Summary
An open chromatin largely devoid of heterochromatin is a hallmark of stem cells, from Planarians to Mammals. It remains unknown whether an open chromatin is necessary for the differentiation potential of stem cells, and what are the molecules that maintain open chromatin in stem cells. Here we show that the chromatin remodeling factor Chd1 is required to maintain the open chromatin state of pluripotent mouse Embryonic Stem (ES) cells. Chd1 is a euchromatin protein that associates with the promoters of active genes, and down-regulation of Chd1 leads to accumulation of heterochromatin in ES cells. Chd1-deficient ES cells are no longer pluripotent, because they are incapable of giving rise to primitive endoderm and have a high propensity for neural differentiation. Furthermore, Chd1 is required for efficient reprogramming of fibroblasts to the pluripotent stem cell state. Our results indicate that Chd1 is essential for open chromatin and pluripotency of ES cells, and for somatic cell reprogramming to the pluripotent state. The data suggest that pluripotent stem cells exist in a dynamic state of opposing epigenetic influences of euchromatin and heterochromatin.
Keywords: ES cells, Chd1, open chromatin, pluripotency, H3K4me3, H3K9me3, heterochromatin, reprogramming
The genome of eukaryotic cells is organized into accessible euchromatin that is permissive for gene activation and packaged heterochromatin that is largely silenced. Different cellular states may be defined at least in part by differential allocation of genomic regions to specific chromatin domains1. Several types of stem cells in organisms ranging from Planarians2 to Mammals3,4 have been reported to have an open chromatin largely devoid of heterochromatin. This phenomenon has been analyzed in greater detail in pluripotent mouse Embryonic Stem (ES) cells. ES cells have an open, “loose” chromatin with high rates of histone protein exchange, and accumulate regions of more rigid heterochromatin upon differentiation5,6. An open chromatin correlates with a globally permissive transcriptional state, and has been proposed to contribute to the developmental plasticity, or pluripotency, of ES cells6. While there is a strong correlation between open chromatin and the undifferentiated state of stem cells, it remains unknown whether open chromatin is necessary for stem cell potential. Furthermore, little is known about the molecules that may regulate open chromatin in stem cells. Here we report the identification of the chromatin remodeler Chd1 as an essential regulator of open chromatin and pluripotency of ES cells, and of somatic cell reprogramming to pluripotency.
Chd1 regulates ES cell self-renewal
We have recently characterized the transcriptional profiles of pluripotent stem cells, including ES cells, and the cells in the mouse embryo from which they are derived7 (and G. Wei, R.-F. Yeh, M. Hebrok and M. R.-S., unpublished results). These studies led to the identification of chromatin remodelers and transcription factors up-regulated in pluripotent cells. In order to test the role of 41 candidate factors in the regulation of pluripotency, we carried out an RNAi screen in ES cells (Supplementary Fig. 1). ES cells expressing GFP under the control of the Oct4 promoter (Oct4-GiP) were infected with a short hairpin RNA (shRNA)-expressing lentiviral vector pSicoR-mCherry8. Using 2–5 shRNAs per candidate target gene, we identified 18 genes that when down-regulated lead to defects in expansion of ES cells and 7 that lead to lower activity of the Oct4 promoter. Chd1 was the only gene with phenotypes in both assays that had not been previously implicated in ES cells (Supplementary Fig. 1).
Chd1 is a chromatin-remodeling enzyme that belongs to the chromodomain family of proteins and contains an ATPase SNF2-like helicase domain9. The two chromodomains in Chd1 are essential to recognize H3K4me2/310, and Chd1 has been implicated in transcriptional activation in yeast11, Drosophila12 and mammalian cells13. Recent transcription factor location studies indicate that the Chd1 gene is bound in mouse ES cells by Oct4, Sox2, Nanog, Smad1, Zfx and E2f1, suggesting that it is a target of multiple regulators of pluripotency and self-renewal14.
RNAi against Chd1 in Oct4-GiP ES cells, using two independent shRNAs targeting different regions of the mRNA, led to a decrease in expansion of ES cells and to lower GFP levels (Fig. 1a, b and Supplementary Fig. 2). Control cells were infected with empty pSicoR-mCherry lentiviral vector or with shRNA targeting GFP (empty and GFPi, respectively), and behaved like uninfected cells (Supplementary Fig. 1d, 2a). Down-regulation of Chd1 mRNA upon RNAi was confirmed by qRT-PCR (Fig. 1c). Endogenous Oct4 down-regulation was confirmed in Chd1-deficient (Chd1i) ES cells (Supplementary Fig. 3a). Oct4 down-regulation in ES cells induces differentiation into the trophectoderm lineage15. Interestingly, knock-down of Chd1 does not induce trophectoderm markers (Cdx2 and Eomes), indicating that the Chd1i phenotype is not simply one of trophectoderm differentiation due to loss of Oct4 (Supplementary Fig. 3b, c, and see below).
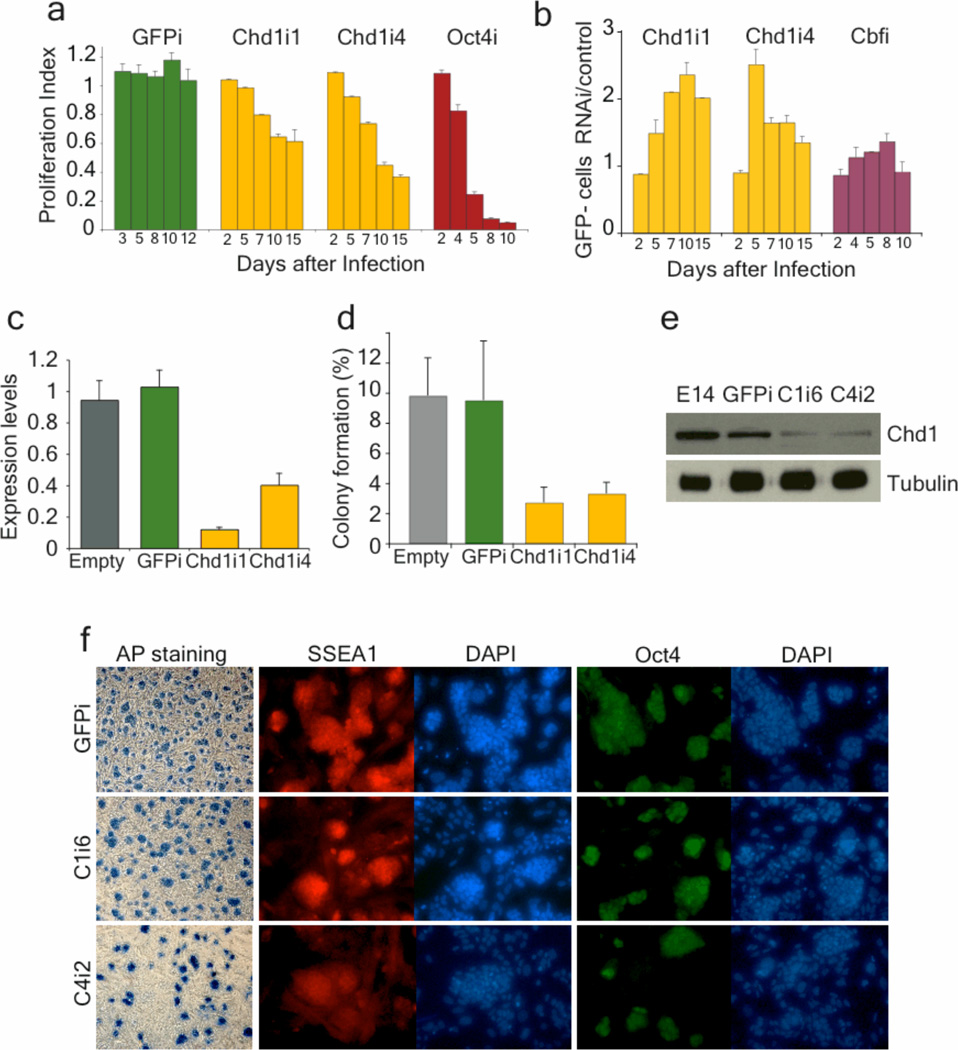
Figure 1. Chd1i ES cells have decreased self-renewal but maintain expression of markers of the undifferentiated state.
a, A competition assay shows that down-regulation of Chd1 in Oct4-GiP ES cells using two independent shRNA (Chd1i1 and Chd1i4) leads to a decreased proliferative capacity. The proliferation index represents the population of mCherry+ cells (which are undergoing RNAi) relative to cells infected with empty vector, at each timepoint. Experiments were done in triplicate and are represented as mean ± s.d. (n=3). b, Down-regulation of Chd1 leads to reduced activity of the Oct4-GFP reporter, as measured by the ratio between percentage of GFP-negative cells in RNAi and in empty vector. Chd1i cells have a 2-fold reduction of Oct4-GFP relative to controls for at least one passage. Down-regulation of a different gene that affects only proliferation (Cbf) has little effect on Oct4-GFP expression. c, Chd1 down-regulation upon RNAi was confirmed by qRT-PCR on cells isolated by FACS for mCherry +. The values are represented as mean of absolute expression ± s.d. (n=3). d, mCherry+ E14 cells isolated by FACS were plated at low density for colony-formation assays. The efficiency of colony formation in Chd1i ES cells was decreased relative to controls (empty and GFPi). The values are represented as mean ± s.d. (n=3), and are representative of two independent experiments. No colonies were recovered when Oct4 was down-regulated. e, Immunoblot with whole cell extracts from 2 control cell lines (parental E14 and GFPi cells) and 2 Chd1i ES cell clones (C1i6 and C4i2), using antibodies against Chd1 or alpha-Tubulin (as a loading control). Chd1 protein is down-regulated upon RNAi. f, Chd1i cells still express markers of undifferentiated ES cells, such as alkaline phosphatase (shown in bright field), and SSEA1 and Oct4 (shown with immunofluorescence).
Chd1 down-regulation decreased clonogenic potential in two independent ES cell lines (Oct4-GiP and E14), but Chd1i cells were still able to form ES-like colonies (Fig. 1d), unlike Oct4i ES cells. ES cell clones constitutively expressing either of the two shRNAs against Chd1 were established, and sustained Chd1 down-regulation was verified by qRT-PCR (see below Supplementary Fig. 5) and western blot (Fig. 1e). Control lines were also established using empty and GFPi viruses. As described below, the two shRNAs targeting Chd1 lead to identical phenotypes in marker gene expression, transcriptional profile, differentiation potential and chromatin state, relative to controls. Results were validated in two independent ES cell lines, Oct4-GiP and E14. The data below are from analyses in standard E14 ES cells not expressing GFP. Chd1i ES cells, even though they have a self-renewal defect, form compact colonies and express markers of ES cells, such as SSEA1, Alkaline Phosphatase and Oct4 (Fig. 1f), indicating that they maintain at least some aspects of the undifferentiated state.
Chd1 is required for ES cell pluripotency
To gain insight into the state of Chd1i ES cells, we determined their global gene expression profiles. Affymetrix mouse Gene 1.0 ST microarray experiments were performed using the parental E14 cells, two control cell lines (empty and GFPi), and four Chd1i ES cells (three clones from the shRNA Chd1i1 – C1i5, C1i6, Ci19; and one clone from the shRNA Chd1i4 – C4i2; Fig. 2a). Hierarchical clustering revealed that the transcriptional profiles of the four Chd1i ES cell lines cluster together and separately from the controls (Fig. 2a). We anticipated that we would find a pattern of down-regulated genes in Chd1i cells, because Chd1 is known to be associated with active transcription13. As expected, both Chd1 and Oct4 were detected as down-regulated in Chd1i ES cells. Surprisingly, however, very few other genes were significantly down-regulated (only 25 genes were down-regulated by more than 2-fold and none by more than 3-fold at 90% confidence, Fig. 2b and Supplementary Data 1). These data indicate that, at least with the low levels of Chd1 still present in Chd1i ES cells, there is a global maintenance of the ES cell transcriptome. On the other hand, a larger group of genes was up-regulated in Chd1i ES cells (Fig. 2b and Supplementary Data 1). A Gene Ontology (GO) analysis of the list of up-regulated transcripts revealed a significant enrichment for genes involved in neurogenesis (Fig. 2c), such as Nestin and Blbp. The expression of neural markers in Chd1i ES cells was confirmed by qRT-PCR (Supplementary Fig. 4).
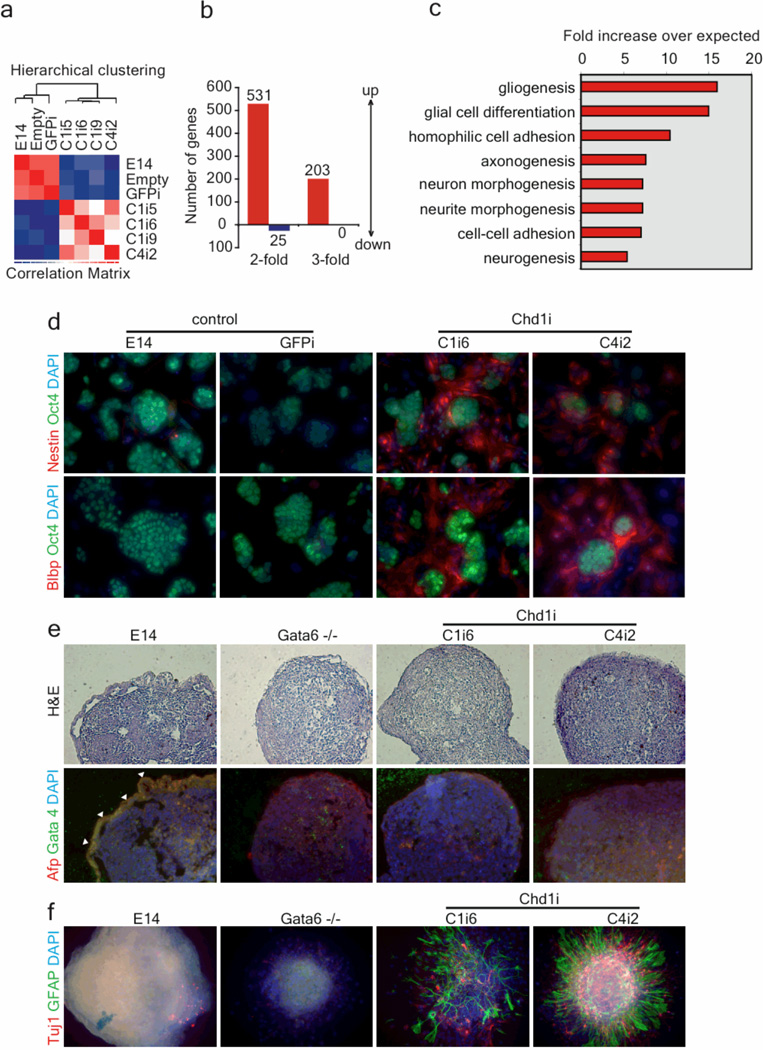
Figure 2. Chd1 is required for ES cell pluripotency.
a, Microarray analysis of 4 Chd1i clones, three from Chd1i1 (C1i5, C1i6 and C1i9) and one from Chd1i4 (C4i2), and 3 control cell lines, E14, empty virus-infected, and GFPi. The transcriptional profiles of Chd1i clones cluster together and are distinct from the controls. b, Few genes are downregulated in Chd1i ES cells relative to controls, but a significantly larger subset of genes are up-regulated. c, The subset of up-regulated genes is enriched for genes with roles in neurogenesis, as determined by Gene Ontology term analysis. All terms shown have p-values adjusted for multiple testing ≤0.01. d, Immunofluorescence analysis of Chd1i cells shows expression of both Nestin and Blbp, but in a population not expressing the ES cell marker Oct4. e, ES cells were cultured in non-attachment conditions and without LIF to form embryoid bodies (EBs). Lack of primitive endoderm development in Chd1i cells was observed by staining paraffin sections of 6 day EBs for Afp and Gata4, and hematoxilineosin. The loss of primitive endoderm layer (highlighted in control GFPi with white arrows) in Chd1i EBs is similar to that observed in EBs mutant for Gata6. f, A significant increase in neural differentiation is observed, as detected by staining EBs (plated on matrigel) for astrocytes (GFAP) and neurons (Tuj1), in 12 day Chd1i EBs, relative to controls.
The maintenance of the ES cell transcriptome and the unexpected expression of neural markers were further analyzed by immunofluorescence for Nestin, Blbp and Oct4. Oct4 was detected in Chd1i colonies, but cells between the colonies stained strongly for Nestin and Blbp (Fig. 2d). No staining for Nestin or Blbp was detected in control ES cells. FACS using the ES cell marker SSEA1 revealed that although SSEA1+ Chd1i cells already express Blbp mRNA, its expression is dramatically induced in SSEA1− Chd1 cells, relative to control GFPi cells (Supplementary Fig. 5). These data also revealed a very pronounced down-regulation of Oct4 in SSEA1− Chd1i cells. In summary, Chd1i ES cells can be propagated with many of the hallmarks of the undifferentiated state, but have a high propensity for neuronal differentiation.
We next tested the differentiation potential of Chd1i ES cells in vitro by formation of Embryoid Bodies (EBs). Chd1i EBs did not form the typical outer layer of primitive endoderm, as marked by immunofluorescence of EB sections with Afp and Gata4 (Fig. 2e). Similarly, yolk sac endoderm cysts were reduced or not observed in Chd1i EBs (Supplementary Fig. 5a), which showed down-regulation of primitive endoderm markers (Gata4, Afp, Hnf4 and Lamb) by qRT-PCR (Supplementary Fig. 5b). The loss of primitive endoderm in Chd1i EBs was comparable to that in EBs lacking an essential regulator of primitive endoderm, Gata616 (Fig. 2e). Beating foci, indicative of cardiac mesoderm differentiation, were not detected in Chd1i EBs, whereas they could be readily quantified in control EBs (Supplementary Fig. 5c). However, beating foci were also not observed in Gata6−/− EBs, indicating that the loss of cardiac mesoderm differentiation in Chd1i EBs may be secondary to the loss of primitive endoderm. Immunostaining of Chd1i EBs plated on matrigel showed a dramatic increase of neurons (stained with Tuj1) and astrocytes (stained with GFAP) relative to controls (Fig. 2f). This increase in neural differentiation is not secondary to the loss of primitive endoderm, because Gata6−/− EBs did not show such phenotype (Fig. 2f). In addition, Chd1i ES cells gave rise to teratomas with abundant neuronal differentiation when compared to wild-type ES cells (Supplementary Fig. 6). These results indicate that down-regulation of Chd1 leads to loss of primitive endoderm, with consequential loss of cardiac mesoderm differentiation, and abnormally high levels of neural differentiation that derives from a propensity already detected in the undifferentiated state.
Chd1 is a euchromatin protein in ES cells
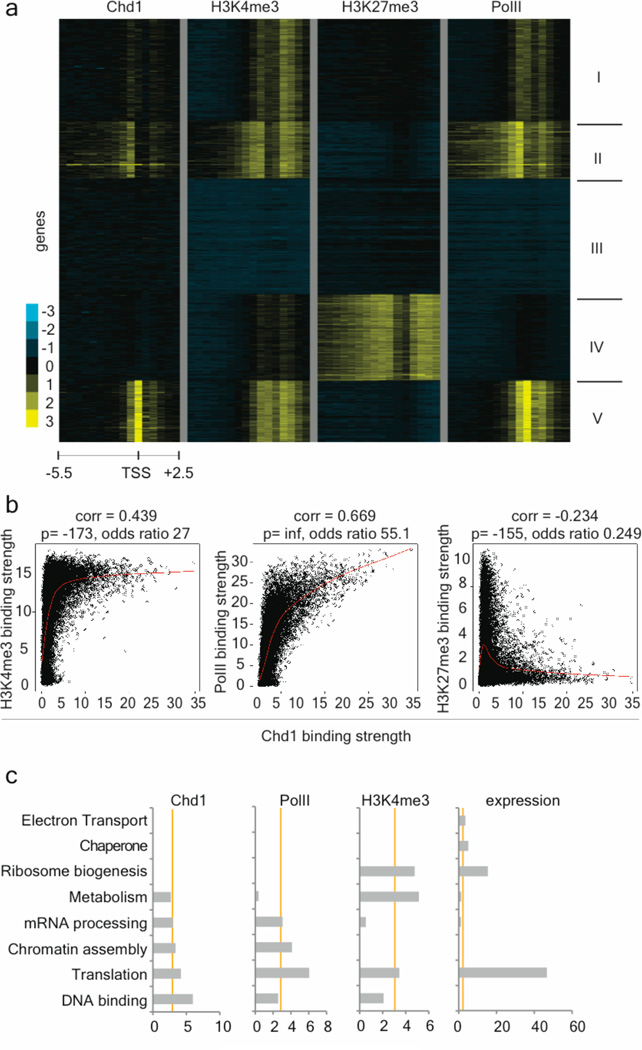
We then sought to understand the potential changes in the chromatin state of Chd1i ES cells that may underlie their differentiation defects. Previous studies11,12,13 indicated that Chd1 associates with euchromatin by binding to H3K4me3, although genome-wide location studies had not been performed. We carried out chromatin immunoprecipitation (ChIP)-chip for Chd1 in wild-type ES cells, and compared the genome-wide location of Chd1 to that of H3K4me3, RNA Polymerase II (PolII) and H3K27me3. These data revealed that Chd1 binding strongly correlates with that of PolII and H3K4me3 (Fig. 3 and Supplementary Fig. 8). Bivalent domains, simultaneously enriched for both the activating H3K4me3 mark and repressive H3K27me3 mark17, are largely devoid of Chd1 (Fig. 3a and Supplementary Fig. 8). Interestingly, GO analysis indicated that the strongest Chd1 and PolII targets are enriched for roles in DNA binding, translation and chromatin assembly genes, and that this enrichment is not strictly correlated with expression levels (Fig. 3c and Supplementary Data 2). Chd1 binding also correlates with H3K4me3 enrichment upon differentiation: during EB formation, the levels of both H3K4me3 and Chd1 are decreased at the Oct4 promoter and increased at the endodermal regulator Gata4 promoter (Supplementary Fig. 9). These data indicate that Chd1 associates globally with euchromatin in ES cells, and may preferentially target genes with roles in chromatin organization and transcription.
Figure 3. Chd1 associates with euchromatic promoter regions in ES cells.
a, Chd1 binding correlates with binding of H3K4me3 and RNA Polymerase II (PolII) but is excluded from bivalent domains in ES cells. K-means clustering of Chd1, H3K4me3, H3K27me3 and RNA PolII binding in ES cells. Each row represents the binding pattern along the −5.5kb to +2.5kb promoter region relative to the transcription start site (TSS), reiterated four times to present the data for each immunoprecipitation. The 8 kb promoter region is divided into sixteen 500bp fragments that display the average log ratio of probe signal intensity with blue, yellow, and grey representing lower-than-average, higher-than-average, and missing values for enrichment due to lack of probes in those regions, respectively. Note lack of Chd1 binding in cluster IV which consists of genes with bivalent domains. b, Genes strongly bound by Chd1 are characterized by high enrichment of H3K4me3 and RNA PolII and lack H3K27me3. Binary correlation of Chd1 binding strength with that of H3K4me3 (left panel), RNA PolII (middle panel) and H3K27me3 (right panel) for each gene (black dot) present on the promoter array. The Pearson correlation value, the –log 10 of the p value of the correlation as determined by Fisher’s exact test and the odds ratio are presented above each plot. Lowess normalization was used to generate the smoother indicated by the red line, revealing the anti-correlation of Chd1 binding with H3K27me3 and positive correlation with RNA PolII and H3K4me3. c, Functional categorization of Chd1 targets. Gene Ontology (GO) terms associated with the 200 genes most strongly bound by Chd1 or RNA PolII, or enriched for H3K4me3, as well as the top 200 genes in expression level in ES cells. Categories above an enrichment score of 3 (X-axes) are considered significantly enriched. 69 genes overlap between the Chd1 and RNA PolII top 200 gene lists, versus 27 for Chd1/expression and 13 for Chd1/H3K4me3.
Chd1 is required for maintenance of open chromatin
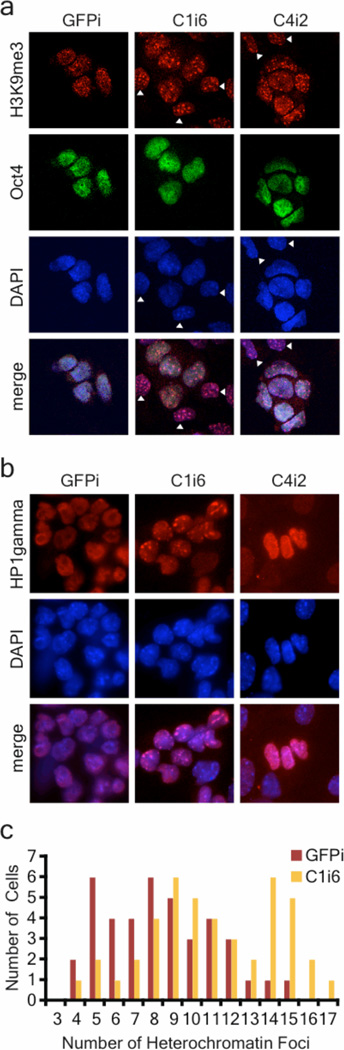
To investigate the effects of Chd1 down-regulation on ES cell chromatin, we performed immunofluorescence for histone marks of euchromatin and heterochromatin. Surprisingly, foci of heterochromatin marks such as H3K9me3 and HP1gamma, which normally appear as dispersed foci in ES cells5, were dramatically increased in Chd1i ES cells (Fig. 4a,b). No obvious differences were observed in staining for H3K4me3 or H3K27me3 between Chd1i ES cells and controls (data not shown). As described above, Chd1i ES cells are prone to spontaneous neural differentiation, and it has been shown that ES cell-derived neural precursors accumulate heterochromatin5. It was therefore important to evaluate whether the accumulation of heterochromatin is a consequence of commitment to the neural lineage, or whether it is present in ES-like cells prior to differentiation. Co-staining with H3K9me3 and Oct4 revealed that Oct4-positive ES-like cells, located in the center of compact colonies that stain for other markers of the undifferentiated state (Fig. 1e), have accumulated high levels of heterochromatin in Chd1i cells (Fig. 4a, quantified in 4c). Moreover, we analyzed the global chromatin dynamics of the Chd1i cells, through a Fluorescence Recovery After Photobleaching (FRAP) assay of a GFP-tagged version of histone H1. H1 is a linker protein involved in condensing nucleosomes that has been shown to rapidly exchange in the hyperdynamic chromatin of undifferentiated ES cells5. H1 showed a significant decrease in recovery in heterochromatin of Chd1i ES cells, indicating that the rapid exchange of H1 is compromised (Supplementary Fig. 10). These results indicate that, despite a global maintenance of the transcriptome, morphology, and marker gene expression of ES cells, Chd1i ES-like cells are not fully undifferentiated: their chromatin is condensed.
Figure 4. Chd1 is required to maintain open chromatin in ES cells.
a, Analysis of H3K9me3 staining by immunofluorescence. Co-staining for H3K9me3 and Oct4 distinguishes between ES-like cells (Oct4-positive) and differentiating cells (Oct4-negative, white arrow). Oct4-positive Chd1i ES-like cells have increased heterochromatin foci. b, Immunofluorescence of Chd1i cells for the heterochromatin mark HP1gamma shows accumulation in heterochromatin foci, whereas localization in the GFPi cells is diffuse throughout the nucleoplasm. c, Quantification of the increase of heterochromatin foci per nucleus in Chd1i ES-like cells, as seen by H3K9me3 staining in Oct4-positive cells, p<0.0005.
Heterochromatin formation is induced by methylation of H3K9 by the enzymes ESET, Suv39H1/2, G9a or Glp18, and reversed by the action of H3K9 demethylases such as Jmjd1a and Jmjd2c. Jmjd1a/2c have been shown to regulate genes expressed in ES cells and to repress differentiation19. All of these H3K9 methyl transferases and demethylases are expressed in Chd1i ES cells at levels similar to control ES cells (Supplementary File 1). Therefore, the accumulation of heterochromatin in Chd1i ES cells is not likely to be due to the differential expression of known H3K9 methyl transferases or demethylases. These results indicate that the capacity to induce heterochromatin formation exists in undifferentiated ES cells, despite the presence of H3K9 demethylases, and that heterochromatinization is countered by Chd1. Our data suggest that ES cells exist in a dynamic state of opposing epigenetic influences of euchromatin and heterochromatin, and that the euchromatin protein Chd1 is required to maintain the heterochromatin-poor pluripotent stem cell state.
Chd1 is required for efficient induction of pluripotency
Given the role of Chd1 in maintaining pluripotency of ES cells, we hypothesized that it may also play a role in re-acquisition of pluripotency during somatic cell reprogramming. Somatic cells can be reprogrammed to become induced Pluripotent Stem (iPS) cells by over-expression of four transcription factors (Oct4, Sox2, Klf4 and cMyc) and drug selection for the expression of an ES cell marker20,21,22,23. We have shown that drug selection is not required for reprogramming, and that nMyc can substitute for cMyc24. We used this assay to analyze the effect of Chd1 down-regulation (Fig. 5a) in reprogramming of Oct4-GFP Mouse Embryonic Fibroblasts (MEFs). Down-regulation of Chd1, using two independent shRNAs in three separate experiments, led to a significant reduction in the number of iPS cell colonies, scored both by morphology and GFP expression (Fig. 5b). This was not due to a delay in colony formation, as colony counts at later time points showed the same relative reduction in reprogramming efficiency upon Chd1 RNAi (data not shown). The iPS cell colonies that did form in Chd1 RNAi wells had either not been infected by the RNAi virus or had silenced it, as assessed both by Chd1 qRT-PCR and mCherry fluorescence (Supplementary Fig.s 11 and 12). Down-regulation of Chd1 could potentially affect proliferation of MEFs, which would confound the calculation of reprogramming efficiency. However, no significant changes in MEF growth rates were found between control and Chd1 RNAi (Fig. 5d). In addition, Chd1 RNAi does not affect the expression level of the exogenous reprogramming factors (data not shown). In summary, our data show that down-regulation of Chd1 does not affect the expansion of fibroblasts but inhibits their reprogramming by induction of pluripotency.
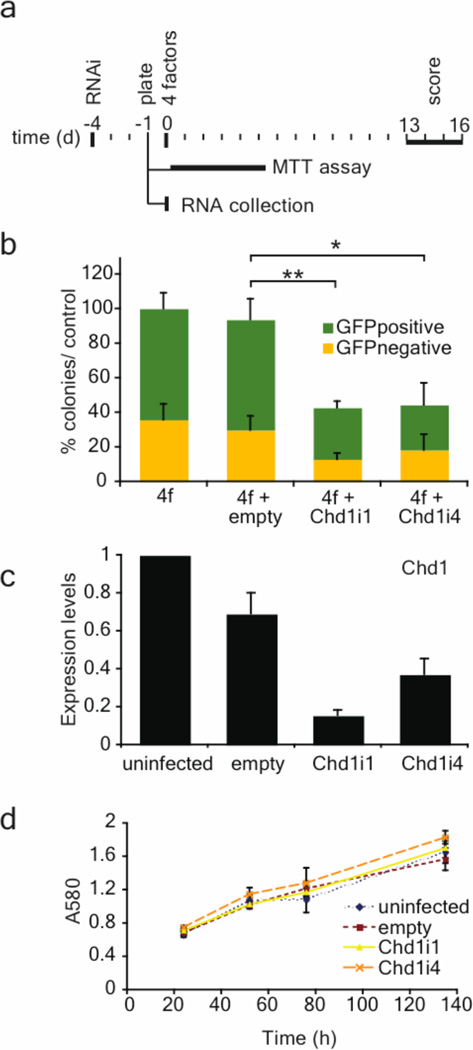
Figure 5. Chd1 is required for efficient induction of pluripotency.
a, Oct4-GFP MEFs were reprogrammed using lentiviral infection (day 0) of four transcription factors (Oct4, Sox2, nMyc and Klf4). RNAi lentiviral vectors (empty, Chd1i1, Chd1i4) were used to infect MEFs 4 days before the addition of the 4 factors. At day −1, MEFs were also plated for a MTT assay to quantify growth rates and for RNA collection for qRT-PCR for Chd1. b, The percentage of reprogrammed colonies was scored both by morphology and GFP expression, and normalized to the total number of colonies obtained in the control (4 factors only). The values are represented as mean ± s.d. of the averages of three independent experiments, each one done in duplicates or triplicates. The total number of colonies in control wells (4f or 4f+empty) in the three separate experiments varied between 200 and 500 per 6-well. The efficiency of induction of pluripotency is significantly reduced upon Chd1 RNAi. Unpaired t-test was performed using the total number of colonies obtained, comparing the control (empty) with RNAi against Chd1 (Chd1i1 and Chd1i4). * represents p<0.0001, ** represents p<0.00001. c, Chd1 down-regulation upon RNAi was confirmed by qRT-PCR. The values are represented as mean of absolute expression ± s.d. (n=3). d, Mean growth rate of MEFs measured by the MTT assay ± s.d. (n=6). No differences in MEF growth rate were observed upon Chd1 RNAi.
Discussion
We show here that Chd1 is required for open chromatin and pluripotency of ES cells. ES cells have been reported to be “poised” for differentiation by the presence of bivalent domains (marked by H3K4me3 and H3K27me3) in developmental regulatory genes17. We speculate that the opposing influences of euchromatin and heterochromatin (marked by H3K9me3) may be an additional mechanism for maintaining ES cells in a state “poised” for differentiation. Chd1 is also highly expressed in human ES cells relative to differentiated cells (Supplementary Fig. 13), suggesting that its role in pluripotent stem cells may be conserved. Interestingly, other stem cells may maintain their differentiation potential using a similar mechanism, since Chd1 has also been identified as a gene up-regulated in adult hematopoietic and neural stem/progenitor cells25. In addition, our data show that Chd1 is required for efficient generation of iPS cells. Fibroblasts have much higher levels of heterochromatin than pluripotent stem cells5 (and data not shown), and therefore a global opening of the chromatin is expected to be a component of reprogramming. Our data suggest that Chd1 may contribute to opening the chromatin and enabling transcription factor-mediated reprogramming to occur, although the precise mechanisms remain to be determined. Ongoing efforts to improve the efficiency of induction of pluripotency, which is currently still very low, should in the future allow an analysis of chromatin dynamics at single cell level during reprogramming.
How does Chd1, a protein associated with euchromatin, act to counter heterochromatinization? Recent genetic studies in yeast indicate that euchromatin-associated factors prevent spreading of heterochromatin to euchromatic regions26,27. Chd1 may have a similar role in ES cells, acting by at least two possible mechanisms. Chd1 binding to H3K4me2/3 may prevent methylation of H3K9 and/or formation of higher order heterochromatin fibers. Alternatively, the SNF2-like helicase domain of Chd1 may mediate incorporation of the histone variant H3.3, which is generally associated with active genes and is less prone to H3K9 methylation28. In support of this model, Chd1 has recently been shown to be required in the Drosophila oocyte for incorporation of H3.3 into sperm chromatin, a step necessary for decondensation and development29. Further studies should focus on distinguishing between these models. In addition, an analysis of the genomic locations of heterochromatin in Chd1i ES cells may provide insight into the differential sensitivity of endoderm and neural lineages to heterochromatinization.
Methods
ES cell culture and differentiation
Mouse E14 and Oct4-GiP ES cells30 were plated on 0.1% gelatin-coated plates or on a feeder layer of irradiated mouse embryonic fibroblasts (MEFs), and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 15% knockout serum replacement (Invitrogen), 1mM L-glutamine, 0.1 mM nonessential amino acids, 100µg/mL Penicillin, 100 µg/mL Streptomycin, 1 mM Sodium Pyruvate, 0.1 mM 2-mercaptoethanol and recombinant LIF. Mouse Gata6−/− ES cells16 were grown in identical conditions except that fetal bovine serum was used instead of knockout serum replacement. Embryoid bodies were formed by suspension culture in ES cell medium with fetal bovine serum and in the absence of LIF. Contractile foci were counted under an inverted microscope using triplicates of 10cm dishes per ES cell clone. Data represent averages with standard error bars.
RNAi and competition assay
The genes tested in the RNAi screen were the following: Ap2gamma, Brca1, Cbf, Chd1, Ddx18, Dmrt1, Dppa2, Dppa3 (Stella), Dppa4, Eed, Foxd3, Hells, Mybl2, c-Myc, Mycbp, n-Myc1, Nanog, NFYa, NFYb, Nr0b1, Nr5a2, Pou5f1 (Oct4), Pramel4, Pramel5, Rex1, Rex2, Rbm35a, Sall4, Six4, Sox2, Suz12, Tcfcp2l1, Terf1, Tex292, Utf1, Zic3, A030007L17Rik, E430003D02Rik, and Affymetrix MG_U74Av2 probe sets 135189_f_at, 97154_f_at and 98524_f_at. shRNA sequences were selected according to published criteria31: GFPi- ACAGCCACAACGTCTATAT; Oct4i-GAACCTGGCTAAGCTTCCA; Chd1i1-ACATTATGATGGAGCTAAA; Chd1i4-GTGCTACTACAACCATTTA. All other sequences are available upon request. Oligos coding for the shRNAs were designed and cloned into the lentiviral vector pSicoR-mCherry as described8. pSicoR-mCherry has been described previously7. Lentiviruses were produced as described8. For transduction, 106 ES cells were incubated with virus in 1 mL of ES cell medium (multiplicity of infection 5–10). After 1 hr rotating at 37°C, 2.5–3×105 cells were plated per gelatinized well of a 12-well plate. A competition assay32 was performed by analyzing cells that were passaged every two or three days. FACS analysis was performed on a LSRII and analyzed on Flojo software. Proliferation index was measured, for every passage, by dividing %mCherry+ (shRNA) with %mCherry+ (empty virus). Loss of Oct4-GFP activity was measured by dividing %GFP− cells (shRNA) with %GFP− (empty virus). The calculation of the loss of Oct4/GFP expression was done with total GFP− cells rather than just GFP−/mCherry+ to account for potential silencing of the mCherry construct upon differentiation or non-cell autonomous effects. Proliferation index data are averages of triplicates (n=3), with standard error bars. mCherry+ ES cells were isolated using a FACSDiVa (BD Biosciences) cell sorter.
Colony formation assay and clonal derivation
E14 and Oct4-GiP ES cells were infected with lentiviruses containing shRNAs or empty virus alone, as described above. mCherry+ cells were sorted on day 5 after infection using a FACSDiVa (BD Biosciences) cell sorter. 5000 cells were plated per 10cm dish in triplicates. After 10 days in culture, individual clones were picked per each condition (empty virus, GFPi, Chd1i1 or Chd1i4) and propagated in standard ES cell growth conditions. Plates were stained for Alkaline Phosphatase using a Vector kit and colonies were counted. Results are averages of triplicates, with standard error bars.
Expression microarrays
Uninfected parental E14 ES cells, one clone infected with empty pSicoR-mCherry, one GFPi clone and four Chd1i clones, three from Chd1i1 (C1i5, C1i6 and C1i9) and one from Chd1i4 (C4i2) were grown on gelatin in ES cell culture medium. Total RNA was isolated using the RNeasy kit (Qiagen) with in-column DNAse digestion. 300 ng of total RNA per sample were amplified and hybridized to Affymetrix Mouse Gene 1.0 ST arrays according to the manufacturer’s instructions at the Genomics Core Facility of the Gladstone Institutes. These arrays assay for the expression of about 35,500 transcripts. Data were normalized using Robust Multi-Array (RMA) normalization. Hierarchical clustering and calculations of differential gene expression were done using dChip (www.dchip.org)33. The full normalized data are in Supplementary File 1. The lower bound of the 90% confidence interval of the fold change (LCB) was used as a conservative estimate of the fold change. 531 transcripts with LCB>2 in Chd1i relative to controls were analyzed in MAPPFinder34 for enrichment of gene ontology terms. Terms with p-values adjusted for multiple testing ≤0.01 were considered enriched.
Immunohistochemistry
ES cells or EBs were plated on chamber glass slides pre-coated with matrigel. ES cells were plated on a layer of irradiated MEFs. After 2 days cells were fixed with 4% paraformaldehyde, permeabilized with PBT (PBS + 0.1% TritonX100) and blocked with 2% BSA + 1% goat or donkey serum in PBT. Slides were immunostained with primary antibody in blocking solution. Alternatively, EBs in suspension (at day 6) were fixed, paraffin-embedded, sectioned (50µm), and stained for hematoxylin and eosin or immunostained.
Primary antibodies used: SSEA1 (MC-480; DSHB; 1:200), Oct4 (sc5279; Santa Cruz; 1:100 / sc9081; Santa Cruz; 1:50), Nestin (MAB353; Chemicon; 1:200), BLBP (ab32423; Abcam; 1:200), Afp (sc8977; Santa Cruz; 1:200), Gata4 (sc1237; Santa Cruz; 1:50), Tuj1 (MMS-435P; Covance; 1:250), GFAP (Z0334; Dako; 1:500), H3K4me3 (ab8580; Abcam; 1:200), H3K9me3 (07-449; Upstate; 1:100 / ab8898; Abcam; 1:100), H3K27me3 (07-449; Upstate; 1:100), HP1gamma (MAB3450; Chemicon; 1:500). Secondary antibodies: Alexa Fluor 488/594 conjugated secondary antibodies (anti-mouse, anti-rabbit, or anti-goat, 1:500 dilution – Molecular Probes). Nuclei were counterstained with DAPI. The MC-480 antibody developed by Dr. Davon Solter was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by University of Iowa, Department of Biological Sciences, Iowa City, IA 52242
qRT-PCR
RNA was isolated according to the RNeasy kit (Qiagen), and reverse-transcribed using the iScript first strand cDNAsynthesis kit (BioRad). The cDNA reaction was diluted 1:5 in TE and used in Sybr Green real-time PCR reactions (BioRad). Housekeeping genes used were Ubiquitin-b and Ribosomal protein L7. PCR primer sequences are available upon request. Reactions were run in replicates on a MyiQ qPCR machine (BioRad) according to the manufacturer's instructions. Cycle threshold values were imported into the REST software35 for fold-change calculations, using the housekeeping genes as controls. Values are presented in Log2 scale or in absolute expression levels compared with parental E14 RNA unless indicated.
SSEA1 cell sorting
ES cells (GFPi control and Chd1i clones) were collected by trypsinization, washed in ice-cold PBS, first re-suspended in staining medium (HBSS, Ca/Mg Free, no phenol red, 2% FBS) with primary antibody mouse SSEA1 (MC-480; DSHB; 1:50) for 30min on ice, and then in secondary anti-mouse IgM-PE (406507; BioLegends; 1:100) for 30 min on ice. Propidium Iodide (P3566; Invitrogen) was added before live SSEA1+ and SSEA1-cells were isolated using a FACSDiVa (BD Biosciences) cell sorter.
Western blotting
Whole cell extracts were prepared and measured with Bradford assay (BioRad) for protein content. From ES whole cell extracts 30µg of protein were resolved on SDS-Page gel (10%) using a rabbit antibody against Chd1 (1:2000, from Robert Perry12) and a goat anti-rabbit HRP (1:10000). The loading control used was alpha-Tubulin, with a mouse antibody (1:1000, Sigma T9026). From EB extracts (at day 12), 10µg of protein were resolved on SDS-Page gel (10%) using antibody against Tuj1 (1:1000) and anti-goat HRP (1:10000). Detection was performed using the ECL according to manufacturer’s instructions (Amersham).
FRAP analysis
Transfection of H1-GFP into ES cells and FRAP analysis were performed as described5.
Generation of teratomas
Teratomas were produced by injecting 3×106 cells subcutaneously in the flanks of SCID mice. Tumor tissue samples developed in 12 weeks and were fixed overnight in 4% paraformaldehyde prior to paraffin embedding. Sections were stained with hematoxylin and eosin with standard protocol.
ChIP
Chromatin-Immunoprecipitation was performed essentially as described by Upstate Biotechnology, with some minor changes described below: chromatin was cross-linked by incubating cells on plates with PBS containing 20mM DMP dimethylpimelimidate, (SIGMA) and 0.25%DMSO for 1 hour at room temperature. Cells were re-fixed with 2% paraformaldehyde for another hour at room temperature, scraped and centrifuged at 1,350 × g for 5 minutes. Pellets were resuspended in SDS lysis buffer and sonicated to obtain fragments of ~200–1000 bp as verified on a gel. Reactions were centrifuged at 13,000 × g for 10 minutes and the supernatants were used. Antibodies used (3µg each): Chd1 (PAB-10569; Orbigen), H3K4me3 (ab8580; Abcam) IgG (ab46540; Abcam). DNA was purified by phenol-chloroform extraction, followed by ethanol precipitation. DNA concentration was determined using a Nanodrop spectrophotometer (NanoDrop Technologies) and 5/10ng were used in Sybr Green real-time PCR reactions (see above) ran in duplicates or triplicates. Primer sequences are available upon request. Fold enrichment over input was calculated using the 2ΔCt method corrected with IgG Ct values. The HoxA3 primer set was used as a control gene because it corresponds to a region known to lack H3K4me317.
ChIP-chip
Chromatin immunoprecipitation and hybridization onto Agilent promoter microarrays was performed as described22. Briefly 500ug of crosslinked ES chromatin was immunoprecipitated with 10ug of Chd1 antibody (Allele Biotech PAB-10568) or hypophosphorylated RNA polymerase II (8WG16). The elute was reverse crosslinked, RNase- and proteinase K treated and purified. Equal amounts of input and immunoprecipitated samples were amplified using WGA2 kit (Sigma), labeled with the Bioprime kit (Invitrogen), hybridized onto Agilent mouse promoter arrays (G4490) according to manufacturers instructions. Data were extracted using Agilent Feature extraction and Chip analytics software. Data was visualized using the Cluster 3.0 and Treeview programs. Bound genes were determined using the Young lab algorithm36 and H3K4me3 and H3K27me3 data were previously published22, as well as the algorithm to generate the 500bp window presentation. The odds ratio for binary correlation of Chd1 binding strength was calculated as the ratio of the probability of a gene being bound by Chd1 divided by the probability of it being bound by H3K4me3 (or RNA PolII or H3K27me3) to the probability of a gene being bound by Chd1 divided by the probability of the gene being unbound by H3K4me3 (or RNA PolII or H3K27me3).
Reprogramming
Reprogramming was performed as previously described24, with minor changes described in Figure 5a and Supp. Figure 12. iPS colonies were scored by GFP fluorescence, using a scale according to the number of cells in a colony that were GFP positive, as described in Supp. Figure 12b (GFPpositive refers to all the colonies with any GFP positive cells), or by their morphology under bright field (GFPnegative).
MTT assay
The growth rate of MEFs was measured using an indirect method. Yellow MTT is reduced by mitochondrial enzymes into a purple formazan and the absorbance measured as a result of the number of viable cells. For the MTT assay, MEFs were plated at 5,000 cells per well in a 96-well plate and analyzed 24, 52, 76 and 135 h after plating. At the indicated time points, ES cell medium was replaced with 100 ml 1 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2.5-diphenyltetrazolium bromide (MTT) (Molecular Probes) in DMEM. After incubation at 37°C for 3 h, the MTT solution was removed. 100 ml DMSO was added to dissolve precipitate for 10min at 37°C and 5min at room temperature. Absorbance was recorded at 540 nm using a Spectramax M2 microplate reader (Molecular Devices).
Supplementary Material
Acknowledgements
The authors wish to thank Austin Smith for Oct4-GiP ES cells, Michael S. Parmacek for Gata6−/− ES cells, Robert P. Perry and David G. Stokes for the Chd1 antibody, Marty Bigos and Valerie Stepps at the Flow Cytometry Core Facility and Linda Ta at the Genomics Core Facility of the Gladstone Institutes for expert assistance, Clara Chiu for technical assistance, members of the Santos lab, in particular Marica Grskovic, for advice, and Arnold Kriegstein, Doug Melton, Arturo Alvarez-Buylla, Didier Stainier, Robert Blelloch, Barbara Panning, Jeremy Reiter and Marica Grskovic for discussions and critical reading of the manuscript. A. G.-M. was the recipient of a predoctoral fellowship from the Foundation for Science and Technology (POCI2010/FSE), Portugal. This work was supported by grants from the Sandler Family to M.T.M., CIRM Young Investigator Award and NIH Director’s New Innovator Award to K.P., Israel Science Foundation (ISF 215/07), European Union (IRG-206872) and Alon Fellowship to E. M., and NIH Director’s New Innovator Award, California Institute for Regenerative Medicine and Juvenile Diabetes Research Foundation to M. R.-S.
Bibliography
- 1.Mikkelsen T, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008:16. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddien P, Sanchez-Alvarado A. Fundamentals of Planarian Regenaration. Annu. Rev. Cell. Dev. Biol. 2004;20(1):725. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 3.Terstappen LW, et al. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38- progenitor cells. Blood. 1991;77(6):1218. [PubMed] [Google Scholar]
- 4.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 5.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental Cell. 2006;10(1):105. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efroni S, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2(5):437. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grskovic M, et al. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3(8):e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101(28):10380. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodage T, et al. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94(21):11472. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims RJ, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280(51):41789. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simic R, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22(8):1846. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci USA. 1996;93(14):7137. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sims RJ, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular Cell. 2007;28(4):665. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 16.Morrisey E, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes & Development. 1998;12(22):3579. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein B, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Current Opinion in Cell Biology. 2002;14(3):286. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 19.Loh Y, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate selfrenewal in embryonic stem cells. Genes & Development. 2007;21(20):2545. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 22.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-celllike state. Nature. 2007;448(7151):318. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 24.Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1(3):245. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramalho-Santos M, et al. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298(5593):597. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 26.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32(3):370. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 27.Venkatasubrahmanyam S, et al. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci USA. 2007;104(42):16609. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. PNAS. 2004:6. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konev AY, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317(5841):1087. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods Bibliography
- 30.Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416(6880):545. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 32.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98(1):31. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doniger SW, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.