Abstract
Breast cancer is a heterogeneous disease and genetic factors play an important role in its genesis. Although mutations in tumor suppressors and oncogenes encoded by the nuclear genome are known to play a critical role in breast tumorigenesis, the contribution of the mitochondrial genome to this process is unclear. Like the nuclear genome, the mitochondrial genome also encodes proteins critical for mitochondria functions such as oxidative phosphorylation (OXPHOS), which is known to be defective in cancer including breast cancer. Due to limited repair mechanisms compared to that for nuclear DNA (nDNA), mitochondrial DNA (mtDNA) is more susceptible to mutations. Thus changes in mitochondrial genes could also contribute to the development of breast cancer. In this review we discuss mtDNA mutations that affect OXPHOS. Continuous acquisition of mtDNA mutations and selection of advantageous mutations ultimately leads to generation of cells that propagate uncontrollably to form tumors. Since irreversible damage to OXPHOS leads to a shift in energy metabolism towards enhanced aerobic glycolysis in most cancers, mutations in mtDNA represent an early event during breast tumorigenesis, and thus may serve as potential biomarkers for early detection and prognosis of breast cancer. Because mtDNA mutations lead to defective OXPHOS, development of agents that target OXPHOS will provide specificity for preventative and therapeutic agents against breast cancer with minimal toxicity.
Keywords: Mitochondrial DNA, OXPHOS, breast cancer, Warburg effect, mtDNA mutations, homoplasmy and heteroplasmy
Introduction
Breast cancer remains a major cause of cancer related death in women despite significant progress in understanding its causes, genesis, detection, and treatment [1]. Tremendous progress has been made as a result of extensive research focusing on the nuclear genome. For example, mutations in the BRCA1 and BRCA2 genes encoded by the nuclear DNA (nDNA) are associated with a high risk of developing breast cancer [2, 3]. Similarly, mutations in TP53, PTEN (phosphatase and tensin homolog), and CHEK1 and CHEK2 (checkpoint kinases 1 and 2) are also associated with increased susceptibility to breast cancer development [4]. Current evidence suggest that mitochondrial function is severely impaired in various cancers including breast cancer [5–16] due to genetic defects of oxidative phosphorylation (OXPHOS) system. Proteins that participate in the proper functioning of the OXPHOS system are encoded by both nDNA and mitochondrial DNA (mtDNA). Similar to nDNA, mtDNA deletions and mutations have been shown to play critical roles in breast tumorigenesis [5, 17–23]. Cancer associated mtDNA mutations can be germline or somatic mutations [24, 25]. Currently it is unclear whether mtDNA mutations or copy number determine the fate of cells undergoing transformation and how homoplasmic (cells harboring identical mtDNA genotype) or heteroplasmic (occurrence of more than one mtDNA genotype) states affect breast tumorigenesis. It is important to critically evaluate the relative contributions of nDNA and mtDNA, and the crosstalk between these two genomes in the regulation of OXPHOS. We expect that detailed analysis of the effects of mtDNA mutations on OXPHOS will not only shed light on breast tumorigenesis, but will also be important for early detection and predicting the prognosis of breast cancer patients.
The OXPHOS system
Although cancer cells are not entirely reliant on energy production through OXPHOS, they do contain necessary components of this system and functional OXPHOS (albeit reduced) similar to those of non-cancerous cells [26, 27]. The OXPHOS system is comprised of five large multi-subunit complexes as follows: complex I (NADH dehydrogenase or NADH:ubiquinone oxidoreductase), complex II (succinate dehydrogenase or succinate:ubiquinone oxidoreductase), complex III (the bc1 complex or ubiquinone:cytochrome c oxidoreductase), complex IV (cytochrome c oxidase, cyclooxygenase or reduced cytochrome c:oxygen oxidoreductase), and complex V (FoF1-ATP-synthase). These complexes are localized at the inner mitochondrial membrane and made up of proteins encoded by nDNA and mtDNA. Apart from these five multi-subunit complexes, cytochrome c and ubiquinone (coenzyme Q10) are also required as electron carriers to generate energy in the form of ATP [28–30]. NADH and FADH2 oxidation reactions feed electrons to respiratory chain (complexes I–IV) that are transferred to molecular oxygen to form water as a byproduct and generate a proton (H+) gradient [28–30]. H+ pumping at complexes I, III, and IV from the mitochondrial matrix into the inter membrane space leads to an increase in the H+ gradient across the inner mitochondrial membrane. Finally, the dissipation of the H+ gradient via complex V provides energy for combining the ADP and inorganic phosphate (Pi) to form ATP [28–30]. Several questions remain unanswered about the structural and functional integrity of the OXPHOS system in cancer cells. Do cancer cells have normal OXPHOS system, and if so, why are they dependent more on glycolysis than OXPHOS for ATP generation? Do cancer cells harbor defects in mtDNA and nDNA encoding various proteins that are part of OXPHOS system?
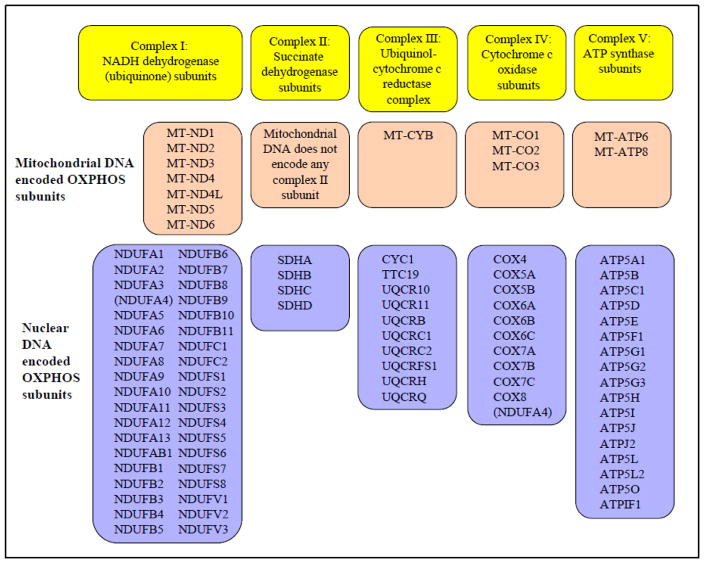
Molecular genetics of the OXPHOS system
OXPHOS system consists of 92 structural subunits (Figure 1) [29, 31]. Most of the proteins of OXPHOS system are encoded by the nDNA except for the following 13 polypeptides that are encoded by the mtDNA: seven subunits of complex I, one subunit of complex III, three subunits of complex IV, and two subunits of complex V. In addition to these 13 polypeptides, the 16.6-kb mitochondrial genome also encodes 22 tRNAs and 2 rRNAs [29, 32]. This suggests that mutations in the regions of mtDNA or nuclear DNA that encode components of the OXPHOS system can affect its function. Indeed, mutations in regions of the mitochondrial and nuclear genes encoding OXPHOS proteins have been reported in a variety of cancers [22, 25, 33–41]. However, little is known about the mtDNA-mediated impairment of mitochondrial function caused by mutations in mtDNA in breast cancer cells.
Figure 1. Components of OXPHOS system.
Mitochondrial OXPHOS complexes are encoded by the nuclear and mitochondrial genome. NDUFA4 was previously classified as a part of complex I, recent findings (see reference 30) suggest that NDUFA4 belongs to complex IV. Therefore, NDUFA4 is mentioned in both complex I and IV.
- MT-ND
- mitochondrially encoded NADH dehydrogenase
- MT-CYB
- mitochondrially encoded cytochrome b
- MT-CO
- mitochondrially encoded cytochrome c oxidase
- MT-ATP
- mitochondrially encoded ATP synthase
- NDUFA
- NADH dehydrogenase (ubiquinone) 1 alpha subcomplex
- NDUFAB1
- NADH dehydrogenase (ubiquinone) 1 alpha/beta subcomplex 1
- NDUFB
- NADH dehydrogenase (ubiquinone) 1 beta subcomplex
- NDUFC
- NADH dehydrogenase (ubiquinone) 1 subcomplex unknown
- NDUFS
- NADH dehydrogenase (ubiquinone) Fe-S protein
- NDUFV
- NADH dehydrogenase (ubiquinone) flavoprotein
- SDHA
- succinate dehydrogenase complex subunit A
- SDHB
- succinate dehydrogenase complex subunit B
- SDHC
- succinate dehydrogenase complex subunit C
- SDHD
- succinate dehydrogenase complex subunit D
- CYC-1
- cytochrome c-1
- TTC19
- tetratricopeptide 19
- UQCR
- ubiquinol-cytochrome c reductase complex III
- UQCRB
- ubiquinol-cytochrome c reductase binding protein
- UQCRC
- ubiquinol-cytochrome c reductase core protein
- UQCRFS1
- ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
- UQCRH
- ubiquinol-cytochrome c reductase hinge protein
- UQCRQ
- ubiquinol-cytochrome c reductase complex III subunit VII
- COX4
- cytochrome c oxidase subunit IV
- COX5A
- cytochrome c oxidase subunit Va
- COX5B
- cytochrome c oxidase subunit Vb
- COX6A
- cytochrome c oxidase subunit VIa
- COX6B
- cytochrome c oxidase subunit VIb
- COX6C
- cytochrome c oxidase subunit VIc
- COX7A
- cytochrome c oxidase subunit VIIa
- COX7B
- cytochrome c oxidase subunit VIIb
- COX7C
- cytochrome c oxidase subunit VIIc
- COX8
- cytochrome c oxidase subunit VIII
- ATP5A1
- ATP synthase H+ transporting mitochondrial F1 complex, alpha subunit 1
- ATP5B
- ATP synthase H+ transporting mitochondrial F1 complex, beta polypeptide
- ATP5C1
- ATP synthase H+ transporting mitochondrial F1 complex, gamma polypeptide 1
- ATP5D
- ATP synthase H+ transporting mitochondrial F1 complex, delta subunit
- ATP5E
- ATP synthase H+ transporting mitochondrial F1 complex, epsilon subunit
- ATP5F1
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1
- ATP5G
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C
- ATP5H
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d
- ATP5I
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E
- ATP5J
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6
- ATP5J2
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2
- ATP5L
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G
- ATP5L2
- ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
- ATP5O
- ATP synthase, H+ transporting, mitochondrial Fo complex, O subunit
- ATPIF1
- ATPase inhibitory factor 1
The copies of mtDNA per mitochondrion and mitochondria per cell vary with cell types. Each mitochondrion contains 2–10 copies of mtDNA, and each cell may harbor hundreds to thousands mitochondria. Damage to mtDNA and change in its copy number can affect OXPHOS function. Multiple factors contribute to the increased vulnerability of mtDNA to damage, and thus mutations. During replication, mtDNA contains a short triple-stranded DNA structure (known as the D-loop), in which a single strand of mtDNA is displaced [42–44]. Unlike nuclear DNA, the mtDNA does not contain the well-known protection mechanisms such as protective histones, harbors limited DNA repair capability, and lacks introns. In addition, since mitochondria are the source of reactive oxygen species (ROS), continued exposure to ROS [45, 46] contributes to the increased susceptibility of mtDNA to mutation.
Multiple copies of mtDNA exist in association with key maintenance proteins in structures known as mitochondrial nucleoids. The mitochondrial D-loop region recruits and binds ATAD3 (ATPase family AAA domain-containing protein 3), a component of many, but not all, mitochondrial nucleoids. The interaction between ATAD3 and D-loop region of mtDNA is required for the purpose of forming and segregating mitochondrial nucleoids [47]. This association is transient, thus ATAD3 may not protect mtDNA as histones do for nDNA. Mitochondrial transcription factor A (Tfam) is the main factor packaging mtDNA into nucleoids or mitochromosome formation and is essential for mtDNA transcription initiation [48]. The interaction between hexameric mtDNA helicase Twinkle [49], tetrameric mitochondrial single-strand DNA binding protein (mtSSB), and the heterotrimeric mtDNA polymerase γ (pol γ) [50] forms mtDNA replisome, which promotes proper mtDNA replication. This suggests that these proteins interact with mtDNA but their function in terms of providing protection is still not clearly defined. There is evidence that a reduced level of Tfam results in increased levels of ROS and mtDNA instability and a subsequent increase in tumor number and growth in the small intestine, suggesting that defects in OXPHOS lead to tumorigenesis [51]. Furthermore, cybrids (a fusion of whole cell with a cytoplasm) with benign mitochondria can reverse oncogenic characteristics of cancer cells [52]. Additional proteins have been co-purified with frog oocyte mtDNA [53] but their roles in mtDNA maintenance are uncertain. Although many details remain to be clarified, it is clear that mtDNA is more susceptible to damage than nDNA, and that mutations of mtDNA could contribute to defects in OXPHOS leading to the development of various disease processes including breast cancer.
Mitochondrial DNA mutations in breast cancer
Although the role of nDNA mutations in cancer development is well established, the importance of mtDNA mutation in the disease process is only now beginning to receive attention. Mutations in mtDNA are expected to destabilize the OXPHOS system because 13 proteins encoded by mtDNA are essential for structural and functional integrity complex I, and III–V. Deficiency of OXPHOS in cancer cells was first described by Otto Warburg who hypothesized that cancer cells rely more on glycolysis for their bioenergetic needs instead of OXPHOS even in the presence of abundant oxygen [54, 55]. However, the Warburg hypothesis does not apply to all types of tumor cell or in all conditions. Several reports suggest that OXPHOS accounts for approximately 40–80 % of the total ATP produced by cells in glucose-enriched medium [26, 56–60]. In addition, lack of oxygen or intermittent hypoxia and/or glucose limitation contributes to a shift toward high glycolysis or glucose-independent respiration in cancer cells in order to generate the required ATP [61–65]. Overall, it is now clear that mtDNA mutation influences OXPHOS function and consequently might play a role in tumor initiation and progression.
Indeed, mtDNA mutations have been identified in various types of cancer [25, 66–69]. Although a number of mtDNA rearrangements and amplifications were reported in acute myeloid leukemia [70], the first somatic mtDNA mutation was detected by Polyak et al., in human colorectal cancer cells [66]. After these initial findings, mtDNA mutations have been reported in esophageal, ovarian, thyroid, head and neck, lung, bladder, renal, and breast cancer cells [25, 67, 71–73]. Mutations in nDNA such as those in p53, BRCA-1 and BRCA-2, PTEN, CHEK2, ATM (ataxia telangiectasia mutated), XPD (xeroderma pigmentosum group D)/ERCC2 (excision repair cross-complementing rodent repair deficiency, complementation group 2 protein), and HER-2 (human epidermal growth factor receptor) have been extensively studied in breast cancer [2–4, 74], whereas mtDNA mutations are only now becoming recognized as an important aspect of breast cancer development and resistance to apoptosis. Studies performed in the last few years have characterized various alterations in mtDNA in breast cancer, including point mutations, mtDNA polymorphisms, mtDNA depletion, microsatellite instability, insertions, changes in mtDNA copy number, homoplasmy, and heteroplasmy of mtDNA [18, 33, 51, 68, 69, 75–99]. Breast nipple aspirate fluid (NAF) with mtDNA mutations at position 204, 207, and 16293 has been suggested to be indicative of breast cancer and mtDNA D-loop mutation has also been proposed as an independent prognostic marker for breast cancer [100, 101]. In addition to a deleterious effect on genes encoding OXPHOS components, mutations in mtDNA could involve tRNAs and rRNA [25, 102, 103] required for the synthesis of peptides that are important in the assembly of various complexes. Therefore, the ultimate outcome of mtDNA mutation is defective OXPHOS function, which leads to a moderate increase in ROS production that in turn may promote tumorigenesis [66, 67, 104].
Germline mtDNA mutations and breast tumorigenesis
DNA instability can be caused by various factors such as lack of DNA repair and proof reading, and instability of nuclear DNA is known to be a major cause of tumorigenesis. Nuclear DNA-encoded proteins are an integral part of the OXPHOS system and, conversely, defects in OXPHOS induce irreversible changes in the nuclear genome [30, 105, 106]. Mitochondria-nucleus crosstalk and mitochondrial retrograde signaling play important role in tumor development [16, 41, 107–111], supporting the notion that mtDNA instability also plays an important role in tumorigenesis. Indeed, various cancer cells including breast cancer cells, harbor instability in the mitochondrial genome [68, 108, 112, 113].
In mtDNA T16189C germ-line mutation, various factors contribute to the substitution of T by C at the nucleotide position (np) 16189, for example, lack of repair and proof reading play a critical role in this transition. Although a large data set is needed, but this study provides evidence that T to C substitution at np 16189 is associated with susceptibility to breast cancer development [114]. The 10398A allele of the NADH dehydrogenase-3 locus (ND3) of mtDNA is associated with increased risk for invasive breast cancer in African-American women [33, 115]. Similarly, 10398A allele is also associated with breast and esophageal cancer in North Indian women [22]. However, other studies demonstrate that mtDNA G10398A polymorphism do not associate with breast cancer in African-American or South Indian women [116, 117]. In contrast, the 10398G polymorphism of ND3 has been shown to increase the risk of breast cancer in European American, Polish, and Malay populations [22, 78, 96, 118, 119]. Therefore, more studies involving larger population sizes are needed to clarify this association. It is also possible that, polymorphisms in the mitochondrial genome could also be influenced by life styles such as alcohol consumption [120]. Chronic alcohol use may cause OXPHOS deficiency and other cellular changes. The mechanism by which the presence of these mutations leads to mitochondrial dysfunction is not clearly defined but the G10398A variant of mtDNA may result in defective complex I function, and thus lead to increased ROS production [22, 121]. Whether ROS produced due to the G10398A polymorphism is sufficient to induce tumor formation remains to be determined, but the presence of other mutations combined with G10398A may contribute to breast tumorigenesis.
Other single nucleotide polymorphisms (SNPs) in mtDNA including G9055A, T16519C, T239C, A263G, and C16207T, also result in increased susceptibility to breast cancer [78, 87]. MtDNA T3197C and G13708A SNPs decrease breast cancer risk [78], and a reduced incidence of mtDNA A73G, C150T, T16183C, T16189C, C16223T, and T16362C SNPs was noted in breast cancer patients compared to database controls [87]. Recent studies have also identified multiple novel mtDNA mutations and polymorphisms in breast cancer patients [79]. Analysis of the sequence of genes encoding complex I in cancer tissues and corresponding normal tissues has led to the discovery of very rare mtDNA polymorphisms, including A4727G, G9947A, A10044G, A10283G, T11233C, and C11503T, that may have implications in breast cancer development [122].
Somatic mtDNA mutations and risk for breast cancer
Although germline mutations have been linked to breast tumorigenesis, the majority of breast cancers are not inherited. In such cases, somatic mtDNA mutations may lead to selective transformation of breast epithelial cells and tumorigenesis. Various somatic mtDNA mutations have been detected in breast cancer [21, 23, 80, 81, 83, 85, 91, 101, 123–127], and here we will discuss a few of these mutations. It is important to note that the prevalence of germline mutations is significantly higher than that of somatic mutations, and most of the patients with germline mutations have multiple mutations [128]. The majority of somatic mutations occur in the D-loop region, which is considered a hot spot for mutation [23] because this region is ~60 times more susceptible to mutations than the coding regions, although others report only a 7-fold increase in susceptibility [100]. The D-loop itself is a non-coding region of mtDNA, but mutations in this area contribute to alterations in the coding sequence of mtDNA and modification of transcription, and thus affecting the expression and function of 13 core proteins of the OXPHOS system.
Somatic mutations of mtDNA can be point mutations, deletions or insertions, or missense mutations. In a study on invasive ductal carcinoma, 11 out of 18 tumor samples showed somatic mtDNA mutations [127]. Similarly, somatic mutations were observed in 14 out of 19 tumor samples, of which only 4 mutations were in polypeptide coding regions and one of these was a missense mutation [23, 129]. In another study, 15 among a total of 45 mutations detected in 15 tumor samples were missense mutations [100, 130]. By targeting MnlI restriction site between nucleotides 16,106 and 16,437 within the D-loop region of mtDNA, Ye et al. [131] showed that somatic mutation at this site occurred more frequently in breast cancer cells than in benign epithelial cells. Their study involved 501 patients with breast cancer and 203 with benign breast disease, and the findings suggest that somatic mutations at the MnlI site in mtDNA may lead to the development of breast cancer. In another study, screening of the D-loop region demonstrated that 8 out of 22 cancer tissues had somatic mutations; of these 6 had a single somatic mutation and 2 had 7 somatic mutations [124].
Deletion of 4977 base pairs (ΔmtDNA4977 mutation) was also observed in breast cancer tissues and surrounding normal breast cells patients [132]. Although the ΔmtDNA4977 mutation has been implicated in the process of carcinogenesis, its occurrence in breast cancer tissue, breast benign tissues, and surrounding normal tissues suggests that it does not play a significant role in breast tumorigenesis [133]. Other groups have reported conflicting data on the role of ΔmtDNA4977 mutation in breast cancer [101, 130]. David Sidranski’s group analyzed 18 primary breast tissues and observed 12 somatic mutations in 11 tumor samples. Out of the 12 mutations, 5 were deletions or insertions within the D310 repeat region of the D-loop, whereas the remaining 7 were single base substitutions in the coding regions and D-loop of mtDNA. The mutations detected in coding regions lead to alterations in the polypeptides encoded by mtDNA. Using D310 as a clonal marker, they concluded that D310 alteration in primary breast cancer may represent a clinical marker for breast tumorigenesis [127]. Recent studies also detected a high frequency of D310 mutations (32.5%) in Asian Indian patients with breast cancer [76], supporting the role of mtDNA instability at D310 as a marker for breast cancer.
Because a normal cell contains multiple copies of mtDNA compared with two copies of nDNA, determination of mtDNA mutations in blood or biofluids with low cellularity such as NAF, which contains shed ductal epithelial cells may represent a better biomarker for breast cancer. In this regard, several studies have been performed in this direction to establish mtDNA somatic mutation as a biological marker for breast tumorigenesis [81, 95, 100, 124, 127, 134–137]. Sauter and colleagues detected mtDNA mutations in primary breast tumor and matched breast NAF [100]. Similarly, other studies have shown a correlation between mtDNA mutations in breast tumor tissues and in biological fluids including serum [81, 95, 99, 127, 137]. However, in some studies, mtDNA mutations were undetectable or follow-up was not available [134], therefore, further studies with more advanced techniques and long-term follow-up are necessary to robustly establish whether mtDNA mutations in blood, NAF, or other biological fluids could be used for early detection of breast cancer.
Reduced level of mtDNA and breast tumorigenesis
Although OXPHOS dysfunction is known to be associated with tumorigenesis in several types of cancer, including breast cancer [9, 11, 138–140], some studies also suggest that OXPHOS dysfunction could play a causative role in cancer development [5, 141, 142]. Because OXPHOS deficiency suppresses p53 expression/function and p53 function declines with age [141, 142], increased mtDNA mutations or reduced mtDNA content-mediated OXPHOS dysfunction may facilitate the suppression of p53 function causing higher tumor incidence. There is evidence that either mtDNA mutation or a reduced mtDNA content in tumor cells can contribute to OXPHOS dysfunction and tumorigenesis [25, 30, 103, 138, 143]. A reduced level of mtDNA associates with increased resistance, invasiveness, and aggressive disease in multiple types of cancer including breast, renal, and thyroid [5, 10, 19, 82, 84, 109, 144–147]. In addition, reduced level or lack of mtDNA has also been associated with resistance to apoptosis, increased proliferation, and increased metastasis [45, 145, 146, 148–150]. The aberration of mitochondrial complex I function has been associated with the enhanced aggressiveness of human breast cancer cells [103].
Although a reduced mtDNA content has been associated with higher histological grade in breast cancer patients in some studies [82, 101]; no correlation between the reduced mtDNA content and tumor grade or metastasis was observed in other studies [144, 151]. A study demonstrated high mtDNA copy number in whole blood cells of breast cancer patients compared to control healthy subjects [75]. High mtDNA content was also associated with decreased endogenous antioxidant system including total glutathione, Cu-Zn superoxide dismutase (SOD1), and catalase [75]. Our recent findings suggest that a reduced level of mtDNA is associated with high incidence and poor prognosis of prostate cancer [152]. Decreased levels of mtDNA in the peripheral blood were reported in breast cancer patients with stage I disease [147], suggesting that a reduction in mtDNA represents an early stage in breast tumorigenesis, which could have significance in early detection of breast cancer.
Comparison of mtDNA content with other prognostic markers such as estrogen receptor (ER) demonstrated a reduced level of mtDNA in ER-negative normal breast tissues compared with ER-positive normal breast tissues [151]. ER localizes to the mitochondria [153, 154], therefore the presence of ER may regulate the level of mtDNA or OXPHOS function [154–156]. Since p53 also localizes to mitochondria and interacts with ER [157], the ER-p53 may regulate mtDNA content, and thus OXPHOS function. Although mtDNA content was highly reduced in breast tumor tissues, the level of mtDNA was similar between ER-negative and ER-positive cancerous breast tissues [151]. These findings indicate that a reduced level of mtDNA alters ER and/or ER-p53 functions during breast tumorigenesis.
Although it is unclear whether the decreased level of mtDNA is due to alterations in copy number per cell or in the number of mitochondria per cell, several lines of evidence suggest decreased mtDNA copy number per cell. Regulation of mtDNA integrity and replication by p53 and pol γ might be defective in cancer cells, causing reduced mtDNA content [105, 158]. Since replication of mtDNA is controlled by the D-loop region, the presence of mutations in the D-loop region in various cancers, including in breast cancer, could also contribute to a reduced copy number of mtDNA [42, 113, 159, 160]. Additionally, mtDNA is especially susceptible to damage caused by the high levels of ROS production in mitochondria of cancer cells, which may lead to degradation of damaged mtDNA [159, 161, 162].
Homoplasmic and heteroplasmic mutations in breast cancer
The number of mtDNA molecules in a typical cancer cell is in the range of thousands and these molecules can be present in homoplasmic or heteroplasmic states. Because the inheritance of mtDNA is maternal with no or minimal intermolecular recombination events [163], the vast majority of mtDNA copies are identical (either wild-type or mutant) at birth, which is called a state of homoplasmy. As mitochondria are the key source of ROS, mtDNA is highly susceptible to damage and mutations from ROS, and as a result multiple subpopulations of mtDNA could exist, creating a heteroplasmic condition in cancer cells [113]. Thus, heteroplasmic state could arise in cells during their life span but most breast tumors whether mitochondrial MnlI restriction site variants or mitochondrial microsatellite instability (MSI) were reported to be homoplasmic [164]. Other groups have also provided evidence that most mtDNA mutations are homoplasmic [66]. For example, Tan et al. [23] identified 27 mutations in 19 tumor samples. Of the 27 mutations, 17 were homoplasmic and the remaining 10 were heteroplasmic. It is unclear how certain mutations are selected towards homoplasmic state during breast cancer development. Because cancer is an age-related disease and takes a long time to develop, it is reasonable to hypothesize that during the course of breast tumorigenesis a particular mutation may be selected and propagated through multiple cell divisions, creating a homoplasmic condition. Two main hypotheses have been proposed to explain the dominance of homoplasmy over heteroplasmy [66, 165–167]. In the first, some of the mtDNA mutations confer a selective growth advantage, allowing the mtDNA with that particular mutation to outgrow others in the mitochondrial genome. Ultimately, cells harboring this selective advantageous mtDNA mutation overpopulate the tumor [166, 168]. In the second hypothesis, mtDNA with the selective mutant over-replicates to maintain OXPHOS function and over multiple cycles of cell division the over-replicated mtDNA replaces the original mtDNA in cells and tissues thus creating a homoplasmic condition [24, 66, 67, 166]. During the course of multiple generations mtDNA may acquire various mutations, some of which may not be advantageous for cell survival and will be eliminated. Mutations in the mtDNA that complement the nuclear genome function may dominate and lead to homoplasmy [169]. Apoptotic sensitivity of proliferating cells is often reduced by impaired OXPHOS [149, 170–172]. Therefore, it is possible that mtDNA mutations impairing OXPHOS system may be positively selected because they will be relatively protected from apoptosis, a key mechanism of cellular culling that provides protection against tumorigenesis.
Conclusions and future perspectives
MtDNA plays an important role in regulating OXPHOS function. Defective OXPHOS will lead to production of ROS, which may enhance cell transformation and ultimately lead to tumor initiation, promotion, and progression. It has also been suggested that the function of an early moderate increase in ROS is to provide protection, which could lead to enhanced survival of cancer cells [173]. Various proapoptotic BH3-only and antiapoptotic Bcl-2 family proteins also regulate glycolysis and respiration [174], thus mtDNA-mediated OXPHOS defects could lead to suppression of apoptosis in cancer cells. Although based on our current knowledge, we can not conclude that mtDNA mutation is a determining factor in breast tumorigenesis, the evidence suggests that OXPHOS dysfunction caused by mtDNA mutations and/or reduced mtDNA copy numbers might suppress p53 function [141], and result in breast tumorigenesis. Additionally, OXPHOS deficiency is known to cause constitutive activation of AKT that is linked with tumorigenesis [175]. Current findings using an animal model derived from inactivation of SUV3, a mitochondrial helicase, suggest that increased mtDNA mutations and decreased mtDNA copy numbers can predispose mice to tumor development in various tissues [5]. Germline introduction of a tumor-associated somatic G13997A mutation in ND6 subunit of complex I predispose mice to lymphoma formation and enhanced metastasis [176]. Future technical advances may allow us to elucidate the effect of key mtDNA mutations and mtDNA copy numbers on OXPHOS dysfunction and cancer, including breast tumorigenesis. Polymorphism in mtDNA has been detected in certain populations. Therefore, it is tempting to assume that the presence of germline mtDNA polymorphisms/mutations might predict susceptibility to developing breast cancer. Since somatic mtDNA mutation is an early event during breast tumorigenesis, such mutations could be used as early detection marker for breast tumor formation and might also predict disease outcome. Although further studies with large data sets and more sensitive methods are needed to determine whether mtDNA mutations and/or mtDNA content in biofluids could be used as biomarkers for early detection of breast cancer, the followings points should be considered with regard to biomarker development. While multiple germline and somatic mutations have been detected, there is no consensus to conclusively identify that a particular mutation is a potential biomarker for breast cancer. Based on the accumulating evidence and our current understanding, reduced mtDNA content in breast tumor compared with matched normal breast tissues could be a viable biomarker for early detection, although available studies related to mtDNA content in biological fluids such as blood, urine, and NAF are not conclusive. Therefore, a reasonable approach may be to combine mtDNA mutations in biolfluids such as blood or NAF with mtDNA content analysis of biopsy tissues for early detection of breast cancer.
Acknowledgments
We thank Dr. Nagendra Yadava for critical reading of this manuscript. We highly appreciate Dr. Keshav Singh for discussion on the early version of manuscript. This work was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA160685 and K01CA123142; and the American Cancer Society Research Scholar Grant RSG-12-214-01 – CCG to DC; and the National Cancer Institute Center Support Grant P30 CA016056 to the Roswell Park Cancer Institute. We apologize to those colleagues whose publications inadvertently could not be cited.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Cancer Society. Cancer: Facts and Figures. 2013. [Google Scholar]
- 2.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes WD, Narod SA. Hereditary breast and ovarian cancer: epidemiology, genetics, screening and predictive testing. Clin Invest Med. 1995;18:473–483. [PubMed] [Google Scholar]
- 4.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 5.Chen PL, Chen CF, Chen Y, Guo XE, Huang CK, Shew JY, Reddick RL, Wallace DC, Lee WH. Mitochondrial genome instability resulting from SUV3 haploinsufficiency leads to tumorigenesis and shortened lifespan. Oncogene. 2013;32:1193–1201. doi: 10.1038/onc.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamarajugadda S, Stemboroski L, Cai Q, Simpson NE, Nayak S, Tan M, Lu J. Glucose oxidation modulates anoikis and tumor metastasis. Molecular and cellular biology. 2012;32:1893–1907. doi: 10.1128/MCB.06248-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putignani L, Raffa S, Pescosolido R, Rizza T, Del Chierico F, Leone L, Aimati L, Signore F, Carrozzo R, Callea F, Torrisi MR, Grammatico P. Preliminary evidences on mitochondrial injury and impaired oxidative metabolism in breast cancer. Mitochondrion. 2012;12:363–369. doi: 10.1016/j.mito.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Cook CC, Kim A, Terao S, Gotoh A, Higuchi M. Consumption of oxygen: a mitochondrial-generated progression signal of advanced cancer. Cell death & disease. 2012;3:e258. doi: 10.1038/cddis.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owens KM, Kulawiec M, Desouki MM, Vanniarajan A, Singh KK. Impaired OXPHOS complex III in breast cancer. PloS one. 2011;6:e23846. doi: 10.1371/journal.pone.0023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayeed A, Meng Z, Luciani G, Chen LC, Bennington JL, Dairkee SH. Negative regulation of UCP2 by TGFbeta signaling characterizes low and intermediate-grade primary breast cancer. Cell death & disease. 2010;1:e53. doi: 10.1038/cddis.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaipparettu BA, Ma Y, Wong LJ. Functional effects of cancer mitochondria on energy metabolism and tumorigenesis: utility of transmitochondrial cybrids. Annals of the New York Academy of Sciences. 2010;1201:137–146. doi: 10.1111/j.1749-6632.2010.05621.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solazzo M, Fantappie O, D’Amico M, Sassoli C, Tani A, Cipriani G, Bogani C, Formigli L, Mazzanti R. Mitochondrial expression and functional activity of breast cancer resistance protein in different multiple drug-resistant cell lines. Cancer Res. 2009;69:7235–7242. doi: 10.1158/0008-5472.CAN-08-4315. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Bai RK, Trieu R, Wong LJ. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta. 2010;1797:29–37. doi: 10.1016/j.bbabio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L, Wei J, Chen T, He J, Qu J, He X, Jiang L, Qu Y, Fang H, Chen G, Lu J, Bai Y. Evaluating mitochondrial DNA in patients with breast cancer and benign breast disease. J Cancer Res Clin Oncol. 2011;137:669–675. doi: 10.1007/s00432-010-0912-x. [DOI] [PubMed] [Google Scholar]
- 18.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. Journal of human genetics. 2009;54:516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M, Shi Y, Wei X, Yang Y, Zang F, Niu R. Mitochondrial DNA depletion promotes impaired oxidative status and adaptive resistance to apoptosis in T47D breast cancer cells. Eur J Cancer Prev. 2009;18:445–457. doi: 10.1097/CEJ.0b013e32832f9bd6. [DOI] [PubMed] [Google Scholar]
- 20.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer biology & therapy. 2009;8:1378–1385. doi: 10.4161/cbt.8.14.8751. [DOI] [PubMed] [Google Scholar]
- 21.Gochhait S, Bhatt A, Sharma S, Singh YP, Gupta P, Bamezai RN. Concomitant presence of mutations in mitochondrial genome and p53 in cancer development - a study in north Indian sporadic breast and esophageal cancer patients. Int J Cancer. 2008;123:2580–2586. doi: 10.1002/ijc.23817. [DOI] [PubMed] [Google Scholar]
- 22.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62:972–976. [PubMed] [Google Scholar]
- 24.Hofhaus G, Gattermann N. Mitochondria harbouring mutant mtDNA--a cuckoo in the nest? Biological chemistry. 1999;380:871–877. doi: 10.1515/BC.1999.107. [DOI] [PubMed] [Google Scholar]
- 25.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 27.Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutrition & metabolism. 2010;7:7. doi: 10.1186/1743-7075-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 29.Koopman WJ, Distelmaier F, Smeitink JA, Willems PH. OXPHOS mutations and neurodegeneration. The EMBO journal. 2013;32:9–29. doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2011;1807:620–625. doi: 10.1016/j.bbabio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landazuri MO, Enriquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell metabolism. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 290(1981):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 33.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 34.Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY, Wong LC, Nagley P, Ngan HY. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Human mutation. 2003;22:173–174. doi: 10.1002/humu.10244. [DOI] [PubMed] [Google Scholar]
- 35.Herrmann PC, Gillespie JW, Charboneau L, Bichsel VE, Paweletz CP, Calvert VS, Kohn EC, Emmert-Buck MR, Liotta LA, Petricoin EF., 3rd Mitochondrial proteome: altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics. 2003;3:1801–1810. doi: 10.1002/pmic.200300461. [DOI] [PubMed] [Google Scholar]
- 36.Krieg RC, Knuechel R, Schiffmann E, Liotta LA, Petricoin EF, 3rd, Herrmann PC. Mitochondrial proteome: cancer-altered metabolism associated with cytochrome c oxidase subunit level variation. Proteomics. 2004;4:2789–2795. doi: 10.1002/pmic.200300796. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Zaera M, Abril J, Gonzalez L, Aguilo F, Condom E, Nadal M, Nunes V. Identification of somatic and germline mitochondrial DNA sequence variants in prostate cancer patients. Mutat Res. 2006;595:42–51. doi: 10.1016/j.mrfmmm.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Yin PH, Wu CC, Lin JC, Chi CW, Wei YH, Lee HC. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion. 2010;10:174–182. doi: 10.1016/j.mito.2009.12.147. [DOI] [PubMed] [Google Scholar]
- 39.Tzen CY, Mau BL, Wu TY. ND4 mutation in transitional cell carcinoma: Does mitochondrial mutation occur before tumorigenesis? Mitochondrion. 2007;7:273–278. doi: 10.1016/j.mito.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harbor symposia on quantitative biology. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 42.Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod. 2000;15(Suppl 2):11–17. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 43.Arnberg A, van Bruggen EF, Borst P. The presence of DNA molecules with a displacement loop in standard mitochondrial DNA preparations. Biochim Biophys Acta. 1971;246:353–357. doi: 10.1016/0005-2787(71)90147-x. [DOI] [PubMed] [Google Scholar]
- 44.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc Natl Acad Sci U S A. 1971;68:2252–2257. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higuchi M. Regulation of mitochondrial DNA content and cancer. Mitochondrion. 2007;7:53–57. doi: 10.1016/j.mito.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 47.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, Reichelt S, Spelbrink JN, Walker JE, Holt IJ. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 50.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo DK, Green PD, Santos JH, D’Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. The American journal of pathology. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benny Abraham Kaipparettu YM, Park Junhyoung, Lee Tin-Lap, Zhang Yiqun, Patricia Yotnda CJC, Chan Wai-Yee, Wong Lee-Jun C. Cross talk from noncancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PloS one. 2013 doi: 10.1371/journal.pone.0061747. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 55.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 57.Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. 2002;364:309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakashima RA, Paggi MG, Pedersen PL. Contributions of glycolysis and oxidative phosphorylation to adenosine 5′-triphosphate production in AS-30D hepatoma cells. Cancer Res. 1984;44:5702–5706. [PubMed] [Google Scholar]
- 59.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 60.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 61.Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, Dusetti NJ, Loncle C, Calvo E, Turrini O, Iovanna JL, Tomasini R, Vasseur S. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiche J, Brahimi-Horn MC, Pouyssegur J. Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. Journal of cellular and molecular medicine. 2010;14:771–794. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell metabolism. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakajima EC, Van Houten B. Metabolic symbiosis in cancer: refocusing the Warburg lens. Molecular carcinogenesis. 2013;52:329–337. doi: 10.1002/mc.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z, Ogasawara M, Keating MJ, Kondo S, Huang P. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286:32843–32853. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 67.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 68.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu M. Somatic mitochondrial DNA mutations in human cancers. Advances in clinical chemistry. 2012;57:99–138. doi: 10.1016/b978-0-12-394384-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 70.Boultwood J, Fidler C, Mills KI, Frodsham PM, Kusec R, Gaiger A, Gale RE, Linch DC, Littlewood TJ, Moss PA, Wainscoat JS. Amplification of mitochondrial DNA in acute myeloid leukaemia. Br J Haematol. 1996;95:426–431. doi: 10.1046/j.1365-2141.1996.d01-1922.x. [DOI] [PubMed] [Google Scholar]
- 71.Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61:5998–6001. [PubMed] [Google Scholar]
- 72.Nagy A, Wilhelm M, Sukosd F, Ljungberg B, Kovacs G. Somatic mitochondrial DNA mutations in human chromophobe renal cell carcinomas. Genes Chromosomes Cancer. 2002;35:256–260. doi: 10.1002/gcc.10118. [DOI] [PubMed] [Google Scholar]
- 73.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 74.Debniak T, Scott RJ, Huzarski T, Byrski T, Masojc B, van de Wetering T, Serrano-Fernandez P, Gorski B, Cybulski C, Gronwald J, Debniak B, Maleszka R, Kladny J, Bieniek A, Nagay L, Haus O, Grzybowska E, Wandzel P, Niepsuj S, Narod SA, Lubinski J. XPD common variants and their association with melanoma and breast cancer risk. Breast Cancer Res Treat. 2006;98:209–215. doi: 10.1007/s10549-005-9151-2. [DOI] [PubMed] [Google Scholar]
- 75.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–68. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alhomidi MA, Vedicherla B, Movva S, Rao PK, Ahuja YR, Hasan Q. Mitochondrial D310 instability in Asian Indian breast cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013 doi: 10.1007/s13277-013-0793-0. [DOI] [PubMed] [Google Scholar]
- 77.Imanishi H, Hattori K, Wada R, Ishikawa K, Fukuda S, Takenaga K, Nakada K, Hayashi J. Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PloS one. 2011;6:e23401. doi: 10.1371/journal.pone.0023401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 79.Tipirisetti NR, Lakshmi RK, Govatati S, Govatati S, Vuree S, Singh L, Raghunadha Rao D, Bhanoori M, Vishnupriya S. Mitochondrial genome variations in advanced stage breast cancer: A case-control study. Mitochondrion. 2013 doi: 10.1016/j.mito.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Ye C, Shu XO, Pierce L, Wen W, Courtney R, Gao YT, Zheng W, Cai Q. Mutations in the mitochondrial DNA D-loop region and breast cancer risk. Breast Cancer Res Treat. 2010;119:431–436. doi: 10.1007/s10549-009-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai FF, Kohler C, Zhang B, Chen WJ, Barekati Z, Garritsen HS, Lenner P, Toniolo P, Zhang JJ, Zhong XY. Mutations of mitochondrial DNA as potential biomarkers in breast cancer. Anticancer research. 2011;31:4267–4271. [PubMed] [Google Scholar]
- 82.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 83.Tseng LM, Yin PH, Yang CW, Tsai YF, Hsu CY, Chi CW, Lee HC. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer. 2011;50:800–811. doi: 10.1002/gcc.20901. [DOI] [PubMed] [Google Scholar]
- 84.Kulawiec M, Safina A, Desouki MM, Still I, Matsui S, Bakin A, Singh KK. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer biology & therapy. 2008;7:1732–1743. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alhomidi MA, Vedicherla B, Movva S, Rao PK, Ahuja YR, Hasan Q. Mitochondrial D310 instability in Asian Indian breast cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:2427–2432. doi: 10.1007/s13277-013-0793-0. [DOI] [PubMed] [Google Scholar]
- 86.Bai RK, Chang J, Yeh KT, Lou MA, Lu JF, Tan DJ, Liu H, Wong LJ. Mitochondrial DNA content varies with pathological characteristics of breast cancer. Journal of oncology. 2011;2011:496189. doi: 10.1155/2011/496189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Czarnecka AM, Krawczyk T, Plak K, Klemba A, Zdrozny M, Arnold RS, Kofler B, Golik P, Szybinska A, Lubinski J, Mossakowska M, Bartnik E, Petros JA. Mitochondrial genotype and breast cancer predisposition. Oncol Rep. 2010;24:1521–1534. doi: 10.3892/or_00001014. [DOI] [PubMed] [Google Scholar]
- 88.De Vitto H, Mendonca BS, Elseth KM, Vesper BJ, Portari EA, Gallo CV, Paradise WA, Rumjanek FD, Radosevich JA. Part II. Mitochondrial mutational status of high nitric oxide adapted cell line BT-20 (BT-20-HNO) as it relates to human primary breast tumors. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:337–347. doi: 10.1007/s13277-012-0555-4. [DOI] [PubMed] [Google Scholar]
- 89.Fang H, Shen L, Chen T, He J, Ding Z, Wei J, Qu J, Chen G, Lu J, Bai Y. Cancer type-specific modulation of mitochondrial haplogroups in breast, colorectal and thyroid cancer. BMC cancer. 2010;10:421. doi: 10.1186/1471-2407-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutierrez Povedano C, Salgado J, Gil C, Robles M, Patino-Garcia A, Garcia-Foncillas J. Analysis of BRCA1 and mtDNA haplotypes and mtDNA polymorphism in familial breast cancer. Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.825773. [DOI] [PubMed] [Google Scholar]
- 91.Kuo SJ, Chen M, Ma GC, Chen ST, Chang SP, Lin WY, Chen YC, Lee TH, Lin TT, Liu CS. Number of somatic mutations in the mitochondrial D-loop region indicates poor prognosis in breast cancer, independent of TP53 mutation. Cancer genetics and cytogenetics. 2010;201:94–101. doi: 10.1016/j.cancergencyto.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Nie H, Shu H, Vartak R, Milstein AC, Mo Y, Hu X, Fang H, Shen L, Ding Z, Lu J, Bai Y. Mitochondrial common deletion, a potential biomarker for cancer occurrence, is selected against in cancer background: a meta-analysis of 38 studies. PloS one. 2013;8:e67953. doi: 10.1371/journal.pone.0067953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rahmani B, Azimi C, Omranipour R, Raoofian R, Zendehdel K, Saee-Rad S, Heidari M. Mutation screening in the mitochondrial D-loop region of tumoral and non-tumoral breast cancer in Iranian patients. Acta medica Iranica. 2012;50:447–453. [PubMed] [Google Scholar]
- 94.Rohan TE, Wong LJ, Wang T, Haines J, Kabat GC. Do alterations in mitochondrial DNA play a role in breast carcinogenesis? Journal of oncology. 2010;2010:604304. doi: 10.1155/2010/604304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sultana GN, Rahman A, Shahinuzzaman AD, Begum RA, Hossain CF. Mitochondrial DNA mutations---candidate biomarkers for breast cancer diagnosis in Bangladesh. Chinese journal of cancer. 2012;31:449–454. doi: 10.5732/cjc.012.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tengku Baharudin N, Jaafar H, Zainuddin Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. The Malaysian journal of medical sciences: MJMS. 2012;19:36–42. [PMC free article] [PubMed] [Google Scholar]
- 97.Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh WP, Yuan JM. Mitochondrial DNA copy number is associated with breast cancer risk. PloS one. 2013;8:e65968. doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tipirisetti NR, Lakshmi RK, Govatati S, Govatati S, Vuree S, Singh L, Raghunadha Rao D, Bhanoori M, Vishnupriya S. Mitochondrial genome variations in advanced stage breast cancer: a case-control study. Mitochondrion. 2013;13:372–378. doi: 10.1016/j.mito.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 99.Xu C, Tran-Thanh D, Ma C, May K, Jung J, Vecchiarelli J, Done SJ. Mitochondrial D310 mutations in the early development of breast cancer. Br J Cancer. 2012;106:1506–1511. doi: 10.1038/bjc.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–152. doi: 10.1093/carcin/bgh282. [DOI] [PubMed] [Google Scholar]
- 101.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 102.Grzybowska-Szatkowska L, Slaska B. Polymorphisms in genes encoding mt-tRNA in female breast cancer in Poland. Mitochondrial DNA. 2012;23:106–111. doi: 10.3109/19401736.2012.660925. [DOI] [PubMed] [Google Scholar]
- 103.Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, Yagi T, Felding-Habermann B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. The Journal of clinical investigation. 2013;123:1068–1081. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 105.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. Journal of carcinogenesis. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh KK. Mitochondria damage checkpoint, aging, and cancer. Annals of the New York Academy of Sciences. 2006;1067:182–190. doi: 10.1196/annals.1354.022. [DOI] [PubMed] [Google Scholar]
- 107.Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PloS one. 2013;8:e61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer biology & therapy. 2005;4:1367–1373. doi: 10.4161/cbt.4.12.2233. [DOI] [PubMed] [Google Scholar]
- 110.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, Matsui S. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–146. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 111.Kulawiec M, Arnouk H, Desouki MM, Kazim L, Still I, Singh KK. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer. Cancer biology & therapy. 2006;5:967–975. doi: 10.4161/cbt.5.8.2880. [DOI] [PubMed] [Google Scholar]
- 112.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell research. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila) 2011;4:638–654. doi: 10.1158/1940-6207.CAPR-10-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Liu VW, Tsang PC, Chiu PM, Cheung AN, Khoo US, Nagley P, Ngan HY. Microsatellite instability in mitochondrial genome of common female cancers. Int J Gynecol Cancer. 2006;16(Suppl 1):259–266. doi: 10.1111/j.1525-1438.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 115.Hall IJ, Moorman PG, Millikan RC, Newman B. Comparative analysis of breast cancer risk factors among African-American women and White women. Am J Epidemiol. 2005;161:40–51. doi: 10.1093/aje/kwh331. [DOI] [PubMed] [Google Scholar]
- 116.Setiawan VW, Chu LH, John EM, Ding YC, Ingles SA, Bernstein L, Press MF, Ursin G, Haiman CA, Neuhausen SL. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer genetics and cytogenetics. 2008;181:16–19. doi: 10.1016/j.cancergencyto.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Francis A, Pooja S, Rajender S, Govindaraj P, Tipirisetti NR, Surekha D, Rao DR, Rao L, Ramachandra L, Vishnupriya S, Ramalingam K, Satyamoorthy K, Thangaraj K. A mitochondrial DNA variant 10398G>A in breast cancer among South Indians: An original study with meta-analysis. Mitochondrion. 2013;13:559–565. doi: 10.1016/j.mito.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 118.Czarnecka AM, Krawczyk T, Zdrozny M, Lubinski J, Arnold RS, Kukwa W, Scinska A, Golik P, Bartnik E, Petros JA. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res Treat. 2010;121:511–518. doi: 10.1007/s10549-009-0358-5. [DOI] [PubMed] [Google Scholar]
- 119.Covarrubias D, Bai RK, Wong LJ, Leal SM. Mitochondrial DNA variant interactions modify breast cancer risk. Journal of human genetics. 2008;53:924–928. doi: 10.1007/s10038-008-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pezzotti A, Kraft P, Hankinson SE, Hunter DJ, Buring J, Cox DG. The mitochondrial A10398G polymorphism, interaction with alcohol consumption, and breast cancer risk. PloS one. 2009;4:e5356. doi: 10.1371/journal.pone.0005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhat A, Koul A, Sharma S, Rai E, Bukhari SI, Dhar MK, Bamezai RN. The possible role of 10398A and 16189C mtDNA variants in providing susceptibility to T2DM in two North Indian populations: a replicative study. Hum Genet. 2007;120:821–826. doi: 10.1007/s00439-006-0272-4. [DOI] [PubMed] [Google Scholar]
- 122.Czarnecka AM, Klemba A, Krawczyk T, Zdrozny M, Arnold RS, Bartnik E, Petros JA. Mitochondrial NADH-dehydrogenase polymorphisms as sporadic breast cancer risk factor. Oncol Rep. 2010;23:531–535. [PubMed] [Google Scholar]
- 123.Fendt L, Niederstatter H, Huber G, Zelger B, Dunser M, Seifarth C, Rock A, Schafer G, Klocker H, Parson W. Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue microdissection. Breast Cancer Res Treat. 2011;128:327–336. doi: 10.1007/s10549-010-1092-8. [DOI] [PubMed] [Google Scholar]
- 124.Barekati Z, Radpour R, Kohler C, Zhang B, Toniolo P, Lenner P, Lv Q, Zheng H, Zhong XY. Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum Mol Genet. 2010;19:2936–2946. doi: 10.1093/hmg/ddq199. [DOI] [PubMed] [Google Scholar]
- 125.Platek ME, Shields PG, Tan D, Marian C, Bonner MR, McCann SE, Nie J, Wilding GE, Ambrosone C, Millen AE, Trevisan M, Russell M, Nochajski TH, Edge SB, Winston J, Freudenheim JL. Alcohol consumption and breast tumor mitochondrial DNA mutations. Breast Cancer Res Treat. 2010;121:453–460. doi: 10.1007/s10549-009-0587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang CY, Wang HW, Yao YG, Kong QP, Zhang YP. Somatic mutations of mitochondrial genome in early stage breast cancer. Int J Cancer. 2007;121:1253–1256. doi: 10.1002/ijc.22822. [DOI] [PubMed] [Google Scholar]
- 127.Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- 128.Barekati Z, Radpour R, Kohler C, Zhang B, Toniolo P, Lenner P, Lv Q, Zheng H, Zhong XY. Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum Mol Genet. 19:2936–2946. doi: 10.1093/hmg/ddq199. [DOI] [PubMed] [Google Scholar]
- 129.Jakupciak JP, Dakubo GD, Maragh S, Parr RL. Analysis of potential cancer biomarkers in mitochondrial DNA. Curr Opin Mol Ther. 2006;8:500–506. [PubMed] [Google Scholar]
- 130.Zhu W, Qin W, Sauter ER. Large-scale mitochondrial DNA deletion mutations and nuclear genome instability in human breast cancer. Cancer Detect Prev. 2004;28:119–126. doi: 10.1016/j.cdp.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 131.Ye C, Shu XO, Pierce L, Wen W, Courtney R, Gao YT, Zheng W, Cai Q. Mutations in the mitochondrial DNA D-loop region and breast cancer risk. Breast Cancer Res Treat. 119:431–436. doi: 10.1007/s10549-009-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bianchi MS, Bianchi NO, Bailliet G. Mitochondrial DNA mutations in normal and tumor tissues from breast cancer patients. Cytogenet Cell Genet. 1995;71:99–103. doi: 10.1159/000134072. [DOI] [PubMed] [Google Scholar]
- 133.Ye C, Shu XO, Wen W, Pierce L, Courtney R, Gao YT, Zheng W, Cai Q. Quantitative analysis of mitochondrial DNA 4977-bp deletion in sporadic breast cancer and benign breast diseases. Breast Cancer Res Treat. 2008;108:427–434. doi: 10.1007/s10549-007-9613-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maggrah A, Robinson K, Creed J, Wittock R, Gehman K, Gehman T, Brown H, Harbottle A, Froberg MK, Klein D, Reguly B, Parr R. Paired ductal carcinoma in situ and invasive breast cancer lesions in the D-loop of the mitochondrial genome indicate a cancerization field effect. BioMed research international. 2013;2013:379438. doi: 10.1155/2013/379438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jakupciak JP, Maggrah A, Maragh S, Maki J, Reguly B, Maki K, Wittock R, Robinson K, Wagner PD, Thayer RE, Gehman K, Gehman T, Srivastava S, Ngom A, Dakubo GD, Parr RL. Facile whole mitochondrial genome resequencing from nipple aspirate fluid using MitoChip v2.0. BMC cancer. 2008;8:95. doi: 10.1186/1471-2407-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Parra MD, Martinez de Morentin BE, Martinez JA. Postprandial insulin response and mitochondrial oxidation in obese men nutritionally treated to lose weight. European journal of clinical nutrition. 2005;59:334–340. doi: 10.1038/sj.ejcn.1602078. [DOI] [PubMed] [Google Scholar]
- 137.Isaacs C, Cavalli LR, Cohen Y, Pennanen M, Shankar LK, Freedman M, Singh B, Liu M, Gallagher A, Rone JD, Dickson RB, Sidransky D, Haddad BR. Detection of LOH and mitochondrial DNA alterations in ductal lavage and nipple aspirate fluids from hngh-risk patients. Breast Cancer Res Treat. 2004;84:99–105. doi: 10.1023/B:BREA.0000018406.03679.2e. [DOI] [PubMed] [Google Scholar]
- 138.Yadava N, Schneider SS, Jerry DJ, Kim C. Impaired mitochondrial metabolism and mammary carcinogenesis. Journal of mammary gland biology and neoplasia. 2013;18:75–87. doi: 10.1007/s10911-012-9271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P. Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat. 2008;110:439–452. doi: 10.1007/s10549-007-9738-x. [DOI] [PubMed] [Google Scholar]
- 140.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. Journal of human genetics. 2009;54:647–654. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Compton S, Kim C, Griner NB, Potluri P, Scheffler IE, Sen S, Jerry DJ, Schneider S, Yadava N. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function. J Biol Chem. 2011;286:20297–20312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Larman TC, DePalma SR, Hadjipanayis AG, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109:14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D. Tumor-specific changes in mtDNA content in human cancer. Int J Cancer. 2005;116:920–924. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- 145.Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 146.Hsu CW, Yin PH, Lee HC, Chi CW, Tseng LM. Mitochondrial DNA content as a potential marker to predict response to anthracycline in breast cancer patients. The breast journal. 2010;16:264–270. doi: 10.1111/j.1524-4741.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- 147.Xia P, An HX, Dang CX, Radpour R, Kohler C, Fokas E, Engenhart-Cabillic R, Holzgreve W, Zhong XY. Decreased mitochondrial DNA content in blood samples of patients with stage I breast cancer. BMC cancer. 2009;9:454. doi: 10.1186/1471-2407-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 149.Chandra D, Liu JW, Tang DG. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J Biol Chem. 2002;277:50842–50854. doi: 10.1074/jbc.M207622200. [DOI] [PubMed] [Google Scholar]
- 150.Xiao D, Powolny AA, Singh SV. Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger reactive oxygen species-dependent apoptosis in human breast cancer cells. J Biol Chem. 2008;283:30151–30163. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fan AX, Radpour R, Haghighi MM, Kohler C, Xia P, Hahn S, Holzgreve W, Zhong XY. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J Cancer Res Clin Oncol. 2009;135:983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PubMed] [Google Scholar]
- 152.Koochekpour S, Marlowe T, Singh KK, Attwood K, Chandra D. Reduced Mitochondrial DNA Content Associates with Poor Prognosis of Prostate Cancer in African American Men. PloS one. 2013;8:e74688. doi: 10.1371/journal.pone.0074688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. American journal of physiology Endocrinology and metabolism. 2004;286:E1011–1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- 154.Chen JQ, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochim Biophys Acta. 2005;1746:1–17. doi: 10.1016/j.bbamcr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 155.Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim Biophys Acta. 2009;1793:1540–1570. doi: 10.1016/j.bbamcr.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yager JD, Chen JQ. Mitochondrial estrogen receptors--new insights into specific functions. Trends Endocrinol Metab. 2007;18:89–91. doi: 10.1016/j.tem.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 157.Konduri SD, Medisetty R, Liu W, Kaipparettu BA, Srivastava P, Brauch H, Fritz P, Swetzig WM, Gardner AE, Khan SA, Das GM. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci U S A. 2010;107:15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. The EMBO journal. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. Journal of genetics and genomics = Yi chuan xue bao. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annual review of cell biology. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 161.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. Herpes simplex virus eliminates host mitochondrial DNA. EMBO reports. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 163.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Richard SM, Bailliet G, Paez GL, Bianchi MS, Peltomaki P, Bianchi NO. Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res. 2000;60:4231–4237. [PubMed] [Google Scholar]
- 165.Cavelier L, Jazin E, Jalonen P, Gyllensten U. MtDNA substitution rate and segregation of heteroplasmy in coding and noncoding regions. Hum Genet. 2000;107:45–50. doi: 10.1007/s004390000305. [DOI] [PubMed] [Google Scholar]
- 166.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 167.Vega A, Salas A, Gamborino E, Sobrido MJ, Macaulay V, Carracedo A. mtDNA mutations in tumors of the central nervous system reflect the neutral evolution of mtDNA in populations. Oncogene. 2004;23:1314–1320. doi: 10.1038/sj.onc.1207214. [DOI] [PubMed] [Google Scholar]
- 168.Shay JW, Werbin H. Are mitochondrial DNA mutations involved in the carcinogenic process? Mutat Res. 1987;186:149–160. doi: 10.1016/0165-1110(87)90028-5. [DOI] [PubMed] [Google Scholar]
- 169.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chauvin C, De Oliveira F, Ronot X, Mousseau M, Leverve X, Fontaine E. Rotenone inhibits the mitochondrial permeability transition-induced cell death in U937 and KB cells. J Biol Chem. 2001;276:41394–41398. doi: 10.1074/jbc.M106417200. [DOI] [PubMed] [Google Scholar]
- 171.Higuchi M, Aggarwal BB, Yeh ET. Activation of CPP32-like protease in tumor necrosis factor-induced apoptosis is dependent on mitochondrial function. The Journal of clinical investigation. 1997;99:1751–1758. doi: 10.1172/JCI119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.McClintock DS, Santore MT, Lee VY, Brunelle J, Budinger GR, Zong WX, Thompson CB, Hay N, Chandel NS. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Molecular and cellular biology. 2002;22:94–104. doi: 10.1128/MCB.22.1.94-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Naviaux RK. Oxidative shielding or oxidative stress? The Journal of pharmacology and experimental therapeutics. 2012;342:608–618. doi: 10.1124/jpet.112.192120. [DOI] [PubMed] [Google Scholar]
- 174.Andersen JL, Kornbluth S. The tangled circuitry of metabolism and apoptosis. Molecular cell. 2013;49:399–410. doi: 10.1016/j.molcel.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, Zhang W, Plunkett W, Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hashizume O, Shimizu A, Yokota M, Sugiyama A, Nakada K, Miyoshi H, Itami M, Ohira M, Nagase H, Takenaga K, Hayashi J. Specific mitochondrial DNA mutation in mice regulates diabetes and lymphoma development. Proc Natl Acad Sci U S A. 2012;109:10528–10533. doi: 10.1073/pnas.1202367109. [DOI] [PMC free article] [PubMed] [Google Scholar]