Preface
Candida species cause frequent infections due to their ability to form biofilms – surface-associated microbial communities – primarily on implanted medical devices. Increasingly, mechanistic studies have identified the gene products that participate directly in Candida albicans biofilm formation, as well as the regulatory circuitry and networks that control their expression and activity. These studies have revealed new mechanisms and signals that govern C. albicans biofilm formation and associated drug resistance, thus providing biological insight and therapeutic foresight.
Introduction
The medically relevant Candida species 1 are mainly commensal fungi that reside on mucosal surfaces and in the gastrointestinal (GI) and genitourinary (GU) tracts. Although these organisms are generally benign, they can cause infection if immune function is impaired or if an environmental niche becomes available 2. Many Candida infections arise as a result of the organism’s ability to grow as a biofilm on implanted medical devices 3–6. The use of these devices, such as venous catheters, urinary catheters, and artificial joints, is routine, with well over 10 million recipients per year 7. Device-associated Candida infections have mortality rates as high as 30% 7, 8, and the annual cost of antifungal therapies in the United States alone is estimated at $2.6 billion 9. Like the biofilms of bacterial pathogens, Candida biofilms are resistant to many antimicrobial agents, so treatment may require surgical removal and later replacement of the infected device 5, 7. Here, we review Candida biofilm formation with a focus on Candida albicans, the most frequently isolated Candida pathogen 10.
Overview of C. albicans Biofilm Formation
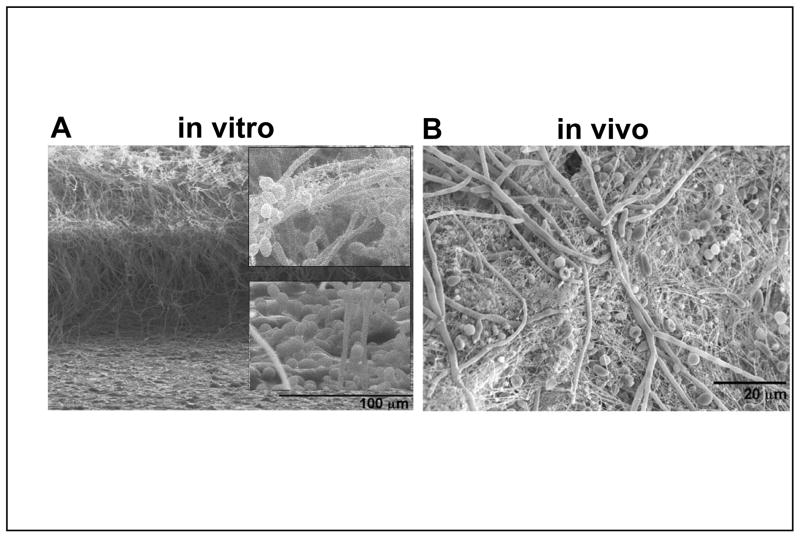
C. albicans biofilms comprise mainly two kinds of cells: small oval yeast-form cells (also called blastospores), and long tubular hyphal cells. Biofilms grown in vitro often have a foundation of yeast cells, from which a hyphal layer emanates (Fig. 1A) 5. Extracellular matrix material is clearly evident as well, bound to both yeast and hyphal cells. It is typically interspersed throughout the biofilm, though it is apparent primarily at the top of this sample. Biofilms from in vivo catheter infection models appear more complex, with yeast and hyphae interspersed (Fig. 1B) 11. Genetic analysis indicates that both yeast cells and hyphae are critical for biofilm formation, which suggests that each cell type has unique roles in the process 5.
Figure 1. C. albicans biofilm structure in vitro and in vivo.
A) Scanning electron micrograph (SEM) of an in vitro biofilm. The biofilm sample was sliced to reveal three layers in a cross-sectional view (J. S. Finkel, J. Suhan, and A. P. Mitchell, unpublished results). The basal layer includes primarily yeast cells, as evident in the lower enlarged inset. The central layer is mainly hyphae. The upper layer has yeast cells budding from the hyphae. The upper enlarged inset shows extracellular matrix material, which appears fibrous in this preparation. B) SEM of an in vivo biofilm from the rat catheter model 11. Yeast cells, hyphae, and some pseudohyphal cells are evident, along with extracellular matrix material. (This image was provided by J. Nett and D. Andes.)
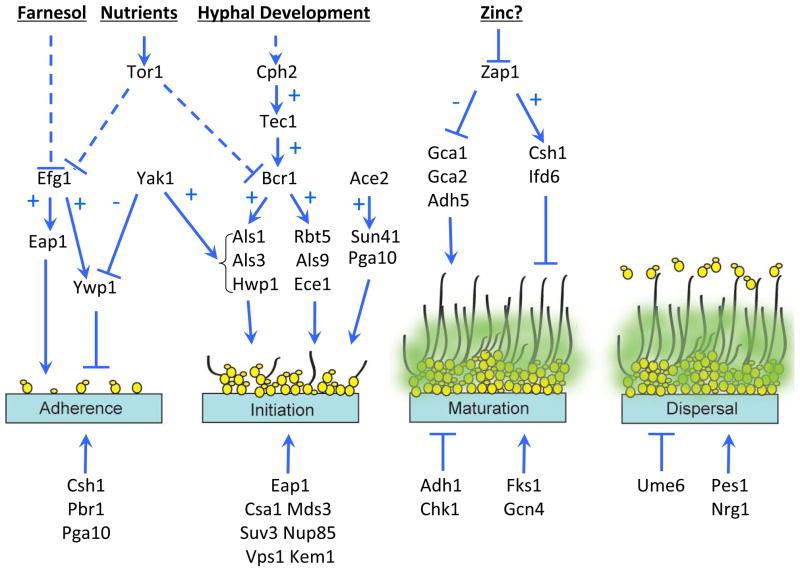
In vitro experiments allow C. albicans biofilm formation to be viewed as a series of sequential steps (Fig. 2) 5, 12. Biofilm formation begins with adherence of yeast cells to a substrate (adherence step) (Fig. 2). Soon afterwards, the yeast cells proliferate across the surface and produce elongated projections that grow into filamentous forms, including hyphae or and pseudohyphae (initiation step). Extracellular matrix accumulates as the biofilm matures, and high-level drug resistance is also acquired (maturation step). Finally, non-adherent yeast cells are released from the biofilm into the surrounding medium (dispersal step) (Fig. 2). While these steps may occur concurrently rather than sequentially during natural biofilm formation in vivo, they provide a useful framework to guide mechanistic analysis.
Figure 2. Gene function in biofilm formation.
In the adherence step, yeast-form cells adhere to the substrate. In the initiation step, the cells propagate to form microcolonies and germ tubes form to yield hyphae. In the maturation step, the biofilm biomass expands, the extracellular matrix accumulates and drug resistance increases. In the dispersal step, yeast-form cells are released to colonize the surrounding environment. The upper half of the diagram depicts several known pathway relationships. The bottom half includes additional genes that function in a specific step, but may not be connected to a known pathway. For simplicity, some known pathway relationships have been omitted. A few genes are presented more than once if they have roles in more then one step of biofilm formation. Arrows represent positive relationships; T-shaped bars represent negative relationships. A “+” indicates that the upstream gene/signal stimulates expression of the downstream target; a “−” indicates that the upstream gene/signal inhibits expression of the downstream target. Dashed lines indicate repression by an indirect mechanism.
Simple Inferences from Biofilm Genetics
Recent strides in expression profiling and genetic manipulation have driven our understanding of the regulatory pathways and mechanisms that govern biofilm formation and biofilm-based drug resistance. In addition, such analyses have pointed to an intriguing relationship between biofilm formation and mating. Of course, genes can have net positive or negative roles in biofilm formation, based on known mutant phenotypes. This distinction is useful in thinking about gene relationships because, for example, a negative gene product can act through inhibition of a positive gene product. On the other hand, it is worth bearing in mind that “dispersal” is an unraveling of the steps in biofilm formation. Therefore, for example, the negative function of Ywp1 in the adherence step that leads to biofilm formation might go hand in hand with a positive function in biofilm dispersal.
The genes that govern C. albicans biofilm formation (Table 1) fit into several broad functional categories. Many of these genes are required for production of hyphae (filamentation). Some of the first C. albicans biofilm genetic studies indicated that hyphae are required for stable biofilm formation 13, 14. In addition, several biofilm genes are involved in the response to the quorum-sensing molecule farnesol 15, 16. Farnesol is an inhibitor of filamentation 15, 16 and, as one would thus expect, inhibits biofilm formation in vitro 17. In fact, several additional quorum sensing molecules accumulate in mature biofilms (see Box 1), and addition of such molecules to biofilm cultures in vitro suggests that they may promote biofilm dispersal 18–20.
Table 1.
| Molecular function of gene producta | Role of gene product | Gene | Reference |
|---|---|---|---|
| Transcription Factor | Positive | ACE2c, BCR1, CPH1, CZF1c, EFG1c, FLO8c, GCN4, TEC1c, UME6c, NRG1c | 14, 27, 28, 90, 98, 99, 118–122 |
| Negative | ZAP1 | 29 | |
| Cell Wall Related Protein | Positive | ALS1, ALS2c, ALS3, ALS4, ALS5, ALS7, ALS9, CSA1, EAP1, FKS1, HWP1, HWP2, OCH1, PGA1, PGA10c, PMT1c, PMT2c, PMT4, PMT6, RBT1, RBT5, SUN41c | 22, 23, 28, 38, 56, 67, 71, 73–76, 97, 123–125 |
| Negative | YWP1 | 126 | |
| Alcohol dehydrogenase | Positive | ADH5 | 127 |
| Negative | ADH1, CSH1, IFD6 | 29, 30 | |
| Protein Kinase | Positive | CBK1c, GIN4c, IRE1c, MKC1, YAK1c | 64, 78, 128 |
| Negative | CHK1, TOR1, | 129, 130 | |
| Drug efflux pump | Positive | CDR1, CDR2, MDR1 | 89 |
| Gluco-amylase | Positive | GCA1, GCA2 | 29 |
| Otherb | Positive | CAT2, ECE1, KEM1c, MDS3c, NDH51, NUP85c, PBR1, PES1, PDX1, RIX7, SUV3c, VAM3c, VPS1c | 28, 68, 98, 131–136 |
Molecular functions have been inferred from protein sequence homology in most cases.
Other refers to gene product functions that does not fit into any of the above categories. For specific functions refer to supplemental table 1.
Indicates a regulator of filamentation
Box 1. Quorum sensing.
Quorum sensing phenomena are those in which microbial behaviors or responses are governed by cell density. Such community behaviors are generally determined by secreted signaling molecules, whose accumulation is a measure of cell density 100. Quorum sensing plays a pivotal role in biofilms of all kinds 101, 102. The most well studied quorum sensing molecule in C. albicans is E,E-farnesol, an inhibitor of hyphal formation. Exogenous farnesol inhibits biofilm formation if provided early, during the time of adherence 17, 18. The limited biofilms that form in the presence of farnesol comprise mainly yeast and pseudohyphal cells, rather than hyphae. Farnesol accumulates in supernatants of mature biofilms 20, where stimulation of yeast cell production may promote biofilm dispersal. Tyrosol, an alcohol derived from tyrosine, has an activity opposite that of farnesol: it stimulates hyphal formation. Exogenous tyrosol addition does not have a measurable effect on overall biofilm formation, but can partially overcome biofilm inhibition by exogenous farnesol 18. Tyrosol also accumulates in mature biofilm supernatants 18, 20, and the overall inhibition of hyphal formation by such supernatants 17, 18, 20 seems to reflect the dominant activity of farnesol 18. Several additional small molecules are detectable in biofilm supernatants, including phenylethyl alcohol, dodecanol, and nerolidol 20. Each isolated molecule can inhibit hyphal formation, and thus all may aid in biofilm dispersal by promoting yeast cell formation. It will be of interest to block synthesis of or response to individual molecules in order to assess their biological functions, and to test their roles in biofilms formed in vivo.
Several noteworthy classes of gene products govern biofilm properties. Many biofilm genes encode known or predicted cell wall proteins. These proteins are of special interest because they may play a direct role in cell-substrate or cell-cell adherence. Indeed, heterologous expression studies indicate that Eap1, Hwp1, Als1, and Als3 have such roles 21–23. Surface proteins are of further interest as accessible therapeutic targets. Finally, it has become increasingly evident that cell heterogeneity is a critical feature of biofilms 24. This attribute is obvious from the different cell types evident in C. albicans biofilms (Fig. 1), and fungal cell wall protein genes are subject to both genetic and epigenetic mechanisms that further contribute to cell heterogeneity 25, 26.
Many of the C. albicans genes involved in biofilm formation encode predicted transcription factors or protein kinases. These regulatory proteins must act indirectly to control biofilm properties, but can be informative indicators of the internal and external signals that influence biofilm development. For example, the transcription factor Bcr1 is required for biofilm formation and is upregulated in hyphae, thus suggesting that Bcr1-dependent gene products may be the hyphal components that are required for biofilm formation 27, 28. Similarly, the zinc-responsive transcription factor Zap1 is a regulator of extracellular matrix accumulation, suggesting that ambient zinc levels may alter matrix levels 29. The signals that influence the activity of many of the other biofilm regulators listed in Table 1 are not well understood, thus presenting an opportunity for future study.
Several alcohol dehydrogenase and aryl-alcohol dehydrogenase genes have an impact on biofilm formation. The fact that both positive and negative roles for these genes have been deduced may suggest that substrate specificity is critical for their biological function 29, 30. Although it is possible that their substrates and products act primarily through effects on intermediary metabolism, we note that the aryl alcohol dehydrogenases in particular have been implicated in synthesis of amino acid-derived alcohols that may function in quorum sensing 19, 31, 32.
We have assembled a model that connects steps in biofilm formation, biofilm genes, and regulatory pathways (Fig. 2). The regulatory relationships diagrammed are based on diverse lines of evidence, including mutant phenotypes, genetic epistasis tests, microarray analysis, gene overexpression phenotypes, and chromatin immunoprecipitation. Some pathway relationships are thus indirect or tentative. The model is intended as a framework for the identification of new areas of inquiry and for interpretation of future studies.
Perhaps the greatest utility of a model for gene function in biofilm development is the application to in vivo models for biofilm formation on implanted devices 11, 33–35. In vivo models are critically important because the nature of the device surface, the presence of host derived conditioning film, the levels of oxygen and carbon dioxide, and liquid flow all affect biofilm development 12, 36–38. It is thus impossible to duplicate all relevant in vivo conditions in vitro.
Diversity of Biofilm Environments and Cohabitants
Many studies have focused on C. albicans biofilm formation on implanted vascular catheters, a major source of infection 7. However, biofilm formation occurs on many other devices. A rat denture biofilm model has been described recently that recapitulates features of denture stomatitis 33. Microscopic and microbiological analysis revealed the signature features of biofilm formation – adherent cells, presence of extracellular matrix material, and high-level drug resistance. One other recently described rat model, in which biofilms form on catheters implanted subcutaneously, also has characteristic matrix material and an abundant hyphal population 34. The latter provides both ethical and technical advantages in that a single animal can be used to culture numerous biofilms.
Biofilms also form on tissue surfaces, such as infections of the oral and vaginal mucosa. In such infection models, C. albicans produces dense three-dimensional biofilms embedded in extracellular matrix material 39, 40. The level of biofilm drug resistance has not been tested directly, though Noverr and colleagues point out that drug resistance is seldom a clinical problem with vaginal candidiasis 40.
Is biofilm formation in these novel environments mechanistically distinct from that the more commonly studied in vitro and in vivo models? There are certainly some conserved features. For example, biofilm formation in all of the models described above depends upon transcription factor Bcr1 (Filler, personal communication, Dongari-Bagtzoglou, personal communication) 33, 34, 40. Where tested, filamentation-defective strains were biofilm-defective as well 34, 40. However, it is likely that distinct genetic requirements and mechanisms will emerge for each system, based on precedents from systematic manipulation of in vitro biofilm environments 38 and the pronounced gene expression responses of C. albicans to distinct host niches 41.
For any one type of biofilm, the environment may be altered by presence of co-infecting microbes. The overall frequency of mixed species biofilm infections has not been reported, but over 20% of Candida bloodstream infections are polymicrobial 42. A recent analysis of 24 implanted device-associated endocarditis cases reported that ~25% of those infections were polymicrobial 43. Within biofilms grown in vitro, the interactions between bacteria and C. albicans are diverse 44. Symbiotic interactions include augmented adherence or antibiotic resistance 45, 46. However, most known interactions are inhibitory. Among the most intriguing examples are those that arise from transkingdom quorum sensing molecule responses. For example, farnesol produced by fungi inhibits biofilm formation of Staphylococcus aureus, and increases its antibiotic susceptibility 47, 48. Bacteria fight back though; for example, the Pseudomonas aeruginosa quorum sensing molecule homoserine lactone mimics farnesol and inhibits C. albicans filamentation, thus preventing biofilm formation by C. albicans 49. Other inhibitory interactions arise from broader environmental manipulation: vaginal bacteria inhibit C. albicans growth and virulence by producing H2O2 or lactic acid 44, 50. The significance of further study in this area is demonstrated by the fact that the presence of combined infection by both bacteria and C. albicans can result in increased mortality 51, 52.
Adherence and Attachment Responses
Gene products assigned to the adherence step (Fig. 2) have been shown through either null mutant analysis or heterologous expression studies to affect binding to a plastic or protein-coated substrate. One of the most clearly defined biofilm adhesins that mediates surface binding is Eap1 21, 53. Eap1 has sequence features commonly found among fungal cell surface proteins 54, 55, including a signal sequence and a composition rich in the prospective glycosylation acceptors serine and threonine. It also contains internal repeats of a peptide motif, WPCL, that is found in numerous fungal cell surface proteins. Finally, it has a short C-terminal sequence that directs attachment of a glycosylphosphatidylinositol (GPI) anchor. GPI-linked proteins are found in many eukaryotes, where the GPI moiety tethers proteins to the plasma membrane. However, C. albicans and many other fungi can cleave this anchor, and then transfer the cleavage product and attached protein to a covalent linkage with β glucan in the cell wall 54, 55. Several approaches indicate that Eap1 is indeed a GPI-linked cell wall protein 53. Eap1 functions directly in biofilm adherence, as indicated by three observations: its expression in a non-adherent Saccharomyces cerevisiae strain confers adherence to polystyrene; a C. albicans eap1Δ/Δ deletion mutant has reduced adherence to polystyrene; and a C. albicans eap1Δ/Δ deletion mutant is defective in biofilm formation, as assayed both in vitro and in an in vivo catheter model 53, 56.
The highly related cell wall proteins Als1 and Als3 also may function in biofilm surface attachment 57. Expression of Als1 or Als3 in S. cerevisiae promotes binding to several different protein-coated substrates 58, which may resemble the conditioned surface of an implanted device. In addition, a C. albicans als1Δ/Δ als3Δ/Δ double mutant is defective in biofilm formation in vitro and in vivo 22. In particular, catheter surfaces inoculated with the double mutant are virtually devoid of cells after incubation in vivo 22, as expected if the mutant has a severe substrate adherence defect.
The idea that Eap1 and Als1 might function in the initial adherence step is consistent with the fact that expression of both genes is detectable in cells grown as either yeast or hyphal cell types 56, 59. This is not the case for Als3, which is expressed primarily or exclusively in hyphae 59. It is possible that the initial adherence step leading to biofilm formation in vivo can be carried out by either yeast-form cells, which express Als1, or by hyphae, which express Als3.
Adherence itself may activate a gene expression response. Murillo and colleagues conducted a microarray comparison of planktonic cells and substrate-adherent cells 60. Interestingly, as little as 30 min after adherence, a change in gene expression was established that, for several genes, was maintained for hours. In addition, Mateus and colleagues found that GFP fusions to the drug efflux proteins Cdr1 and Mdr1 were up-regulated within a few minutes after adherence to a glass slide 61. Thus C. albicans may sense and respond to surface contact. The regulators that promote attachment responses are unknown, but the transmembrane protein Dfi1 and MAP kinases Mkc1 and Cek1 are mediators of other surface-dependent responses 62–64 and are thus excellent candidates. Mkc1 is required for normal biofilm formation 64, and it would be interesting if this requirement reflected effects on adherence-induced gene expression.
Adherence is also highly regulated through a novel mating factor-response pathway (Box 2). Interestingly, the pathway operates in cells that do not mate, but rather assist in mating through formation of a biofilm 65–68. The responsive cells are of genotype MTLa/MTLa, and thus have the potential to mate with MTLα/MTLα cells. They are unable to do so because they have not made the epigenetic transition from the “white” mating-incompetent state to the “opaque” mating-competent state 69. They nonetheless respond to α mating factor through a newly evolved hybrid signal transduction pathway 66 to create a biofilm. Four white cell α factor-induced genes are required for full adherence of this biofilm, those encoding the cell surface proteins Eap1 and Pga10, the predicted secreted protein Pbr1, and the putative aryl-alcohol dehydrogenase Csh1 68. (Csh1 has been detected on the C. albicans cell surface 70 so the nature of its role in adherence is uncertain.) Most C. albicans isolates are MTLa/MTLα and do not secrete or respond to mating factor; MTLa/MTLα strains have been used for the bulk of the biofilm studies described in this review. However, the fact that Eap1, Pga10, and Csh1 all have roles in MTLa/MTLα biofilm formation (discussed above and below) argues that both kinds of C. albicans biofilms may employ similar gene products 68.
Box 2. C. albicans mating.
One of the most exciting C. albicans biological discoveries in recent years is the finding that this organism, long considered asexual, can mate. A mating type locus called MTL determines sexual identity through regulatory relationships with some similarity to those of S. cerevisiae: MTLa/MTLacells can mate with MTLa/MTLα cells, and MTLa/MTLα cells cannot mate 103. The mating response is induced by secreted mating pheromones: MTLa/MTLacells secrete a factor; MTLa/MTLα cells secrete α factor 104. Mating involves more than just MTL-specified sexual identity, though; cells must switch from the mating-incompetent “white” cell type to the mating-competent “opaque” cell type 105–109. The epigenetic white-opaque switch responds to numerous genetic and environmental signals, but is not regulated by mating factors as far as we know now110, 111.
Mating of two C. albicans diploid cells yields a tetraploid that breaks down through chromosome loss to yield recombinant diploid progeny 112–115. Normal functioning of the chromosome loss pathway depends upon Spo11 116, whose orthologs function in meiosis in other organisms, but there is no evidence that C. albicans has a complete meiotic pathway.
Although white cells do not mate, they do respond to mating pheromone. The response can be assayed through changes in gene expression as well as increases cell-cell and cell-substrate adherence, yielding biofilm formation 65–68, 104, 117 Interestingly, only a portion of the opaque pheromone-response pathway is utilized in white cells; they employ a hybrid pathway with novel downstream components 66–68. Opaque cells may be rare in many niches, and the biological role of this white cell biofilm seems to be to facilitate mating among disperse opaque cells 65, 110, 115.
Biofilm Initiation, Filamentation and Cell-Cell Adhesion
Gene products assigned to the initiation step (Fig. 2) reflect a range of functions. This diversity reflects in part our broad definition: these are genes in which mutations cause production of only a small, rudimentary biofilm in vitro. A few additional gene products in this group (Rbt5, Als9 and Ece1) are those whose overexpression improves biofilm formation in a bcr1Δ/Δ mutant, which is defective in biofilm initiation (see below). We note that a mutant with a partial adherence defect might be categorized as an initiation mutant; thus our assignment of gene products to this step is tentative.
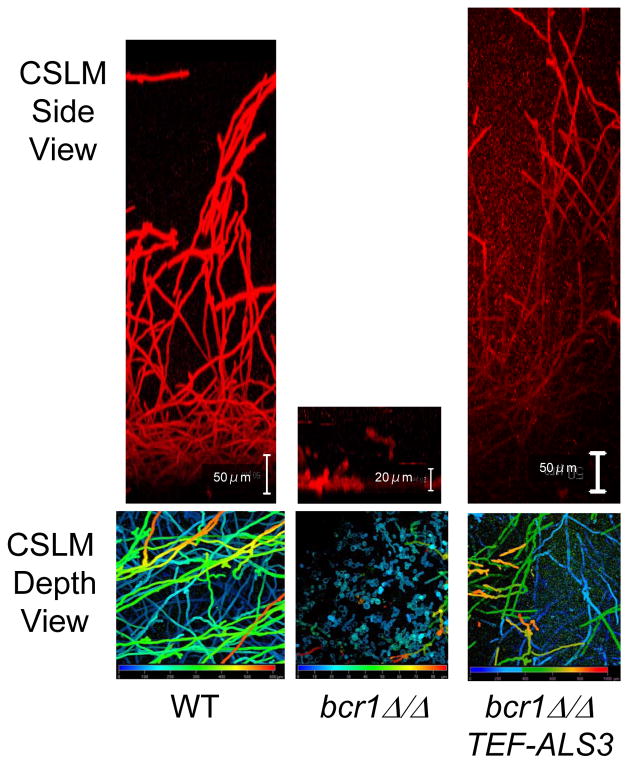
The production of hyphae is a hallmark of initiation, and many initiation-defective mutants grow solely as yeast cells under biofilm conditions (see Table 1). What is the role of hyphae? Insight has come from the transcription factor Bcr1, expression of which is up-regulated in hyphae 27, 28. Bcr1 is required for biofilm initiation, but not for production of morphologically normal hyphae 27, 28. Rather, it is required for full expression of a number of cell-surface proteins, several of which are hyphally induced. The failure to express hyphally induced surface proteins, such as Als3 and Hwp1, is the cause of the bcr1Δ/Δ mutant biofilm defect, because increased expression of ALS3 or HWP1 in the bcr1Δ/Δ mutant restores biofilm formation ability, both in vitro and in vivo (Fig. 3) 23, 28. Moreover, expression of Bcr1 or its target genes can even permit biofilm formation by mutants defective in hyphal morphogenesis: Specifically, increased expression of BCR1 in a hyphal-defective tec1-/- mutant permits in vitro formation of a biofilm, albeit a fragile one 28. In addition, expression of a surface-directed Als3 fusion protein permits biofilm formation in vitro by a hyphal-defective efg1Δ/Δ mutant 71. Therefore, the major way that hyphae promote biofilm formation is through expression of their surface protein complement.
Figure 3. Restoration of biofilm formation in a bcr1Δ/Δ mutant background by overexpression of surface protein gene ALS3.
These panels are confocal scanning laser micrographs of concanavalin A-alexafluor stained biofilms, grown under standard in vitro conditions 28. The top panels are side views; the bottom panels are pseudocolor depth views, in which blue color represents cells closest to the substrate and red color represents cells farthest from the substrate. The wild type biofilm has a dense mixture of yeast cells and hyphae, which gradually becomes predominantly hyphae at the top of the biofilm. The bcr1Δ/Δ biofilm forms a basal layer of yeast cells attached to the substrate with little to no hyphae. Increased expression of ALS3 in the bcr1Δ/Δ strain permits substantial biofilm formation.
Interestingly, the Bcr1 ortholog in biofilm-forming Candida parapsilosis is also required for biofilm formation 72. Because C. parapsilosis does not form hyphae, the regulatory pathway upstream of Bcr1 may be divergent. Nonetheless, this finding points toward the exciting possibility that Bcr1 orthologs in other species may also govern biofilm formation.
What do these hyphal surface proteins do? Hyphae are extremely “sticky”, and both Als3 and Hwp1 are adhesins in some contexts 54, so it seems reasonable that they may promote cell-cell or cell-substrate binding. In fact, Als3 (along with the closely related Als1) and Hwp1 seem to function as complementary cell-cell adhesins, analogous to the mating agglutinins of S. cerevisiae that promote binding between a and α cells. Two main observations support this idea. First, both an hwp1Δ/Δ mutant and a double als1Δ/Δ als3Δ/Δ mutant are defective in biofilm formation, but a mixture of the two mutant strains produces a robust biofilm both in vitro and in vivo 22. This finding indicates that Hwp1 and Als1/3 have distinct and complementary roles in biofilm formation. Second, expression of HWP1 in S. cerevisiae promotes its adherence to hyphae of wild-type C. albicans 22. Adherence is diminished when tested with hyphae of an als1Δ/Δ als3Δ/Δ mutant C. albicans. These findings points toward cell-cell adherence as the function that is mediated by Hwp1 and Als1/3.
At this juncture, one can envision a minimalist pathway of biofilm formation. First, yeast cells express Eap1 and Als1, which mediate cell-substrate binding. Second, surface-bound cells propagate and express Als3 and Hwp1, which mediate cell-cell binding. Hyphal formation might provide a simple pathway that leads to Als3 and Hwp1 accumulation. Als3 would also augment cell-substrate binding, as discussed in the previous section. In addition, it has been shown that Eap1 mediates cell-cell binding as well as cell-substrate binding, so it would participate in both processes. Many gene products required for biofilm initiation are also required for hyphal formation; their functions are explained by the minimalist model as ultimately being required for ALS3 and HWP1 expression.
However, one group of biofilm initiation gene products is less readily explained by the minimalist model: the additional cell surface proteins, including Sun41, Csa1, Pga10, Rbt5, Hwp2, and Rbt1 (Table 1). Analysis has been challenging for several of these gene products because they belong to families with overlapping or compensatory functions (Csa1/Pga10/Rbt5; Hwp2/Rbt1/Hwp1; Sun41/Sun42) 67, 73–76. The requisite construction of multi-gene mutants for study of these genes is non-trivial, even in this era of accelerated C. albicans genetics. In any case, current observations suggest that some of these gene products may function as adhesins. In particular, additive effects of hwp2 and rbt1 mutations with an hwp1 mutation, along with the known role of Hwp1 as an adhesin, suggests that Hwp2 and Rbt1 are adhesins67. They may contribute to a threshold level of cell-cell binding, for example, that is required for biofilm stability. (C. parapsilosis Bcr1 promotes expression of CpRBT1, so perhaps CpRbt1, which has a major role in biofilm formation 72, 77, has assumed a predominant adhesin function in that species.) A second suggestion is that some of these cell-surface proteins may have general roles in cell wall structure, and that perturbation of cell wall architecture impairs adherence through effects on either post-translational modification or expression of adhesins. We note that loss of Sun41 or Pga10 confers hypersensitivity to cell wall inhibitors, an expected consequence of a general cell wall defect 73–76. It is also noteworthy that ALS1 RNA levels are reduced in the biofilm- and cell wall-defective protein kinase mutants gin4-/-, ire1-/-, and cbk1-/-, thus suggesting that adhesin gene expression may be regulated through cell wall regulatory pathways 78. The mechanistic contribution of so many cell surface proteins to biofilm formation is among the major questions to be addressed, particularly because such proteins are inviting therapeutic targets.
Biofilm Maturation and the Extracellular Matrix
Biofilm maturation includes continued growth as well as accumulation of extracellular matrix material. Genes assigned to this category (Fig. 2) include those affecting matrix production or overall biofilm biomass.
The composition of the matrix that is produced in vitro includes carbohydrate, protein, hexosamine, phosphorus and uronic acid 79. One major extracellular carbohydrate constituent is β-1,3 glucan, and increased production is associated with biofilm cells compared to planktonic cells 80. Proteomic analysis has revealed the presence of specific proteins associated with the biofilm cell surface; these proteins may include matrix components 30, 81. Finally, a recent study reports detection of extracellular DNA 82, as has been found in bacterial biofilms 83. Addition of DNase to a mature biofilm partially disrupts the biofilm 79, 82 and the addition of eDNA at the beginning of biofilm development results in mature biofilms with increased biofilm biomass. Thus the matrix eDNA contributes to the structure and stability of a mature biofilm.
The transcription factor Zap1, a regulator of zinc acquisition 84, 85, is a net negative regulator of biofilm matrix production (Fig. 2). A zap1Δ/Δ mutant forms a biofilm with elevated levels of matrix β-1,3 glucan in vitro and in vivo 29. Zap1 activates expression of CSH1 and IFD6, which have inferred negative roles in matrix production, and represses GCA1, GCA2, and ADH5, which have inferred positive roles 29. Gca1 and Gca2 may function through hydrolytic release of soluble β-1,3 glucan fragments from longer glucan chains. The precise functions of the alcohol dehydrogenase-related gene products, Csh1, Ifd6 and Adh5, are unknown, but several similar S. cerevisiae alcohol dehydrogenases function in synthesis of acyl and aryl alcohols 31, 32, 86, 87. These alcohols have roles in quorum sensing and cell signaling (see Box 1), as indicated for example by effects on hyphal growth 15–18, 20, 88. Thus a possible mechanistic role for these dehydrogenases is to promote biogenesis of biofilm-associated acyl and aryl alcohols that in turn would control matrix synthesis. Csh1 and Ifd6 may act preferentially to yield a matrix inhibitory signal, while Adh5 may act preferentially to yield a matrix stimulatory signal 29. The known role of Zap1 in zinc-responsive gene expression suggests that ambient zinc levels may be a critical determinant of biofilm matrix levels.
Zap1 may have a broader role in biofilm maturation than simply to control matrix accumulation. The zap1Δ/Δ mutant has reduced levels of several genes that are normally up-regulated in mature biofilms, including ergosterol biosynthetic genes and putative hexose transporters 29. Thus Zap1 seems to govern several aspects of biofilm maturation. It will be interesting to see if any of these mutant phenotypes reflect the postulated alteration of quorum sensing molecule levels.
A unique feature of mature biofilms, in addition to matrix accumulation, is the acquisition of high-level resistance to antifungals 5, notably the azoles and polyenes that target membrane sterols. The nature of biofilm drug resistance may reflect four distinct mechanisms. First, mature biofilm cells have reduced membrane sterol levels 89 and elevated expression of several ergosterol biosynthetic genes 60, 90, 91, perhaps reflecting hypoxia 38, 77. The ability of mature biofilm cells to survive with low sterol levels, combined with elevated biosynthetic enzyme levels, may contribute to azole and polyene resistance. Indeed, a recent study revealed that a polyene-resistant biofilm cell subpopulation displays substantially increased ergosterol biosynthetic gene expression 92. Second, the azole efflux genes CDR1, CDR2, and MDR1 are induced early in biofilm formation, and may contribute to overall azole resistance. However, their phenotypic contribution is detectable only in early biofilm cells 89. Third, like several bacterial biofilms 93, C. albicans biofilms contain a subpopulation of “persister cells” that are tolerant to a variety of otherwise cidal treatments 94. The significance of this phenomenon is highlighted by the presence of persisters in human oral C. albicans populations 95. Persisters do not have the long-term stability of mutants, but are phenotypic variants that may arise from an epigenetic change or, perhaps, transient aneuploidy 96. Finally, the β-1,3 glucan of biofilm matrix binds to and sequesters azole drugs 80. The physiological significance of this mechanism has been demonstrated through analysis of a strain with reduced β-1,3 glucan biosynthetic capacity (genotype FKS1/fks1Δ) 97. Biofilms of this strain have reduced matrix β-1,3 glucan and reduced azole resistance levels in both in vitro and in vivo models. The dramatic sensitivity of the mutant biofilm cells to azole treatment, particularly in the in vivo biofilm model, suggests that this sequestration mechanism is a major contributor to C. albicans biofilm azole resistance 97.
Cell Dispersal
Ultimately a biofilm releases cells that can initiate formation of new biofilms or disseminate into host tissues. Recent studies examined the quantitative and qualitative properties of cells released from a C. albicans biofilm 98, 99, yielding three important findings. First, the majority of dispersed cells are yeast cells, as depicted in Fig. 2 98. This observation suggests that the transition from yeast to hyphae that occurs during biofilm initiation may be thrown into reverse for dispersal. Second, three new regulators of dispersal, Ume6, Pes1 and Nrg1, were identified. Over-expression of UME6 reduced the release of cells from a biofilm, while over-expression of either PES1 or NRG1 increased release 98, 99. Thus changes in expression or activity of Ume6 or Pes1 during biofilm maturation – perhaps in response to quorum sensing molecule accumulation – may govern cell dispersal. Finally, the study reports the fascinating observation that dispersed cells have distinct phenotypes from planktonic cells; they display elevated adherence, filamentation capacity, and increased pathogenicity in a disseminated infection model. Thus the dispersal step releases cells that are uniquely equipped to seed new biofilms and sites of infection 98, 99.
Concluding Remarks
C. albicans biofilm formation on implanted devices is a major source of disseminated Candida infection. The last decade has seen major advances in the definition of C. albicans genes that govern biofilm formation. In many cases we have moved forward from gene discovery to definition of pathway relationships and, in some cases, mechanistic understanding. In addition, numerous molecules with the potential for quorum-sensing roles in biofilm maturation have been defined. Moreover, there are now several animal models for analysis of biofilm formation in vivo that have validated the significance of key biofilm genes discovered in vitro. Yet a summary of that substantial progress also reveals major gaps in our understanding: How can so many cell wall proteins participate in biofilm formation; are they all adhesins? Which small molecules are actually active in quorum sensing in vivo, and can we harness their activities for therapeutic development? Can we use our understanding of biofilm drug resistance to develop better therapeutics and more focused assays of biological activity? What are the dynamics of formation and key molecular players in mixed-species biofilms? And, perhaps most difficult to answer and most interesting to ponder, what selective pressures caused evolution of biofilm formation ability – was it for mating, or for mucosal surface adherence and persistence in the host? There has never been a more interesting time to study C. albicans biofilms.
Supplementary Material
Acknowledgments
We are grateful to Jill Blankenship, Saranna Fanning, Shantanu Ganguly, Elizabeth Hill, and Carol Woolford for comments on this manuscript. Our studies on biofilm formation have been supported by NIH grant R01 AI067703 (APM) and NIH fellowship F32 AI085521 (JSF).
References
- 1.Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 2.Pappas PG, et al. Guidelines for treatment of candidiasis. Clin Infect Dis. 2004;38:161–89. doi: 10.1086/380796. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Marrie TJ, Costerton JW. Scanning and transmission electron microscopy of in situ bacterial colonization of intravenous and intraarterial catheters. J Clin Microbiol. 1984;19:687–93. doi: 10.1128/jcm.19.5.687-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–6. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 6.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–93. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–67. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viudes A, et al. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur J Clin Microbiol Infect Dis. 2002;21:767–74. doi: 10.1007/s10096-002-0822-1. [DOI] [PubMed] [Google Scholar]
- 9.Wilson LS, et al. The direct cost and incidence of systemic fungal infections. Value Health. 2002;5:26–34. doi: 10.1046/j.1524-4733.2002.51108.x. [DOI] [PubMed] [Google Scholar]
- 10.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andes D, et al. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–31. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra J, et al. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–94. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–9. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- 14.Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002c;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 15.Hornby JM, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67:2982–92. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh KB, Miyazawa H, Naito T, Matsuoka H. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc Natl Acad Sci U S A. 2001;98:4664–8. doi: 10.1073/pnas.071404698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002b;68:5459–63. doi: 10.1128/AEM.68.11.5459-5463.2002. This paper reports that farnesol functions as a quorum sensing molecule in C. albicans biofilms, and that biofilm density and morphology are altered by high concentrations of farnesol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alem MA, Oteef MD, Flowers TH, Douglas LJ. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot Cell. 2006;5:1770–9. doi: 10.1128/EC.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S, Kebaara BW, Atkin AL, Nickerson KW. Regulation of aromatic alcohol production in Candida albicans. Appl Environ Microbiol. 2008;74:7211–8. doi: 10.1128/AEM.01614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins M, et al. Morphogenesis control in Candida albicans and Candida dubliniensis through signaling molecules produced by planktonic and biofilm cells. Eukaryot Cell. 2007;6:2429–36. doi: 10.1128/EC.00252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Palecek SP. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology. 2008;154:1193–203. doi: 10.1099/mic.0.2007/013789-0. [DOI] [PubMed] [Google Scholar]
- 22.Nobile CJ, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–24. doi: 10.1016/j.cub.2008.06.034. This paper reports that Als1/3 and Hwp1 function as complementary adhesins in biofilms both in vivo and in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5:1604–10. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 25.Domergue R, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–70. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- 26.Verstrepen KJ, Fink GR. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu Rev Genet. 2009;43:1–24. doi: 10.1146/annurev-genet-102108-134156. [DOI] [PubMed] [Google Scholar]
- 27.Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–5. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 28.Nobile CJ, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobile CJ, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. This paper showed that transciption factor Zap1 is a key regulator of extracellular matrix production by biofilms in vitro and in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee PK, et al. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect Immun. 2006;74:3804–16. doi: 10.1128/IAI.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;74:2259–66. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006;20:1150–61. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nett JE, Marchillo K, Spiegel CA, Andes DR. Development and Validation of an In vivo Candida albicans Biofilm Denture Model. Infect Immun. 2010 doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricicova M, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–19. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- 35.Schinabeck MK, et al. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Chemother. 2004;48:1727–32. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002a;70:878–88. doi: 10.1128/iai.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baillie GS, Douglas LJ. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J Antimicrob Chemother. 2000;46:397–403. doi: 10.1093/jac/46.3.397. [DOI] [PubMed] [Google Scholar]
- 38.Stichternoth C, Ernst JF. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl Environ Microbiol. 2009;75:3663–72. doi: 10.1128/AEM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS One. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Noverr MC. Candida albicans forms Biofilms on the Vaginal Mucosa. Microbiology. 2010 doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumamoto CA. Niche-specific gene expression during C. albicans infection. Curr Opin Microbiol. 2008;11:325–30. doi: 10.1016/j.mib.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Microbiol Infect Dis. 2007;59:401–6. doi: 10.1016/j.diagmicrobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Chrissoheris MP, et al. Endocarditis complicating central venous catheter bloodstream infections: a unique form of health care associated endocarditis. Clin Cardiol. 2009;32:E48–54. doi: 10.1002/clc.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adam B, Baillie GS, Douglas LJ. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol. 2002;51:344–9. doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 46.Bamford CV, et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother. 2006;50:1463–9. doi: 10.1128/AAC.50.4.1463-1469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroda M, Nagasaki S, Ito R, Ohta T. Sesquiterpene farnesol as a competitive inhibitor of lipase activity of Staphylococcus aureus. FEMS Microbiol Lett. 2007;273:28–34. doi: 10.1111/j.1574-6968.2007.00772.x. [DOI] [PubMed] [Google Scholar]
- 49.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–23. doi: 10.1111/j.1365-2958.2004.04349.x. This paper describes that a molecule produced by P. aeruginosa mimics the actions of the C. albicans quorum sensing molecule farnesol, thus providing P. aeruginosa a competitive advantage in the host. [DOI] [PubMed] [Google Scholar]
- 50.Boris S, Barbes C. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes Infect. 2000;2:543–6. doi: 10.1016/s1286-4579(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 51.Shirtliff ME, et al. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother. 2009;53:2392–401. doi: 10.1128/AAC.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009 doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F, et al. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot Cell. 2007 doi: 10.1128/EC.00049-07. Eap1 is shown to be a glycosylphosphatidylinositol-anchored, glucan-cross-linked cell wall protein that acts as an adhesin required for biofilm formation in vitro as well as in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richard ML, Plaine A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot Cell. 2007;6:119–33. doi: 10.1128/EC.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li F, Palecek SP. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot Cell. 2003;2:1266–73. doi: 10.1128/EC.2.6.1266-1273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–80. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 58.Sheppard DC, et al. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–9. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- 59.Green CB, Zhao X, Yeater KM, Hoyer LL. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology. 2005;151:1051–60. doi: 10.1099/mic.0.27696-0. [DOI] [PubMed] [Google Scholar]
- 60.Murillo LA, et al. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell. 2005;4:1562–73. doi: 10.1128/EC.4.9.1562-1573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mateus C, Crow SA, Jr, Ahearn DG. Adherence of Candida albicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob Agents Chemother. 2004;48:3358–66. doi: 10.1128/AAC.48.9.3358-3366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zucchi PC, Davis TR, Kumamoto CA. A Candida albicans cell wall-linked protein promotes invasive filamentation into semi-solid medium. Mol Microbiol. 2010;76:733–48. doi: 10.1111/j.1365-2958.2010.07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumamoto CA. Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat Rev Microbiol. 2008;6:667–73. doi: 10.1038/nrmicro1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc Natl Acad Sci U S A. 2005;102:5576–81. doi: 10.1073/pnas.0407097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. Embo J. 2006 doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahni N, et al. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 2010;8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ene IV, Bennett RJ. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot Cell. 2009;8:1909–13. doi: 10.1128/EC.00245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sahni N, et al. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 2009;5:e1000601. doi: 10.1371/journal.ppat.1000601. This paper identifies several genes in white cells that are upregulated in the presence of mating pheromone. It also reports that white cells use their pheromone response pathway to produce a mature biofilm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–55. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 70.Singleton DR, Hazen KC. Differential surface localization and temperature-dependent expression of the Candida albicans CSH1 protein. Microbiology. 2004;150:285–92. doi: 10.1099/mic.0.26656-0. [DOI] [PubMed] [Google Scholar]
- 71.Zhao X, et al. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 2006;152:2287–99. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding C, Butler G. Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryot Cell. 2007;6:1310–9. doi: 10.1128/EC.00136-07. This paper reports that transcription factor Bcr1 is a conserved regulator of biofilm formation in C. parapsilosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Firon A, et al. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol Microbiol. 2007;66:1256–75. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- 74.Perez A, et al. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 2006;6:1074–84. doi: 10.1111/j.1567-1364.2006.00131.x. [DOI] [PubMed] [Google Scholar]
- 75.Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot Cell. 2007;6:2046–55. doi: 10.1128/EC.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hiller E, Heine S, Brunner H, Rupp S. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot Cell. 2007;6:2056–65. doi: 10.1128/EC.00285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossignol T, et al. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot Cell. 2009;8:550–9. doi: 10.1128/EC.00350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- 80.Nett J, et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007;51:510–20. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez-Gomariz M, et al. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics. 2009;9:2230–52. doi: 10.1002/pmic.200700594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martins M, et al. Presence of Extracellular DNA in the Candida albicans Biofilm Matrix and its Contribution to Biofilms. Mycopathologia. 2009 doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–6. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 84.Kim WI, Lee WB, Song K, Kim J. Identification of a putative DEAD-box RNA helicase and a zinc-finger protein in Candida albicans by functional complementation of the S. cerevisiae rok1 mutation. Yeast. 2000;16:401–9. doi: 10.1002/(SICI)1097-0061(20000330)16:5<401::AID-YEA531>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 85.Kim MJ, Kil M, Jung JH, Kim J. Roles of Zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic Yeast Candida albicans. J Microbiol Biotechnol. 2008;18:242–7. [PubMed] [Google Scholar]
- 86.Delneri D, Gardner DC, Bruschi CV, Oliver SG. Disruption of seven hypothetical aryl alcohol dehydrogenase genes from Saccharomyces cerevisiae and construction of a multiple knock-out strain. Yeast. 1999;15:1681–9. doi: 10.1002/(SICI)1097-0061(199911)15:15<1681::AID-YEA486>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 87.Dickinson JR, Salgado LE, Hewlins MJ. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8028–34. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Fujita M, Feng Q, Clardy J, Fink GR. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A. 2004;101:5048–52. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71:4333–40. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia-Sanchez S, et al. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 2004;3:536–45. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nett JE, Lepak AJ, Marchillo K, Andes DR. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis. 2009;200:307–13. doi: 10.1086/599838. This study is the first in vivo characterization of C. albicans biofilms through microarray analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khot PD, Suci PA, Miller RL, Nelson RD, Tyler BJ. A small subpopulation of blastospores in candida albicans biofilms exhibit resistance to amphotericin B associated with differential regulation of ergosterol and beta-1,6-glucan pathway genes. Antimicrob Agents Chemother. 2006;50:3708–16. doi: 10.1128/AAC.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis K. Persister Cells. Annu Rev Microbiol. 2010 doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 94.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–46. doi: 10.1128/AAC.00684-06. This paper reports the discovery of persister cells that contribute to C. albicans biofilm-based drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lafleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother. 2010;54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Selmecki A, Forche A, Berman J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell. 2010;9:991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nett JE, Sanchez H, Cain MT, Andes DR. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis. 2010;202:171–5. doi: 10.1086/651200. This paper defines beta-glucan levels as a key determinant of in vivo biofilm-based azole drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uppuluri P, et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. This paper describes a novel assay for biofilm cell dispersal in C. albicans, the unique virulence properties of the dispersed cells, and genetic regulators of dispersal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uppuluri P, et al. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 9:1531–7. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–58. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 101.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 8:340–9. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 102.Molloy S. Quorum sensing: Setting the threshold. Nat Rev Micro. 8:388–389. [Google Scholar]
- 103.Nasmyth KA. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 104.Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–9. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soll DR, Lockhart SR, Zhao R. Relationship between switching and mating in Candida albicans. Eukaryot Cell. 2003;2:390–7. doi: 10.1128/EC.2.3.390-397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lockhart SR, Zhao R, Daniels KJ, Soll DR. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell. 2003;2:847–55. doi: 10.1128/EC.2.5.847-855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 109.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:e1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang G, et al. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–3. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 113.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–10. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 114.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–3. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bennett RJ. A Candida-based view of fungal sex and pathogenesis. Genome Biol. 2009;10:230. doi: 10.1186/gb-2009-10-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Forche A, et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao R, et al. Unique aspects of gene expression during Candida albicans mating and possible G(1) dependency. Eukaryot Cell. 2005;4:1175–90. doi: 10.1128/EC.4.7.1175-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kelly MT, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–83. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 119.Sanchez AA, et al. Relationship between Candida albicans virulence during experimental hematogenously disseminated infection and endothelial cell damage in vitro. Infect Immun. 2004;72:598–601. doi: 10.1128/IAI.72.1.598-601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kumamoto CA, Vinces MD. Alternative Candida albicans Lifestyles: Growth on Surfaces. Annu Rev Microbiol. 2005 doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- 121.Lewis RE, Lo HJ, Raad II, Kontoyiannis DP. Lack of catheter infection by the efg1/efg1 cph1/cph1 double-null mutant a Candida albicans strain that is defective in filamentous growth. Antimicrob Agents Chemother. 2002;46:1153–5. doi: 10.1128/AAC.46.4.1153-1155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cao YY, et al. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother. 2005;49:584–9. doi: 10.1128/AAC.49.2.584-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao X, Oh SH, Yeater KM, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology. 2005;151:1619–30. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hashash R, et al. Characterisation of Pga1, a putative Candida albicans cell wall protein necessary for proper adhesion and biofilm formation. Mycoses. doi: 10.1111/j.1439-0507.2010.01883.x. [DOI] [PubMed] [Google Scholar]
- 125.Peltroche-Llacsahuanga H, Goyard S, d’Enfert C, Prill SK, Ernst JF. Protein O-mannosyltransferase isoforms regulate biofilm formation in Candida albicans. Antimicrob Agents Chemother. 2006;50:3488–91. doi: 10.1128/AAC.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. Yeast wall protein 1 of Candida albicans. Microbiology. 2005;151:1631–44. doi: 10.1099/mic.0.27663-0. [DOI] [PubMed] [Google Scholar]
- 127.Nobile CJ, Mitchell AP. Large-scale gene disruption using the UAU1 cassette. Methods Mol Biol. 2009;499:175–94. doi: 10.1007/978-1-60327-151-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goyard S, et al. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol Biol Cell. 2008;19:2251–66. doi: 10.1091/mbc.E07-09-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kruppa M, et al. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot Cell. 2004;3:1062–5. doi: 10.1128/EC.3.4.1062-1065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 2009;5:e1000294. doi: 10.1371/journal.ppat.1000294. This study connects biofilm adhesin expression to global nutrient sensing via the conserved Tor pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Strijbis K, et al. Carnitine-dependent transport of acetyl coenzyme A in Candida albicans is essential for growth on nonfermentable carbon sources and contributes to biofilm formation. Eukaryot Cell. 2008;7:610–8. doi: 10.1128/EC.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Richard ML, Nobile CJ, Bruno VM, Mitchell AP. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4:1493–502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu G, Vellucci VF, Kyc S, Hostetter MK. Simvastatin inhibits Candida albicans biofilm in vitro. Pediatr Res. 2009;66:600–4. doi: 10.1203/PDR.0b013e3181bd5bf8. [DOI] [PubMed] [Google Scholar]
- 134.Melo AS, et al. The Candida albicans AAA ATPase homologue of Saccharomyces cerevisiae Rix7p (YLL034c) is essential for proper morphology, biofilm formation and activity of secreted aspartyl proteinases. Genet Mol Res. 2006;5:664–87. [PubMed] [Google Scholar]
- 135.Palanisamy SK, Ramirez MA, Lorenz M, Lee SA. Candida albicans PEP12 is required for biofilm integrity and in vivo virulence. Eukaryot Cell. 2009 doi: 10.1128/EC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bernardo SM, Khalique Z, Kot J, Jones JK, Lee SA. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet Biol. 2008;45:861–77. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.