossicida | Tobacco leaves | 10 h | 13 | 88.00 | [2] |
| P. plecoglossicida | Tobacco wastes | 10 h | 5 | 96.10 | [2] |
NM: not mentioned.
The two major bacterial species that degrade nicotine are Pseudomonas and Arthrobacter [6]. Furthermore, recently, a new nicotine-degrading strain, Arthrobacter sp. strain M2012083, was sequenced by Illumina High-Seq 2000, which represented the first sequenced nicotine-degrading Arthrobacter strain [58]. Tang et al. [59] recently sequenced a novel bacterial strain Pseudomonas geniculata N1 which represents the first sequence of the Pseudomonas geniculate group.
3.4. Pigment Production during Nicotine Biodegradation
During nicotine degradation different colours were produced. The Gram-positive bacteria A. nicotinovorans utilized nicotine and changed to yellow pigments and further turned carmine during nicotine degradation [60–62]. In contrast, Hylin [23] reported that A. nicotinovorans did not produce any pigments during nicotine degradation. Arthrobacter sp. produced blue violet colour during nicotine degradation due to the transformation of nicotine into 2,3,6-trihydroxypyridine. The colour change was due to the presence of oxygen and the absence of metabolic catalyzing enzymes [63, 64]. The hydroxylated pyridine ring dimerized and formed a blue pigment during nicotine degradation by Arthrobacter sp. [62]. Interestingly, during nicotine degradation the Gram-negative bacteria such as Pseudomonas sp., Shinella sp. HZN1, and Acinetobacter sp. ND12 changed the colour of the nicotine medium to green and oxidized to blue gray and finally to brown [2, 26, 31, 32, 36, 42, 57]. The formation of green colour was mainly dependent upon the concentration of nicotine [2, 32, 36]. P. geniculata N1 produced golden yellow pigment on the plate during nicotine degradation [59]. However, no pigment was observed during nicotine degradation by P. putida and Ensifer sp. N7 [30, 33].
3.5. Microbial Degradation Procedure
Standard biodegradation methods were employed to treat tobacco wastes [30]. Initially, the tobacco wastes are sterilized under UV light. The resting cells of biodegrading strain are inoculated into tobacco wastes. Then the tobacco wastes are baked at 60°C for 6 days, crushed to a powder, and passed through a sieve with muslin cloth. The tobacco powder is extracted with methanol and the concentration of nicotine is determined as described in [30].
4. Biochemical Pathways That Mediate Nicotine Biodegradation
Biodegradation of nicotine by A. nicotinovorans and A. oxidans has been reported [65, 66]. These Gram-positive bacteria followed pyridine pathway. The nicotine intermediate metabolites of pyridine pathway were identified and characterized [67]. The microbial degradation of nicotine differs among the different strains of species [54]. Previous research suggests that the encoding genes that mediate nicotine degradation are not only located on the bacterial chromosomes but also present in the plasmids [65, 66, 68]. Plasmid-borne genes (160 Kb) in A. nicotinovorans are responsible for nicotine degradation [65, 66]. Similarly, nicotine-degrading genes are located on the outside of the chromosomes of P. convexa [68]. The bacteria such as Arthrobacter sp. (Gram-positive) followed pyridine pathway, which attacked the pyridine ring of the nicotine during degradation. The Gram-negative bacteria Pseudomonas sp. initially attacked pyrrolidine ring and followed pyrrolidine pathway. Fungi employed the demethylation pathway that demethylates methyl group in the pyrrolidine ring of the nicotine. Surprisingly, Agrobacterium sp. followed a new variant pathway of pyridine and pyrrolidine for nicotine degradation. Several nicotine-degrading genes have been reported [6, 45, 69].
4.1. Pyridine Pathways of Nicotine Biodegradation
Pyridine pathways of nicotine degradation by A. nicotinovorans, Nocardioides sp. JS614 are presented in Figure 1. A. nicotinovorans harbors a 160 Kb plasmid which is responsible for nicotine degradation [65, 66]. These bacteria initially attacked 6th position of carbon in the pyridine ring of nicotine by hydroxylation to introduce hydroxyl group and formed 6-hydroxynicotine (6-HN). This hydroxylation step was catalyzed out by nicotine dehydrogenase (NDH). It is a heterotrimeric molybdenum enzyme called nicotine : accepter oxidoreductase [70–73]. This NDH is a heterotrimeric enzyme of xanthine dehydrogenase family containing dinucleotide form of molybdopterin cofactor, a flavin adenine dinucleotide (FAD), and two iron-sulphur clusters [74]. This 6-HN was oxidized to form an optically inactive 6-hydroxy-N-methlymyosmine (6-HMM) by oxidation of pyrrolidine ring at the 2nd position of carbon. Interestingly, this step was catalyzed by two enzymes, namely, 6-hydroxy-L-nicotine oxidase (6-HLNO) and 6-hydroxy-D-nicotine oxidase (6HDNO). The dimeric form of 6-HLNO and monomeric form of 6HDNO contained FAD noncovalently bound to 47 kDa and covalently to 49 kDa polypeptide, respectively [75]. The third step was followed by hydration of 6-HMM that spontaneously leads to the opening of the pyrrolidine ring and tautamerization of ketone moiety to form 6-hydroxypseudooxynicotine (6-HPON) [76]. The 2nd position of the carbon of pyridine ring of this metabolite was further hydroxylated by ketone oxidase or ketone dehydrogenase enzyme similar to nicotine dehydrogenase to form 2,6-dihydroxypseudooxynicotine (2,6-DHPON) [77]. The subsequent cleavage of side chain of 2,6-DHPON to form γ-N-methylaminobutyrate (MGABA) and 2,6-dihydroxypyridine (2,6-DHP) by the action of 2-6-dihyroxypseudooxynicotine hydrolase (2,6-DHPONH) [61, 78–80]. 2,6-DHP was further hydroxylated to form 2,3,6-trihydroxypyridine (2,3-6-THP) by the addition of hydroxyl group to the 3rd position of carbon in the pyridine ring. This step was catalyzed by an enzyme 2,6-dihydroxypyridine-3-hydroxylase (2,6-DHPH) in a strictly nicotinamide adenine dinucleotide hydrogen (NADH)-dependent manner. Dimeric flavoprotein of this enzyme is tightly bound noncovalently to a FAD subunit and inhibited by 2,3-dihydroxypyridine and 2,6-dimethoxypyridine. Each subunit consists of 397 amino acids and mass of 43.4 kDa with addition of one FAD molecule [81]. In the presence of oxygen, spontaneous oxidation of 2,3,6-THP and dimerization of this hydroxylated pyridine ring moiety formed nicotine blue colour [63]. However, Sachelaru et al. [82] reported that mobA gene that encodes MobA protein with molybdenum cofactor cytidylate transferase was responsible for formation of nicotine blue colour in A. nicotinovorans.
Figure 1.
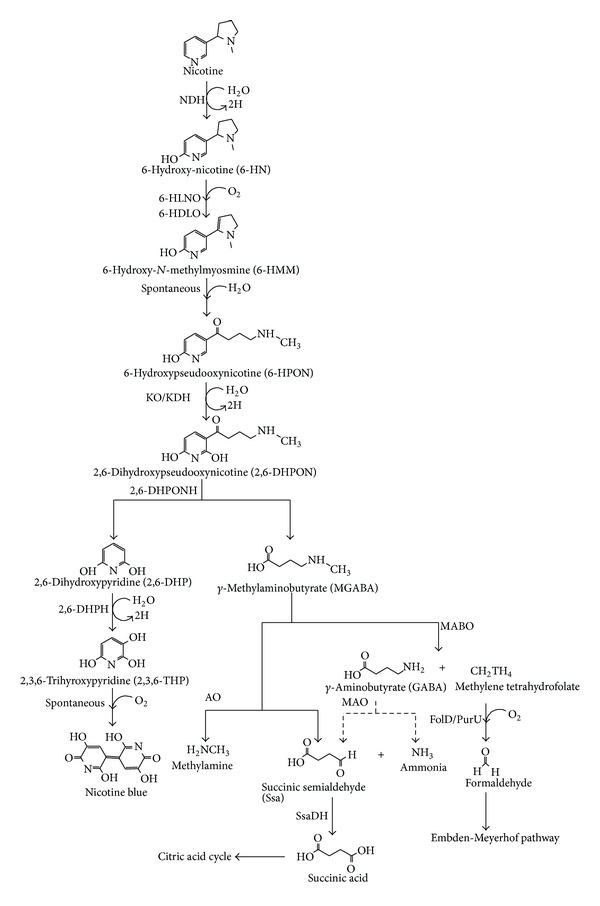
The pyridine pathway of nicotine degradation by A. nicotinovorans and Nocardioides sp. NDH, nicotine dehydrogenase; 6-HLNO, 6-hydroxy-L-nicotine oxidase; 6HDNO, 6-hydroxy-D-nicotine oxidase; KO, ketone oxidase; KDH, ketone dehydrogenase; 2,6-DHPONH, 2-6-dihyroxypseudooxynicotine hydrolase; 2,6-DHPH, 2,6-dihydroxypyridine-3-hydroxylase; MABO, γ-N-methylaminobutyrate oxidase; MAO, monoamine oxidase; AO, amine oxidase; FolD, methylenetetrahydrofolate dehydrogenase/cyclohydrolase; PurU, formyltetrahydrofolate deformylase; SsaDH, succinic semialdehyde dehydrogenase.
The degradation of MGABA was regulated by cluster of genes encoded by plasmid pAO1. This cluster of genes contained purU-mabO-folD operon that transcribed only in the presence of nicotine and was regulated by transcriptional activator pmfR [83]. MGABA was further degraded into two pathways. The first pathway started with an enzyme γ-N-methylaminobutyrate oxidase (MABO), which catalyzed MGABA to form γ-aminobutyrate (GABA) and methylenetetrahydrofolate. The methylene group of methylenetetrahydrofolate was further oxidized by two enzymes, namely, methylene-tetrahydrofolate dehydrogenase/cyclohydrolase (FolD) and formyltetrahydrofolate deformylase (PurU) to formaldehyde [77]. These two enzymes are nicotinamide adenine dinucleotide hydrogen phosphate (NADPH) dependent and formaldehyde may be assimilated in the Embden-Meyerhof pathway [84, 85]. Another metabolite, GABA metabolized into an ammonia and succinic semialdehyde (Ssa) by monoamine oxidase (MAO), is an amine oxidase (AO) enzyme family. Ssa is further oxidized to succinic acid by an enzyme called nicotinamide adenine dinucleotide phosphate (NADP+) dependent succinic semialdehyde dehydrogenase (SsaDH) [86, 87]. This succinic acid entered into citric acid cycle and this catabolic pathway of γ-aminobutyrate was normally found in bacteria [88–90]. A newly discovered second degradation pathway of MGABA is deaminated into Ssa and methylamine by amine oxidase (AO) with reduction of FAD to FADH2. It produced succinic acid, which entered citric acid cycle [83, 91].
4.2. Pyrrolidine Pathways of Nicotine Biodegradation
The extrachromosomal genes of Pseudomonas sp. that degrades organic pollutants such as octane, camphor, toluene, methyl benzoate, salicylate, naphthalene, and xylene are responsible for nicotine degradation [68]. A purple crystalline substance, N-methylmyosmine, was isolated from nicotine containing medium during the degradation of nicotine by A. oxydans [92, 93]. The transformation of intermediate metabolites of nicotine such as 3-nicotinoylpropionic acid, pseudooxynicotine, and N-methylmyosmine produced by Pseudomonas sp. has been reported [25, 94–96]. Thacker et al. [68] reported that P. convexa Pc1 degraded nicotine to 2,5-dihydroxypyridine (2,5-DHP) via pseudooxynicotine, 3-succinoyl pyridine (SP), and 6-hydroxy-3-succinoyl pyridine (HSP) by nicotine-degrading enzyme 2,5-dihydroxypyridine oxygenase. P. plecoglossicida TND35 degraded nicotine to 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB) via N-methylmyosmine [2].
The predominant nicotine-degrading Gram-negative Pseudomonas sp. followed pyrrolidine pathway [97]. However, the mechanism of this pathway has been studied poorly [98]. Pseudomonas sp. followed mainly four different pathways of pyrrolidine (Figure 2). Pyrrolidine pathway of nicotine degradation of P. putida S16 has been reported [28] (Figure 2). The intermediate metabolites and its nicotine-degrading genes of pyrrolidine pathways of other Pseudomonas sp. have not been fully elucidated or thoroughly characterized. Ruan et al. [26] reported that the Pseudomonas sp. HF-1 followed three other incomplete pyrrolidine pathways such as nicotine to cotinine, nicotyrine, and nornicotine (Figure 2). However, Pseudomonas sp. Nic22 degraded nicotine via cotinine and nornicotine pathways of pyrrolidine [29].
Figure 2.
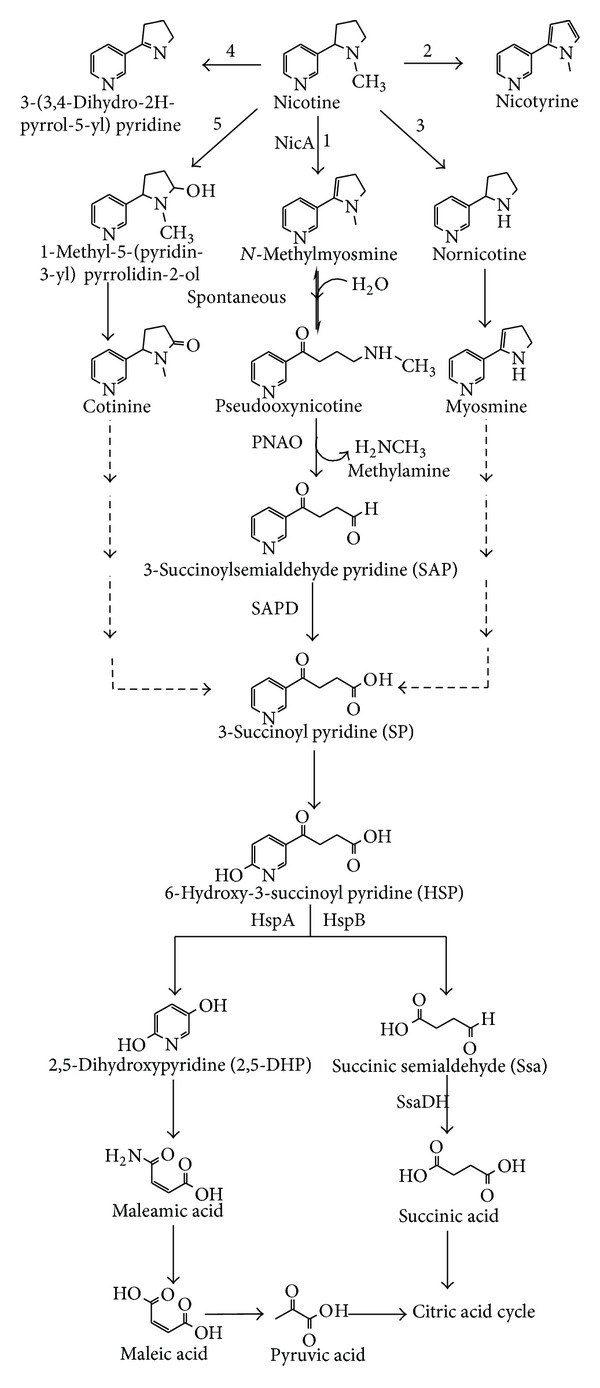
Various pyrrolidine pathways of nicotine degradation followed by 1. Pseudomonas sp. HZN6 and P. putida S16; 2. Pseudomonas sp. HF-1; 3. Pseudomonas sp. HF-1 and Pseudomonas sp. Nic22; 4. Pseudomonas sp. HF-1, Pseudomonas sp. Nic22, and Pseudomonas sp. CS3; 5. Pseudomonas sp. CS3. NicA, nicotine oxidoreductase; PNAO, pseudooxynicotine amine oxidase; SAPD, 3-succinoylsemialdehyde pyridine dehydrogenase; HspA and HspB, 6-hydroxy-3-succinoyl pyridine hydroxylase.
The pyrrolidine pathway of P. putida S16 initially attacked the pyrrolidine ring of nicotine to give N-methylmyosmine by the formation of a double bond. NicA enzyme was involved in the first step of dehydrogenation and it was believed that nicA gene encodes nicotine oxidoreductase that plays an important role in the initial steps of pyrrolidine pathway and involved in the degradation of nicotine to SP [99]. The reversible second step was carried out in the presence of water and a double bond of N-methylmyosmine was spontaneously hydrolyzed to form pseudooxynicotine, a direct precursor of a potent tobacco-specific lung carcinogen [99–101]. This carcinogenic intermediate metabolite pseudooxynicotine was further dehydrogenated to give methylamine and SP by NicA enzyme. Tang et al. [99] hypothesized that two unstable compounds were produced during the conversion of pseudooxynicotine to SP. The 6th position of carbon of SP was further hydrolyzed to yield HSP by an unknown enzyme. The hsp gene encodes an enzyme 6-hydroxy-3-succinoyl pyridine hydroxylase (HSP hydroxylase). The hsp gene attacked on the 3rd position of HSP to form 2,5-DHP and succinic semialdehyde (Ssa) [28, 97, 102]. The 2,5-DHP was further degraded to maleamic acid, which deaminates by hydrolysis and produces maleic acid [103]. The cleavage of 5th and 6th position of carbon of maleic acid gave pyruvic acid, which entered into citric acid cycle [102]. On the other side, Ssa easily converted into succinic acid by succinic semialdehyde dehydrogenase (SsaDH) enzyme that is widely present in Pseudomonas sp. [104–106].
Interestingly, the two genes hspA and hspB that encode HSP hydroxylase are involved in the conversion of HSP to 2,5-DHP [97, 99, 107]. The hydroxylase enzyme is involved in oxidation reactions in which one of the two atoms of molecular oxygen is integrated into the substrate and another is used to oxidize NADH or NADPH [108–110]. In P. putida S16 4879 bp nic gene cluster encoded three open reading frames (ORF), namely, ORF1 (1853 bp), ORF2 (936 bp), and ORF3 (582 bp). The ORF1 encoded nicA gene and ORF2 encoded hspA gene, whereas the function of remaining ORF3 is unknown [97, 99, 107]. Nevertheless, the newly identified hspB gene was located on 30 Kb of DNA away from the nic gene cluster [107]. The molecular mass of HspA has 38 kDa and NADH dependent, while HspB has 40 kDa, a dimer, and a prosthetic group FAD dependent. The deletion of hspB gene in the mutant strain could not degrade HSP, suggesting that hspB gene plays an important role in the conversion of HSP to 2,5-DHP [107]. The molecular mass of flavin adenine mononucleotide (FMN) dependent enzyme NicA is approximately 65 kDa that degraded nicotine into N-methylmyosmine, pseudooxynicotine, and SP in P. putida S16. These three nicotine intermediate metabolites were confirmed by electrospray ionization quadrupole time of flight mass spectrometry (ESI-Q-TOF-MS) analysis [99]. All three genes have been cloned and overexpressed in Escherichia coli [97, 99, 107]. However, the Pseudomonas sp. Nic22 bacteria followed other pyrrolidine pathways that produced myosmine, 2,3′-dipyridyl, and cotinine during biodegradation of nicotine [29] (Figure 2). Shinella sp. HZN1 produced three nicotine intermediate metabolites during biodegradation, which were characterized and identified as cotinine, myosmine, and nicotyrine using gas chromatography-mass spectrometry (GC-MS) analysis [36]. Four major nicotine intermediate metabolites pseudooxynicotine, SP, an unstable compound 3-succinoylsemialdehyde pyridine (SAP), and HSP and three nicotine-degrading genes sir A2, pao, and sap were identified in Pseudomonas sp. HZN6 [35, 98]. Sir A2 protein was encoded by Sir A2 and a sulfurtransferase homologue gene is responsible for the degradation of SP [98]. An unstable nicotine intermediate metabolite SAP produced during the conversion of pseudooxynicotine by pseudooxynicotine amine oxidase (PNAO) was encoded by pna gene. This enzyme oxidatively deaminates pseudooxynicotine by noncovalently bound FAD, O2, and H2O and forms SAP, methylamine, and H2O2. Another gene sap encodes NADP+ dependent enzyme 3-succinoylsemialdehyde pyridine dehydrogenase (SAPD) dehydrogenates SAP to SP [35]. Another nicotine-degrading bacterium Pseudomonas CS3 produced three new nicotine intermediate metabolites [111] (Figure 2). In the initial step, demethylation of nicotine forms a metabolite 3-(3,4-dihydro-2H-pyrrol-5-yl) pyridine. Concurrently, further degradation was initiated by hydroxylation of 2nd position of pyrrolidine ring of nicotine to form 1-methyl-5-(3-pyridyl) pyrroline-2-ol which is further transformed to cotinine.
Wang et al. [112] identified nicotine-degrading gene hsp in the plasmid pMF1 (21 Kb) of Pseudomonas sp. HF-1 and reported that nicotine degradation is regulated by plasmid not chromosomal DNA. Ketopantoate hydroxymethyltransferase encoded by panB gene of P. putida J5 is involved in nicotine catabolism. Pyruvic acid the end product of pyrrolidine pathway is a precursor of ketoisovalerate, which undergoes catabolism using the enzyme ketopantoate hydroxymethyltransferase leading to synthesis of vitamin pantothenate [113].
Raman et al. [2] recently reported that P. plecoglossicida TND35 followed a variant of pyrrolidine pathway, which is different from pathways of other bacteria and fungi (Figure 3). Strain TND35 oxidized pyrrolidine ring moiety of nicotine to form N-methylmyosmine. This intermediate was further hydroxylated at 2nd position of pyrrolidine ring to form a new cotinine analogue metabolite, 2,3-dihydro-1-methyl-5-(pyridin-3-yl)-1H-pyrrol-2-ol (IM2). In addition, this metabolite was demethylated and hydroxyl group of 2,3-dihydro-1-methyl-5-(pyridin-3-yl)-1H-pyrrol-2-ol was further oxidized to form another new cotinine analogue metabolite, 5-(pyridin-3-yl)-1H-pyrrol-2(3H)-one (IM3). Concurrently, 2,3-dihydro-1-methyl-5-(pyridin-3-yl)-1H-pyrrol-2-ol was further oxidized and released methylamine. This reaction further leads to opening of the pyrrolidine ring to form an end product 4-hydroxy-1-(3-pyridyl)-1-butanone (IM4). Interestingly, new metabolite 3,5-bis (1-methylpyrrolidin-2-yl) (IM5) was observed during nicotine degradation. This may be due to the cleavage of bond between pyridine and pyrrolidine ring of the nicotine. This N-methyl pyrrolidine ring attacked the pyridine moiety of nicotine and formed this metabolite.
Figure 3.
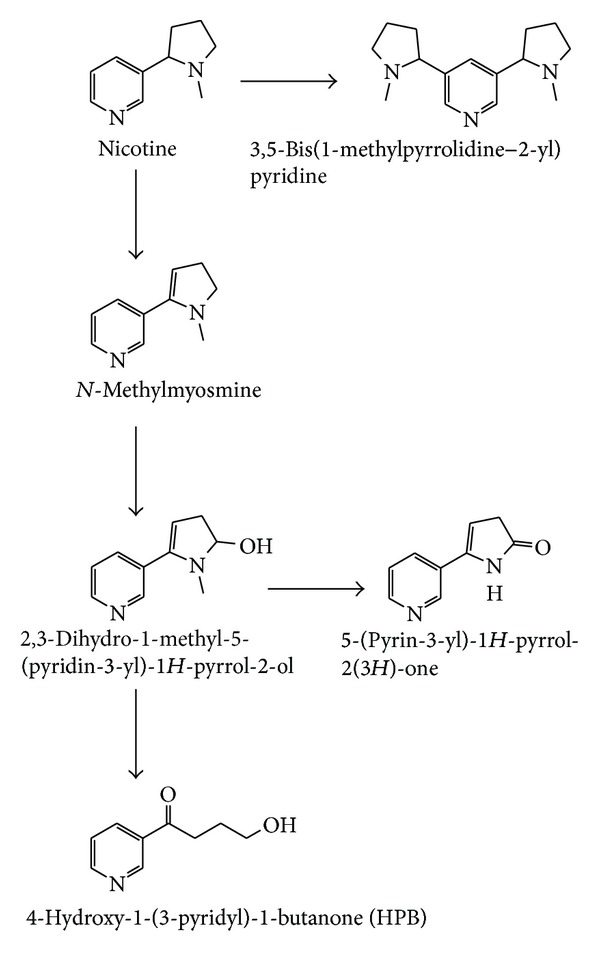
A variant pyrrolidine pathway of nicotine biodegradation by P. plecoglossicida TND35.
4.3. Variant Pathway of Pyridine and Pyrrolidine for Nicotine Biodegradation
The bacterium A. tumefaciens S33 followed a variant of pyridine and pyrrolidine pathway (Figure 4). This bacterium partially shared both the pathways and produced its intermediate metabolites [114]. Similarly, this bacterium also produced bright green colour initially and oxidized to brown colour at concentration above 3 g/L nicotine. The production of colour depends upon the concentration of nicotine, oxygen content, and pH of the medium [114]. Pathway of pyridine intermediate metabolites 6-HN, 6-HMM, and 6-HPON and pyrrolidine pathway metabolites HSP and 2,5-DHP were identified by ultraviolet-visible (UV-Vis) spectroscopy, thin layer chromatography (TLC), high performance liquid chromatography (HPLC), gas chromatography-high resolution-mass spectrometry (GC-HR-MS), and ESI-Q-TOF-MS analyses during nicotine degradation of A. tumefaciens S33. The catabolic nicotine-degrading enzymes NDH, 6-HLNO, and HSP hydroxylase were also observed in this bacterium. The initial step of pyridine pathway followed hydroxylation of nicotine to 6-HN by NDH. This metabolite was further oxidized and transformed into 6-HMM by 6-HLNO. The occurrence of spontaneous hydrolysis of 6-HMM into 6-HPON makes entry into pyrrolidine pathway. The metabolite 6-HPON is dehydrogenated and spontaneous hydrolysis leads to the production of HSP and removal of methylamine. However, similar mechanism was also observed during catabolic degradation of chlorine and aromatic amine, 2-phenethylamine [115, 116]. The HSP was further catabolized into 2,5-DHP. In contrast, N-methylmyosmine, pseudooxynicotine, and SP were not found during this degradation [114].
Figure 4.
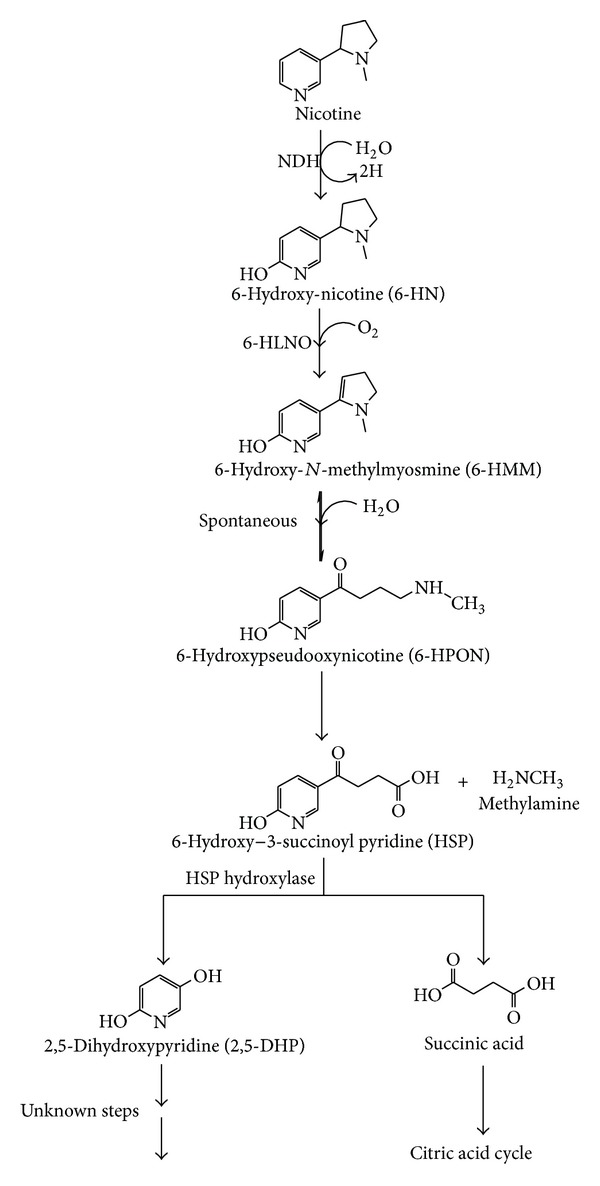
A variant of pyridine and pyrrolidine pathway of nicotine degradation by A. tumefaciens S33. NDH, nicotine dehydrogenase; 6-HLNO, 6-hydroxy-L-nicotine oxidase; HSP hydroxylase, 6-hydroxy-3-succinoyl pyridine hydroxylase.
4.4. Demethylation Pathway for Nicotine Biodegradation
Fungi such as M. gypseum, P. filamentosa JTS-208, and C. echinulata have been reported for nicotine degradation. These fungi are involved in the initial step, demethylation of nicotine that leads to the formation of nornicotine [21, 117] (Figure 5). Recently, Meng et al. [22] reported nicotine intermediate metabolites, namely, nornicotine, N-methylnicotinamide, 2,3-DHP, 2-hydroxy-N-methylnicotinamide, acetic acid, carbomic acid, and succinic acid on the basis of TLC, GC-MS, nuclear magnetic resonance (NMR), and Fourier transform infrared (FT-IR) analyses and proposed a hypothetical demethylation pathway of nicotine degradation by A. oryzae 112822 (Figure 5). The primary step of this hypothetical pathway is the elimination of the removal of methyl group in the pyrrolidine ring of nicotine to form nornicotine, which is further converted into myosmine by the formation of double bond in the pyrrolidine ring (Figure 5). The subsequent cleavage of pyrrolidine ring resulted in the formation of unknown intermediate metabolite. The hydrolytic attack on the postulated unknown intermediate metabolite resulted in N-methylnicotinamide and acetic acid. The N-methylnicotinamide was hydroxylated to form 2-hydroxy-N-methylnicotinamide, which was catabolized to a new nicotine intermediate metabolite 2,3-DHP with the formation of aminomethyl. The aminomethyl was further transformed into carbomic acid. The opening of ring in 2,3-DHP leads to the formation of succinic acid, which enters into citric acid cycle [22]. Nevertheless, no catabolic enzymes were identified during the degradation of nicotine by fungi.
Figure 5.
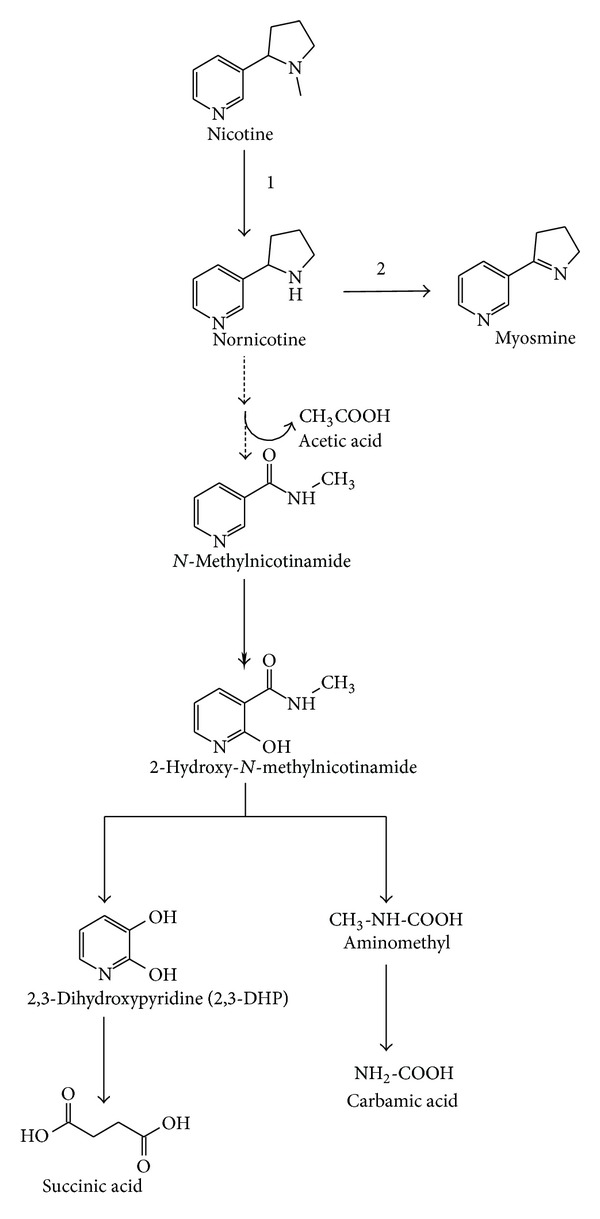
Demethylation pathway of nicotine degradation by fungi. 1. A. oryzae 11282; 2. M. gypseum, P. filamentosa JTS-208, and C. echinulata.
5. Biotechnological Applications of Nicotine Intermediate Metabolites
Biotransformation or biocatalysis involves the use of microorganisms to catalyze the conversion of one metabolite into another. These metabolites were catalyzed by whole microbial cells, cellular extracts, or enzymes [118]. Biotransformation is a promising tool, used in the synthesis of bulk chemicals for synthesis of pharmaceutical, food and agrochemical ingredients in the industry [119]. Nicotine is used as a starting material for the biocatalytic production of functionalized pyridines from renewable sources [120]. The easiest and friendliest ways of biotransformation approach were used to transfer toxic nicotine into valuable compounds such as HSP and DHP [110, 119, 121, 122]. Biotransformation intermediates of nicotine are widely used in anticancer therapies, antimalarial and analgesics drug development, and treatment of Parkinson's disease, hypertension and disorders of central nervous system [123]. Nicotine intermediate metabolites are precursors in the synthesis of drug such as analogues of epibatidine, an extremely effective analgesic molecule that is used in pharmaceutical industry [121]. Hydroxylated pyridine intermediates are used as precursors for the synthesis of drugs and instecticides via chemical methods [114]. The biologically active metabolites, 2,5- or 3,5-disubstituted pyridines, are catabolized from 6HLN and HSP, which is used for the synthesis of insecticide imidacloprid; SIB-1508Y is an anti-Parkinson's agent [119, 121, 122]. The important nicotine intermediate metabolite, 2,5-DHP, can be used as the initial material for the chemical synthesis of universal precursor, aminolevulinic acid. This precursor is used to synthesize plant growth regulators, herbicides, and drugs used in cancer therapies and to synthesize porphyrins such as heme and chlorophyll [110]. The biotransformation nicotine intermediate metabolite HPB has been widely used as a biomarker for tobacco related lung cancer studies [2].
6. Conclusions and Future Perspectives
Environmental pollution is one of the major problems in the world. Tobacco industries produced enormous amount of nicotine. The nonreadily degradable nicotine causes environmental problems and human health when directly entered into soil. Major microbes degrade the toxic compound, nicotine. In this paper, we have discussed all the metabolic pathways and the genes involved in nicotine degradation. These microbes produce various intermediate metabolic compounds of pharmaceutical importance during nicotine degradation. Bioremediation is one of the promising tools used to convert the toxic compounds into valuable compounds. These nicotine-degrading microbes can be used for bioremediation of nicotine-polluted environments. Large scale production of these intermediate metabolites of nicotine could be of great use in pharmaceutical industries.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgments
This work was supported by UGC-Major Project and UGC-SAP Programme coordinated by Professor N. Sakthivel and UGC-Rajiv Gandhi National Fellowship awarded to Dr. G. Raman.
References
- 1.ERC Statistics International PLC. World Cigarettes 2005. Newmarket, UK: ERC Statistics International PLC; 2005. [Google Scholar]
- 2.Raman G, Mohan K, Manohar V, Sakthivel N. Biodegradation of nicotine by a novel nicotine-degrading bacterium, Pseudomonas plecoglossicida TND35 and its new biotransformation intermediates. Biodegradation. 2013 doi: 10.1007/s10532-013-9643-4. [DOI] [PubMed] [Google Scholar]
- 3.Dani JA, Ji D, Zhou F. Synaptic plasticity and nicotine addiction. Neuron. 2001;31(3):349–352. doi: 10.1016/s0896-6273(01)00379-8. [DOI] [PubMed] [Google Scholar]
- 4.Bush L, Hempfling WP, Burton H. Biosynthesis of nicotine and related compounds. In: Gorrod JW, Jacob P III, editors. Analytical Determination of Nicotine and Related Compounds and Their Metabolites. Amsterdam, The Netherlans: Elsevier; 1999. pp. 13–44. [Google Scholar]
- 5.Sloan FA, Gelband H. Cancer Control Opportunities in Low- and Middle-Income Countries. Washington, DC, USA: Institute of Medicine (U. S.) Committee on Cancer Control in Low- and Middle-Income Countries, National Academies Press; [PubMed] [Google Scholar]
- 6.Li H, Li X, Duan Y, Zhang K, Yang J. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Applied Microbiology and Biotechnology. 2010;86(1):11–17. doi: 10.1007/s00253-009-2427-4. [DOI] [PubMed] [Google Scholar]
- 7.Civilini M, Domenis C, Sebastianutto N, de Bertoldi M. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Management and Research. 1997;15(4):349–358. [Google Scholar]
- 8.Novotny TE, Zhao F. Consumption and production waste: another externality of tobacco use. Tobacco Control. 1999;8(1):75–80. doi: 10.1136/tc.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong DW, Wang X, Ercal N. Enantiomeric composition of nicotine in smokeless tobacco, medicinal products and commercial reagents. Chirality. 1998;10:587–591. [Google Scholar]
- 10.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins BT, Abbruscato TJ, Egleton RD, et al. Nicotine increases in vivo blood-brain barrier permeability and alters cerebral microvascular tight junction protein distribution. Brain Research. 2004;1027(1-2):48–58. doi: 10.1016/j.brainres.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 12.Cogo K, Franz Montan M, Bergamaschi CDC, Andrade ED, Rosalen PL, Groppo FC. In vitro evaluation of the effect of nicotine, cotinine, and caffeine on oral microorganisms. Canadian Journal of Microbiology. 2008;54(6):501–508. doi: 10.1139/w08-032. [DOI] [PubMed] [Google Scholar]
- 13.Yildiz D. Nicotine, its metabolism and an overview of its biological effects. Toxicon. 2004;43(6):619–632. doi: 10.1016/j.toxicon.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Brunnemann KD, Prokopczyk B, Djordjevic MV, Hoffmann D. Formation and analysis of tobacco-specific N-nitrosamines. Critical Reviews in Toxicology. 1996;26(2):121–137. doi: 10.3109/10408449609017926. [DOI] [PubMed] [Google Scholar]
- 15.Campain JA. Nicotine: potentially a multifunctional carcinogen? Toxicological Sciences. 2004;79(1):1–3. doi: 10.1093/toxsci/kfh106. [DOI] [PubMed] [Google Scholar]
- 16.Yildiz D, Ercal N, Armstrong DW. Nicotine enantiomers and oxidative stress. Toxicology. 1998;130(2-3):155–165. doi: 10.1016/s0300-483x(98)00105-x. [DOI] [PubMed] [Google Scholar]
- 17.Qiao D, Seidler FJ, Slotkin TA. Oxidative mechanisms contributing to the developmental neurotoxicity of nicotine and chlorpyrifos. Toxicology and Applied Pharmacology. 2005;206(1):17–26. doi: 10.1016/j.taap.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YD, Luo CR, Wang HL, Lu GF. Advances in microbial degradation of nicotine and its application. Tobacco and Science Technology. 2003;12:3–7. [Google Scholar]
- 19.Schievelbein H. Nicotine, resorption and fate. Pharmacology and Therapeutics. 1982;18(2):233–248. doi: 10.1016/0163-7258(82)90068-7. [DOI] [PubMed] [Google Scholar]
- 20.Sabha M, Tanus-Santos JE, Toledo JCY, Cittadino M, Rocha JC, Moreno H., Jr. Pharmacodynamics and drug action: transdermal nicotine mimics the smoking-induced endothelial dysfunction. Clinical Pharmacology and Therapeutics. 2000;68(2):167–174. doi: 10.1067/mcp.2000.108851. [DOI] [PubMed] [Google Scholar]
- 21.Uchida S, Maeda S, Kisaki T. Conversion of nicotine into nornicotine and N-methylmyosmine by fungi. Agricultural and Biological Chemistry. 1983;47(9):1949–1953. [Google Scholar]
- 22.Meng XJ, Lu LL, Gu GF, Xiao M. A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Research in Microbiology. 2010;161(7):626–633. doi: 10.1016/j.resmic.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Hylin JW. Microbial degradation of nicotine. I. Morphology and physiology of Achromobacter nicotinophagum n. sp. Journal of Bacteriology. 1958;76(1):36–40. doi: 10.1128/jb.76.1.36-40.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruan A, Min H, Zhu W. Studies on biodegradation of nicotine by Arthrobacter sp. strain HF-2. Journal of Environmental Science and Health B. 2006;41(7):1159–1170. doi: 10.1080/03601230600856934. [DOI] [PubMed] [Google Scholar]
- 25.Wada E, Yamasaki K. Mechanism of microbial degradation of nicotine. Science. 1953;117(3033):152–153. doi: 10.1126/science.117.3033.152. [DOI] [PubMed] [Google Scholar]
- 26.Ruan A, Min H, Peng X, Huang Z. Isolation and characterization of Pseudomonas sp. strain HF-1, capable of degrading nicotine. Research in Microbiology. 2005;156(5-6):700–706. doi: 10.1016/j.resmic.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Yuan YJ, Lu ZX, Huang LJ, Bie XM, Lü FX, Li Y. Optimization of a medium for enhancing nicotine biodegradation by Ochrobactrum intermedium DN2. Journal of Applied Microbiology. 2006;101(3):691–697. doi: 10.1111/j.1365-2672.2006.02929.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang SN, Liu Z, Tang HZ, Meng J, Xu P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology. 2007;153(5):1556–1565. doi: 10.1099/mic.0.2006/005223-0. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Li X, Yang J, Gong X, Li B, Zhang K. Isolation of nicotine-degrading bacterium Pseudomonas sp. Nic22, and its potential application in tobacco processing. International Biodeterioration and Biodegradation. 2008;62(3):226–231. [Google Scholar]
- 30.Wei H, Lei L, Xia Z, Liu S, Liu P, Liu X. Characterisation of a novel aerobic nicotine-biodegrading strain of Pseudomonas putida . Annals of Microbiology. 2008;58(1):41–45. [Google Scholar]
- 31.Gong X, Yang J, Duan Y, et al. Isolation and characterization of Rhodococcus sp. Y22 and its potential application to tobacco processing. Research in Microbiology. 2009;160(3):200–204. doi: 10.1016/j.resmic.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Wang SN, Liu Z, Xu P. Biodegradation of nicotine by a newly isolated Agrobacterium sp. strain S33. Journal of Applied Microbiology. 2009;107(3):838–847. doi: 10.1111/j.1365-2672.2009.04259.x. [DOI] [PubMed] [Google Scholar]
- 33.Lei L, Zhang W, Wei H, Xia Z, Liu X. Characterization of a novel nicotine-degrading Ensifer sp. strain N7 isolated from tobacco rhizosphere. Annals of Microbiology. 2009;59(2):247–252. [Google Scholar]
- 34.Zhong W, Zhu C, Shu M, et al. Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresource Technology. 2010;101(18):6935–6941. doi: 10.1016/j.biortech.2010.03.142. [DOI] [PubMed] [Google Scholar]
- 35.Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Applied Environmantal Microbiology. 2012;78:2154–2160. doi: 10.1128/AEM.07025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang HJ, Ma Y, Qiu GJ, Wu FL, Chen SL. Biodegradation of nicotine by a novel Strain Shinella sp. HZN1 isolated from activated sludge. Journal of Environmental Science and Health B. 2011;46(8):703–708. doi: 10.1080/03601234.2011.594409. [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Yang G, Wang X, Yao Y, Min H, Lu Z. Nicotine degradation by two novel bacterial isolates of Acinetobacter sp. TW and Sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World Journal of Microbiology and Biotechnology. 2011;27(7):1633–1640. [Google Scholar]
- 38.Ma G, Lei L, Xia Z, Gong X, Zhou W, Yang J. Diversity and phylogenetic analyses of nicotine-degrading bacteria isolated from tobacco plantation soils. African Journal of Microbiological Research. 2012;6:6392–6398. [Google Scholar]
- 39.Zhao L, Zhu C, Gao Y, et al. Nicotine degradation enhancement by Pseudomonas stutzeri ZCJ during aging process of tobacco leaves. World Journal of Microbiology and Biotechnology. 2012;28(5):2077–2086. doi: 10.1007/s11274-012-1010-9. [DOI] [PubMed] [Google Scholar]
- 40.Giovannozzi-Sermanni G. Industrial experiments of fermentation with addition of microbic cultures. Tobacco. 1947;51:6–15. [Google Scholar]
- 41.Gravely LE, Lawrence E, Geiss VL, Vernon L, Charles F. Process for reduction of nitrate and nicotine content of tobacco by microbial treatment. US Patent No, 4557280, 1978.
- 42.Li H, Duan Y, Ma G, Lei L, Zhang K, Yang J. Isolation and characterization of Acinetobacter sp. ND12 capable of degrading nicotine. African Journal of Microbiology Research. 2011;5(11):1335–1341. [Google Scholar]
- 43.Niu T, Chen C, Jiatong N, et al. Nicotine dependence and its familial aggregation in Chinese. International Journal of Epidemiology. 2000;29(2):248–252. doi: 10.1093/ije/29.2.248. [DOI] [PubMed] [Google Scholar]
- 44.Zbikowski SM, Swan GE, McClure JB. Cigarette smoking and nicotine dependence. Medical Clinics of North America. 2004;88(6):1453–1465. doi: 10.1016/j.mcna.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Dome P, Lazary J, Kalapos MP, Rihmer Z. Smoking, nicotine and neuropsychiatric disorders. Neuroscience and Biobehavioral Reviews. 2010;34(3):295–342. doi: 10.1016/j.neubiorev.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Lenkey AA. Nicotine removal process and product produced thereby, mixing with alkaline agent in aerobic environment. United States Patent No, 4848373, 1989.
- 47.Beskoski VP, Gojgicc-Cvijovic G, Milic J, et al. Ex situ bioremediation of a soil contaminated by mazut (heavy residual fuel oil): a field experiment. Chemosphere. 2011;83:34–40. doi: 10.1016/j.chemosphere.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 48.Langer U, Böhme L, Böhme F. Classification of soil microorganisms based on growth properties: a critical view of some commonly used terms. Journal of Plant Nutrition and Soil Science. 2004;167(3):267–269. [Google Scholar]
- 49.Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends in Biotechnology. 2008;26(9):483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Briski F, Horgas N, Vukovic M, Gomzi Z. Aerobic composting of tobacco industry solid waste-simulation of the process. Clean Technologies and Environmental Policy. 2003;5:295–301. [Google Scholar]
- 51.Meher KK, Panchwagh AM, Rangrass S, Gollakota KG. Biomethanation of tobacco waste. Environmental Pollution. 1995;90(2):199–202. doi: 10.1016/0269-7491(94)00107-o. [DOI] [PubMed] [Google Scholar]
- 52.Schmit CG, Jahan K, Schmit KH, Debik E, Mahendraker V. Activated sludge and other aerobic suspended culture processes. Water Environment Research. 2009;81(10):1127–1193. doi: 10.2175/106143002x140152. [DOI] [PubMed] [Google Scholar]
- 53.Mace S, Mata-Alvarez J. Utilization of SBR technology for wastewater treatment: an overview. Industrial and Engineering Chemistry Research. 2002;41(23):5539–5553. [Google Scholar]
- 54.Eberhardt H. The biological degradation of nicotine by nicotinophilic microorganisms. Beitrage zur Tabakforschung International. 1995;16:119–129. [Google Scholar]
- 55.Batham HN. Nitrification in soil. II. Soil Science. 1927;24:187–203. [Google Scholar]
- 56.Huang J, Yang J, Duan Y, et al. Bacterial diversities on unaged and aging flue-cured tobacco leaves estimated by 16S rRNA sequence analysis. Applied Microbiology and Biotechnology. 2010;88(2):553–562. doi: 10.1007/s00253-010-2763-4. [DOI] [PubMed] [Google Scholar]
- 57.Yuan YJ, Lu ZX, Wu N, Huang LJ, Lü FX, Bie XM. Isolation and preliminary characterization of a novel nicotine-degrading bacterium, Ochrobactrum intermedium DN2. International Biodeterioration and Biodegradation. 2005;56(1):45–50. [Google Scholar]
- 58.Yao Y, Tang H, Ren H, Yu H, Wang L, Xu P. Genome sequence of a nicotine-degrading strain of Arthrobacter . Journal of Bacteriology. 2012;194:5714–5715. doi: 10.1128/JB.01370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang H, Yu H, Tai C, et al. Genome sequence of a novel nicotine-degrading strain, Pseudomonas geniculata N1. Journal of Bacteriology. 2012;194:p. 3553. doi: 10.1128/JB.00625-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giovannozzi-Sermanni G. Arthrobacter nicotianae, a new type of Arthrobacter causing nicotine degradation. Coersta. 1959;3:p. 2595. [Google Scholar]
- 61.Gherna RL, Richardson SH, Rittenberg SC. The bacterial oxidation of nicotine. VI. The metabolism of 2,6-dihydroxypseudooxynicotine. The Journal of Biological Chemistry. 1965;240(9):3669–3674. [PubMed] [Google Scholar]
- 62.Cobzaru C, Brandsch R. Steps in uncovering the key enzyme in the degradation of the pyridine ring in the Arthrobacter nicotinovorans . Genetics and Molecular Biology. 2008;4:1–9. [Google Scholar]
- 63.Holmes PE, Rittenberg SC, Knackmuss HJ. The bacterial oxidation of nicotine. 8. Synthesis of 2,3,6-trihydroxypyridine and accumulation and partial characterization of the product of 2,6-dihydroxypyridine oxidation. The Journal of Biological Chemistry. 1972;247(23):7628–7633. [PubMed] [Google Scholar]
- 64.Igloi GL, Brandsch R. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. Journal of Bacteriology. 2003;185(6):1976–1989. doi: 10.1128/JB.185.6.1976-1986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandsch R, Decker K. Isolation and partial characterization of plasmid DNA from Arthrobacter oxidans . Archives of Microbiology. 1984;138(1):15–17. doi: 10.1007/BF00425400. [DOI] [PubMed] [Google Scholar]
- 66.Decker K, Brandsch R. Flavoproteins with a covalent histidyl(N3)-8α-riboflavin linkage. BioFactors. 1991;3(2):69–81. [PubMed] [Google Scholar]
- 67.Schenk S, Hoelz A, Krauß B, Decker K. Gene structures and properties of enzymes of the plasmid-encoded nicotine catabolism of Arthrobacter nicotinovorans . Journal of Molecular Biology. 1998;284(5):1323–1339. doi: 10.1006/jmbi.1998.2227. [DOI] [PubMed] [Google Scholar]
- 68.Thacker R, Rorvig O, Kahlon P, Gunsalus IC. NIC, a conjugative nicotine-nicotinate degradative plasmid in Pseudomonas convexa . Journal of Bacteriology. 1978;135(1):289–290. doi: 10.1128/jb.135.1.289-290.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brandsch R. Microbiology and biochemistry of nicotine degradation. Applied Microbiology and Biotechnology. 2006;69(5):493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 70.Hochestein LI, Rittenberg SC. The bacterial oxidation of nicotine. I. Nicotine oxidation by cell-free preparations. The Journal of Biological Chemistry. 1959;234(1):151–155. [PubMed] [Google Scholar]
- 71.Hochestein LI, Rittenberg SC. The bacterial oxidation of nicotine. II. The isolation of the first oxidative product and its identification as (1)-6-hydroxynicotine. The Journal of Biological Chemistry. 1959;234(1):156–160. [PubMed] [Google Scholar]
- 72.Grether-Beck S, Igloi GL, Pust S, Schilz E, Decker K, Brandsch R. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans . Molecular Microbiology. 1994;13(5):929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 73.Andreesen JR, Fetzner S. The molybdenum-containing hydroxylases of nicotinate, isonicotinate, and nicotine. Metal Ions in Biological Systems. 2002;39:405–430. [PubMed] [Google Scholar]
- 74.Hänzelmann P, Meyer O. Effect of molybdate and tungstate on the biosynthesis of CO dehydrogenase and the molybdopterin cytosine-dinucleotide-type of molybdenum cofactor in Hydrogenophaga pseudoflava . European Journal of Biochemistry. 1998;255(3):755–765. doi: 10.1046/j.1432-1327.1998.2550755.x. [DOI] [PubMed] [Google Scholar]
- 75.Schenk S, Decker K. Horizontal gene transfer involved in the convergent evolution of the plasmid-encoded enantioselective 6-hydroxynicotine oxidases. Journal of Molecular Evolution. 1999;48(2):178–186. doi: 10.1007/pl00006456. [DOI] [PubMed] [Google Scholar]
- 76.Koetter JWA, Schulz GE. Crystal structure of 6-hydroxy-D-nicotine oxidase from Arthrobacter nicotinovorans . Journal of Molecular Biology. 2005;352(2):418–428. doi: 10.1016/j.jmb.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 77.Chiribau CB, Sandu C, Fraaije M, Schiltz E, Brandsch R. A novel γ-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. European Journal of Biochemistry. 2004;271(23-24):4677–4684. doi: 10.1111/j.1432-1033.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- 78.Gries FA, Decker K, Brühmüller M. Über den Abbau des Nicotins durch Bakterienenzyme. V. Der Abbau des L-6-Hydroxynicotins zu [γ-Methylamino-propyl]-[6-hydroxy-pyr- idyl-(3)]-ketons. Hoppe-Seyler’s Zeitschrift Physiologishe Chemie. 1961;325:229–241. [Google Scholar]
- 79.Gries FA, Decker K, Eberwein H, Brühmüller M. Über den Abbau des Nicotins durch Bakteienenzyme. VI. Die enzymatische Umwandlung des (γ-Methylamino-propyl)-[6-hydroxy- pyridyl-(3)]-ketons. Biochemische Zeitschrift. 1961;335:285–302. [Google Scholar]
- 80.Sachelaru P, Schiltz E, Igloi GL, Brandsch R. An α/β-fold C-C bond hydrolase is involved in a central step of nicotine catabolism by Arthrobacter nicotinovorans . Journal of Bacteriology. 2005;187(24):8516–8519. doi: 10.1128/JB.187.24.8516-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Treiber N, Schulz GE. Structure of 2,6-dihydroxypyridine 3-hydroxylase from a nicotine-degrading pathway. Journal of Molecular Biology. 2008;379(1):94–104. doi: 10.1016/j.jmb.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 82.Sachelaru P, Schiltz E, Brandsch R. A functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans . Applied and Environmental Microbiology. 2006;72(7):5126–5131. doi: 10.1128/AEM.00437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiribau CB, Sandu C, Igloi GL, Brandsch R. Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans . Journal of Bacteriology. 2005;187(9):3062–3070. doi: 10.1128/JB.187.9.3062-3070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levering PR, Binnema DJ, van Dijken JP, Harder W. Enzymatic evidence for a simultaneous operation of two one-carbon assimilation pathways during growth of Arthrobacter P1 on choline. FEMS Microbiology Letters. 1981;12(1):19–25. [Google Scholar]
- 85.Zhang X, Fuller JH, McIntire WS. Cloning, sequencing, expression, and regulation of the structural gene for the copper/topa quinone-containing methylamine oxidase from Arthrobacter strain P1, a gram-positive facultative methylotroph. Journal of Bacteriology. 1993;175(17):5617–5627. doi: 10.1128/jb.175.17.5617-5627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bartsch K, von Johnn-Marteville A, Schulz A. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD) Journal of Bacteriology. 1990;172(12):7035–7042. doi: 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chambliss KL, Caudle DL, Hinson DD, et al. Molecular cloning of the mature NAD+-dependent succinic semialdehyde dehydrogenase from rat and human. cDNA Isolation, evolutionary homology, and tissue expression. The Journal of Biological Chemistry. 1995;270(1):461–467. doi: 10.1074/jbc.270.1.461. [DOI] [PubMed] [Google Scholar]
- 88.Dover S, Halpern YS. Utilization of c-aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. Journal of Bacteriology. 1972;109(2):835–843. doi: 10.1128/jb.109.2.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Metzer E, Halpern YS. In vivo cloning and characterization of the gabCTDP gene cluster of Escherichia coli K-12. Journal of Bacteriology. 1990;172(6):3250–3256. doi: 10.1128/jb.172.6.3250-3256.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niegemann E, Schulz A, Bartsch K. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Archives of Microbiology. 1993;160(6):454–460. doi: 10.1007/BF00245306. [DOI] [PubMed] [Google Scholar]
- 91.Ganas P, Sachelaru P, Mihasan M, Igloi GL, Brandsch R. Two closely related pathways of nicotine catabolism in Arthrobacter nicotinovorans and Nocardioides sp. strain JS614. Archives of Microbiology. 2008;189(5):511–517. doi: 10.1007/s00203-007-0340-8. [DOI] [PubMed] [Google Scholar]
- 92.Wenusch A. Further study of a biological decomposition of nicotine. Zeitschrift für Lebensmittel-Untersuchung und -Forschung. 1942;84:498–501. [Google Scholar]
- 93.Bucherer H. Über den mikrobiellen Abbau von Giftstoffen. I. Mitteilung: Über den mikrobiellen Abbau von Nikotin. Zentralblatt fur Bakteriology. 1942;105:166–173. [Google Scholar]
- 94.Tabuchi T. Microbial degradation of nicotine and nicotinic acid. I. Isolation of nicotine-decomposing bacteria and their morphological and physiological properties. Journal of Agricultural Chemical Society of Japan. 1955;28:806–810. [Google Scholar]
- 95.Tabuchi T. Microbial degradation of nicotine and nicotinic acid. II. Isolation of nicotine-decomposing bacteria and their morphological and physiological properties. Journal of Agricultural Chemical Society of Japan. 1955;29:219–221. [Google Scholar]
- 96.Tabuchi T. Microbial degradation of nicotine and nicotinic acid. III. Isolation of nicotine-decomposing bacteria and their morphological and physiological properties. Journal of Agricultural Chemical Society of Japan. 1955;29:222–225. [Google Scholar]
- 97.Tang H, Wang S, Ma L, et al. A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Applied and Environmental Microbiology. 2008;74(5):1567–1574. doi: 10.1128/AEM.02529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu J, Ma Y, Chen L, Wu L, Wen Y, Liu W. A sirA-like gene, sirA2, is essential for 3-succinoyl-pyridine metabolism in the newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Applied Microbiology and Biotechnology. 2011;92(5):1023–1032. doi: 10.1007/s00253-011-3353-9. [DOI] [PubMed] [Google Scholar]
- 99.Tang H, Wang L, Meng X, et al. Novel nicotine oxidoreductase- encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Applied and Environmental Microbiology. 2009;75(3):772–778. doi: 10.1128/AEM.02300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hecht SS, Hochalter BJ, Villalta PW, Murphy SE. 2′-hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes: formation of a lung carcinogen precursor. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(23):12493–12497. doi: 10.1073/pnas.220207697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hukkanen J, Jacob P, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacological Reviews. 2005;57(1):79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 102.Kaiser J, Feng Y, Bollag J. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiological Reviews. 1996;60(3):483–498. doi: 10.1128/mr.60.3.483-498.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Behrman EJ, Stanier RY. The bacterial oxidation of nicotinic acid. The Journal of Biological Chemistry. 1957;228(2):923–945. [PubMed] [Google Scholar]
- 104.Callewaert DM, Rosemblatt MS, Suzuki K, Tchen TT. Succinic semialdehyde dehydrogenase from a Pseudomonas species. I. Purification and chemical properties. The Journal of Biological Chemistry. 1973;248(17):6009–6013. [PubMed] [Google Scholar]
- 105.Rosemblatt MS, Callewaert DM, Tchen TT. Succinic semialdehyde dehydrogenase from a Pseudomonas species. II. Physical and immunochemical properties of the enzyme. The Journal of Biological Chemistry. 1973;248(17):6014–6018. [PubMed] [Google Scholar]
- 106.Sànchez M, Alvarez MA, Balaña R, Garrido-Pertierra A. Properties and functions of two succinic-semialdehyde dehydrogenases from Pseudomonas putida . Biochimica et Biophysica Acta. 1988;953:249–257. doi: 10.1016/0167-4838(88)90032-5. [DOI] [PubMed] [Google Scholar]
- 107.Tang H, Yao Y, Zhang D, et al. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida . The Journal of Biological Chemistry. 2011;286(45):39179–39187. doi: 10.1074/jbc.M111.283929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baitsch D, Sandu C, Brandsch R, Igloi GL. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. Journal of Bacteriology. 2001;183(18):5262–5267. doi: 10.1128/JB.183.18.5262-5267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hirschberg R, Ensign JC. Oxidation of nicotinic acid by a Bacillus species: source of oxygen atoms for the hydroxylation of nicotinic acid and 6-hydroxynicotinic acid. Journal of Bacteriology. 1971;108(2):757–759. doi: 10.1128/jb.108.2.757-759.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakano H, Wieser M, Hurh B, et al. Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. European Journal of Biochemistry. 1999;260(1):120–126. doi: 10.1046/j.1432-1327.1999.00124.x. [DOI] [PubMed] [Google Scholar]
- 111.Wang HH, Yin B, Peng XX, et al. Biodegradation of nicotine by newly isolated Pseudomonas sp. CS3 and its metabolites. Journal of Applied Microbiology. 2012;112(2):258–268. doi: 10.1111/j.1365-2672.2011.05208.x. [DOI] [PubMed] [Google Scholar]
- 112.Wang M, Yang G, Min H, Lv Z. A novel nicotine catabolic plasmid pMH1 in Pseudomonas sp. strain HF-1. Canadian Journal of Microbiology. 2009;55(3):228–233. doi: 10.1139/w08-135. [DOI] [PubMed] [Google Scholar]
- 113.Wei H, Lei L, Liu S, Xia Z, Liu X, Liu P. PanB is involved in nicotine metabolism in Pseudomonas putida . International Biodeterioration and Biodegradation. 2009;63(8):988–992. [Google Scholar]
- 114.Wang S, Huang H, Xie K, Xu P. Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Applied Microbiology and Biotechnology. 2012;95:1567–1578. doi: 10.1007/s00253-012-4007-2. [DOI] [PubMed] [Google Scholar]
- 115.Diaz E, Ferrández A, Prieto MA, García JL. Biodegradation of aromatic compounds by Escherichia coli . Microbiology and Molecular Biology Review. 2001;65:523–569. doi: 10.1128/MMBR.65.4.523-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xun L, Webster CM. A monooxygenase catalyzes sequential dechlorinations of 2, 4, 6-trichlorophenol by oxidative and hydrolytic reactions. The Journal of Biological Chemistry. 2004;279(8):6696–6700. doi: 10.1074/jbc.M312072200. [DOI] [PubMed] [Google Scholar]
- 117.Sindelar RD, Rosazza JP, Barfknecht CF. N-demethylation of nicotine and reduction of nicotine-1’-N-oxide by Microsporum gypseum . Applied and Environmental Microbiology. 1979;38(5):836–839. doi: 10.1128/aem.38.5.836-839.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ghisalba O, Meyar HP, Wohlgemuth R. Industrial biotransformation. In: Flickinger MC, editor. Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology. Wiley; 2010. pp. 1–18. [Google Scholar]
- 119.Wang SN, Xu P, Tang HZ, Meng J, Liu XL, Ma CQ. ‘Green’ route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environmental Science and Technology. 2005;39(17):6877–6880. doi: 10.1021/es0500759. [DOI] [PubMed] [Google Scholar]
- 120.Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409(6817):258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 121.Spande TF, Garraffo HM, Edwards MW, Yeh HJC, Pannell L, Daly JW. Epibatidine: a novel (chloropyridyl)azabicycloheptane with potent analgesic activity from an ecuadoran poison frog. Journal of the American Chemical Society. 1992;114(9):3475–3478. [Google Scholar]
- 122.Roduit J, Wellig A, Kiener A. Renewable functionalized pyridines derived from microbial metabolites of the alkaloid (5)-nicotine. Heterocycles. 1997;45(9):1687–1702. [Google Scholar]
- 123.Rathbone DA, Bruce NC. Microbial transformation of alkaloids. Current Opinion in Microbiology. 2002;5(3):274–281. doi: 10.1016/s1369-5274(02)00317-x. [DOI] [PubMed] [Google Scholar]


