Abstract
Dravet syndrome (DS) is a catastrophic pediatric epilepsy with severe intellectual disability, impaired social development and persistent drug-resistant seizures. One of its primary monogenic causes are mutations in Nav1.1 (SCN1A), a voltage-gated sodium channel. Here we characterise zebrafish Nav1.1 (scn1Lab) mutants originally identified in a chemical mutagenesis screen. Mutants exhibit spontaneous abnormal electrographic activity, hyperactivity and convulsive behaviors. Although scn1Lab expression is reduced, microarray analysis is remarkable for the small fraction of differentially expressed genes (~3%) and lack of compensatory expression changes in other scn subunits. Ketogenic diet, diazepam, valproate, potassium bromide and stiripentol attenuate mutant seizure activity; seven other antiepileptic drugs have no effect. A phenotype-based screen of 320 compounds identifies a US Food and Drug Administration-approved compound (clemizole) that inhibits convulsive behaviors and electrographic seizures. This approach represents a new direction in modeling pediatric epilepsy and could be used to identify novel therapeutics for any monogenic epilepsy disorder.
Epilepsy can be acquired as a result of an insult to the brain or genetic mutation. Among the genetic epilepsies more than 650 variants have been identified in the SCN1A gene1,2. Missense or frame-shift mutations in this gene are associated with generalized epilepsy with febrile seizures plus (GEFS+)3 as well as a more severe disorder known as Dravet syndrome. Children with DS initially exhibit normal development but often experience febrile seizure episodes within the first year of life with eventual progression to severe spontaneous recurrent seizures, intellectual disability, ataxia, and psychomotor dysfunction. Seizures are inadequately managed using available antiepileptic drugs (AEDs) and these children are poor candidates for neurosurgical resection4.
In mammalian brain there are four main subtypes of voltage-gated sodium channel alpha subunits: NaV1.1, NaV1.2, NaV1.3 and NaV1.6, encoded for by the genes SCN1A, SCN2A, SCN3A, and SCN8A, respectively. Opening of these channels produces a sodium conductance and rapid cell membrane depolarization e.g., features integral to action potential initiation5. In mice, Nav1.1 is widely expressed in the central nervous system including the axon initial segment of parvalbumin-positive hippocampal interneurons and excitatory principal cells7,8. Heterozygous deletion of Nav1.1 in mice leads to a reduction in the firing capability of acutely dissociated fast-spiking interneurons9. Mice with global or interneuron-specific heterozygous deletion of Nav1.1 exhibit temperature-induced and spontaneous seizures, mild ataxia, autism-like behaviors and premature death9–11. Knock-in mouse carrying a premature stop codon in domain III of the Nav1.1 channel also exhibit a decrement in spike amplitude during prolonged interneuron firing and increased sensitivity to temperature-induced seizures6.
Generation and characterization of valid animal models is critical to efforts to understand the pathophysiology of DS, and to aid in identification of novel therapies. While considerable attention has focused on modeling SCN1A mutations in mice these animals have proven difficult to breed and epilepsy phenotypes are strongly influenced by background strain genetics. Induced pluripotent stem cells can be generated from DS patients but individual neurons do not recapitulate the network environment necessary for in vivo seizure generation. Danio rerio (zebrafish), a simple vertebrate species, provide an alternative model system with significant advantages for genetic manipulation, cost-efficient breeding and in vivo drug discovery12–14. Ideally, an animal model should be based on a known genetic cause of the disease (SCN1A mutation), accurately recapitulate key features of the disease (epilepsy), and respond, or not, to therapies commonly used in patients with the disease (pharmacological validation). If successful, such a model could inform our understanding of the disease process and catalyze explorations toward new therapies. In zebrafish, the voltage-gated sodium channel family consists of four sets of duplicated genes: scn1Laa & scn1Lab, scn4aa & scn4ab, scn5Laa & scn5Lab, and scn8aa & scn8ab15. The zebrafish scn1Lab gene shares a 77% identity with human SCN1A and is expressed in the central nervous system. A homozygous zebrafish mutant for this gene (originally termed didys552) was discovered in a chemical mutagenesis screen using the optokinetic response as an assay16. These types of screens are based on inducing random point mutations using the alkylating agent N-ethyl-N-nitrosourea (ENU), resulting mutations are typically loss-of-function and recessive. Although this is a homozygous mutation, scn1Lab zebrafish mutants are relevant for the autosomal dominant human Dravet Syndrome given the genome duplication in zebrafish and the presence of an additional Nav1.1 homologue (scn1Laa). In this manuscript, we characterized scn1Lab mutants at the molecular and behavioral level, demonstrated that mutants exhibit spontaneous drug-resistant seizures, and then used them in a novel high-throughput screening program to identify compounds that ameliorate the epilepsy phenotype. A phenotype-based screen identified clemizole, an FDA-approved compound, as an effective inhibitor of spontaneous convulsive behaviors and electrographic seizures in these mutants.
Results
Developmental scn1Lab expression and characterization
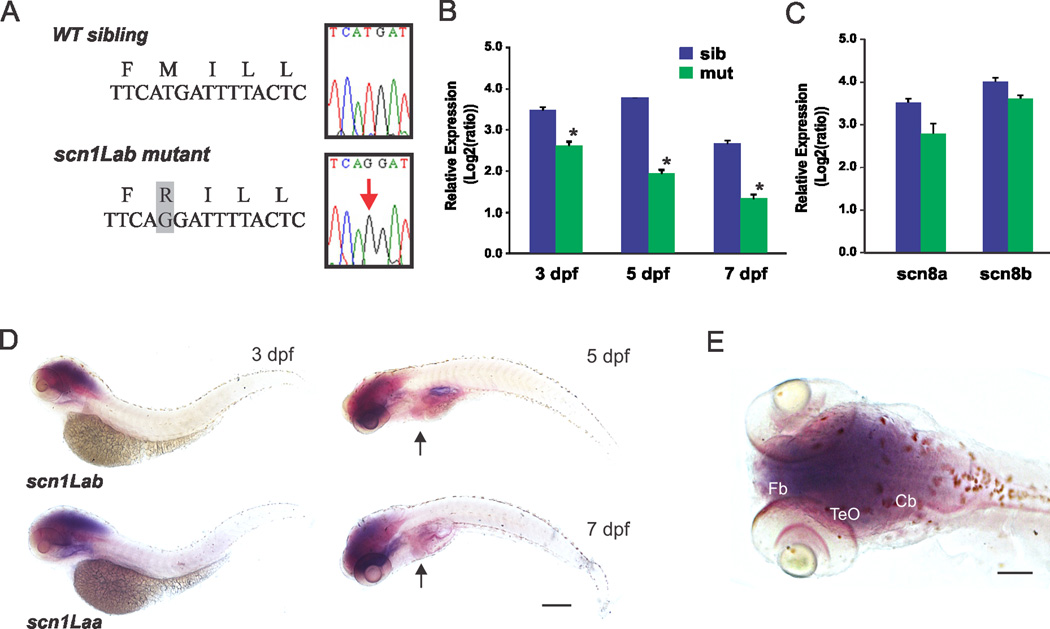
Zebrafish with a mutation in domain III of a voltage-gated sodium channel were identified by Dr. Herwig Baier during a chemical mutagenesis screen16. We backcrossed original scn1Lab mutants onto the Tupfel long (TL) background for 7–10 generations and confirmed a methionine (M) to arginine (R) mutation in our colony (Fig. 1A). Reverse transcriptase (RT) and quantitative (q) PCR revealed a decrease in mRNA expression for scn1Lab in mutant larvae at 3, 5 and 7 days post-fertilization (dpf)(Fig. 1B); antibodies recognizing this protein in zebrafish are not available. As expected15, scn1Lab is prominently expressed during early stages of larval development (Fig. 1B) and specifically in the central nervous system at 3 dpf (Figs. 1D, E). Whole-mount in situ hybridization revealed diffuse but prominent expression in brain regions corresponding to forebrain (telencephalon), optic tectum and cerebellum. A similar expression pattern was observed for scn1Laa at 3 dpf. At 5 and 7 dpf, CNS expression remained prominent and faint scn1Lab signal was also noted in the heart (Fig. 1D). Relative expression of scn8aa or scn8ab (Nav1.6) e.g., a subunit thought to act as a genetic modifier of DS17, failed to reveal a significant difference in expression between mutants and sibling controls at 5 dpf (Fig. 1C). Similarly, microarray analysis at 5 dpf also failed to detect a compensatory change in the mRNA expression of thirteen different zebrafish scn subunits (Table I) including the other homolog (scn1Laa). These results demonstrate a selective defect in a zebrafish Nav1.1 gene expressed in the CNS during early development.
Figure 1. Molecular characterization of scn1Lab zebrafish mutants.
(A) Sequencing confirmed a T-to-G mutation in scn1Lab mutant cDNA. (B) Verification of reduced expression in scn1Lab mutants compared to sibling controls at 3, 5 and 7 dpf using qPCR. Data presented as mean ± S.E.M; *significance taken as p < 0.05 student’s t-test. Data were normalized to internal reference gene β-actin. Values represent averages from five independent biological samples (1 sample = 10 pooled larvae) for each of the 3 developmental stages. Data presented as mean ± S.E.M; *significance taken as p < 0.05 student’s t-test. (C) Relative expression of scn8aa and scn8ab in Nav1.1 mutants (n = 5) and sibling controls (n = 5) at 5 dpf. Data presented as in B. (D) Whole-mount in situ hybridization for scn1Lab in larval zebrafish at 3, 5 and 7 dpf. Wild-type larvae are shown in lateral views; expression is shown in dark purple. Scn1Laa expression at 3 dpf is shown for comparison. Heart indicated by arrowheads in 5 and 7 dpf panels. (E) Dorsal view of scb1Laa expression at 3 dpf; note prominent expression in regions corresponding to the larval zebrafish CNS. Abbreviations: Tel, telencephalon; TeO, optic tectum; Cb, cerebellum. Scale bars = 0.35 mm in D, 0.2 mm in E.
Table I.
| Accession | Gene | Description | Fold | M |
|---|---|---|---|---|
| NM_200132 | scn1Laa | Danio rerio sodium channel, voltage-gated, type I, alpha, mRNA | 0.919 | −0.12 |
| NM_001077539 | scn1ba | Danio rerio sodium channel, voltage-gated, type I, beta a, mRNA | 1.293 | 0.37 |
| NM_001128156 | scn1bb | Danio rerio sodium channel, voltage-gated, type I, beta b, mRNA | 1.004 | 0.01 |
| NM_001077629 | scn2b | Danio rerio sodium channel, voltage-gated, type II, beta, mRNA | 1.032 | 0.05 |
| NM_001080802 | scn3b | Danio rerio sodium channel, voltage-gated, type III, beta, mRNA | 1.115 | 0.16 |
| NM_001039825 | scn4aa | Danio rerio sodium channel, voltage-gated, type IV, alpha a, mRNA | 1.063 | 0.09 |
| NM_001045065 | scn4ab | Danio rerio sodium channel, voltage-gated, type IV, alpha b, mRNA | 0.738 | −0.44 |
| NM_001077570 | scn4ba | Danio rerio sodium channel, voltage-gated, type IV, beta a, mRNA | 1.115 | 0.16 |
| NM_001077573 | scn4bb | Danio rerio sodium channel, voltage-gated, type IV, beta b, mRNA | 0.956 | −0.06 |
| NM_131628 | scn8aa | Danio rerio sodium channel, voltage-gated, type VIII, alpha a, mRNA | 1.351 | 0.43 |
| NM_001045183 | scn8ab | Danio rerio sodium channel, voltage-gated, type VIII, alpha b, mRNA | 1.246 | 0.32 |
| NM_001044922 | scn12aa | Danio rerio sodium channel, voltage gated, type XII, alpha a, mRNA | 1.123 | 0.17 |
| NM_001045123 | scn12ab | Danio rerio sodium channel, voltage gated, type XII, alpha b, mRNA | 1.003 | 0.00 |
Large-scale transcriptomic analysis of scn1Lab mutants
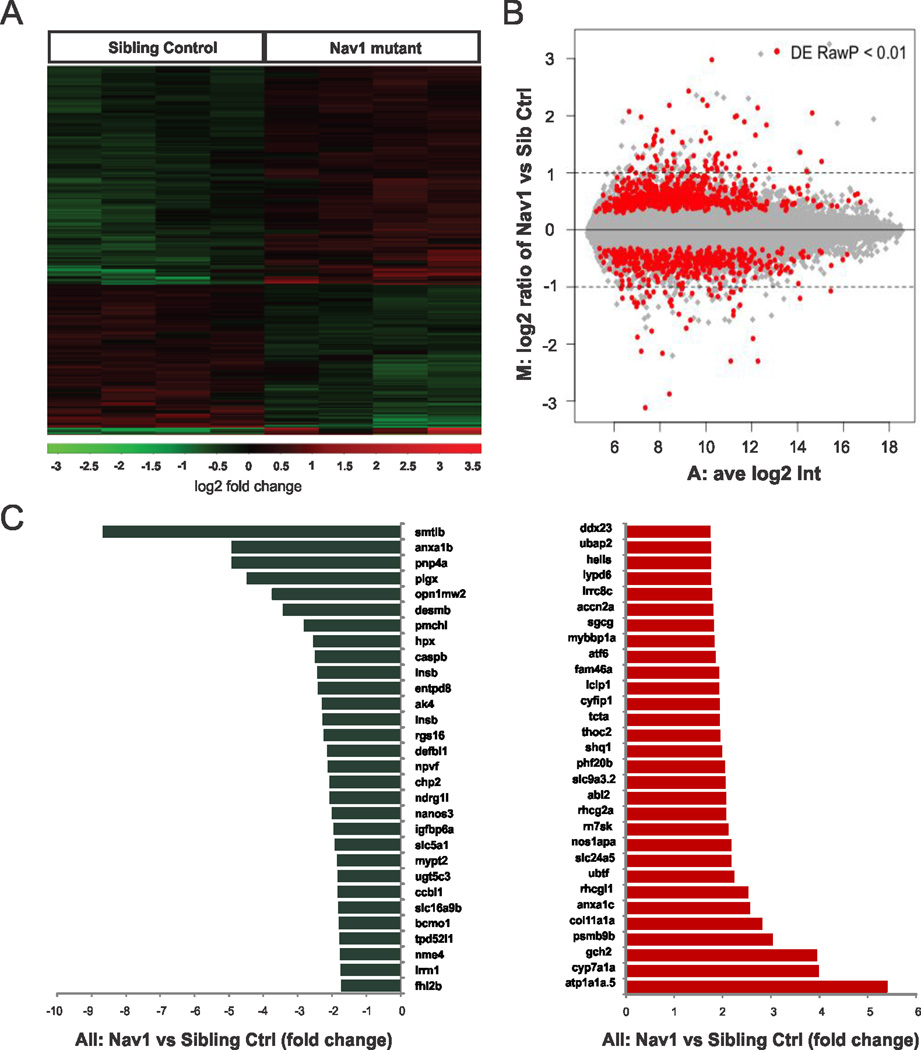
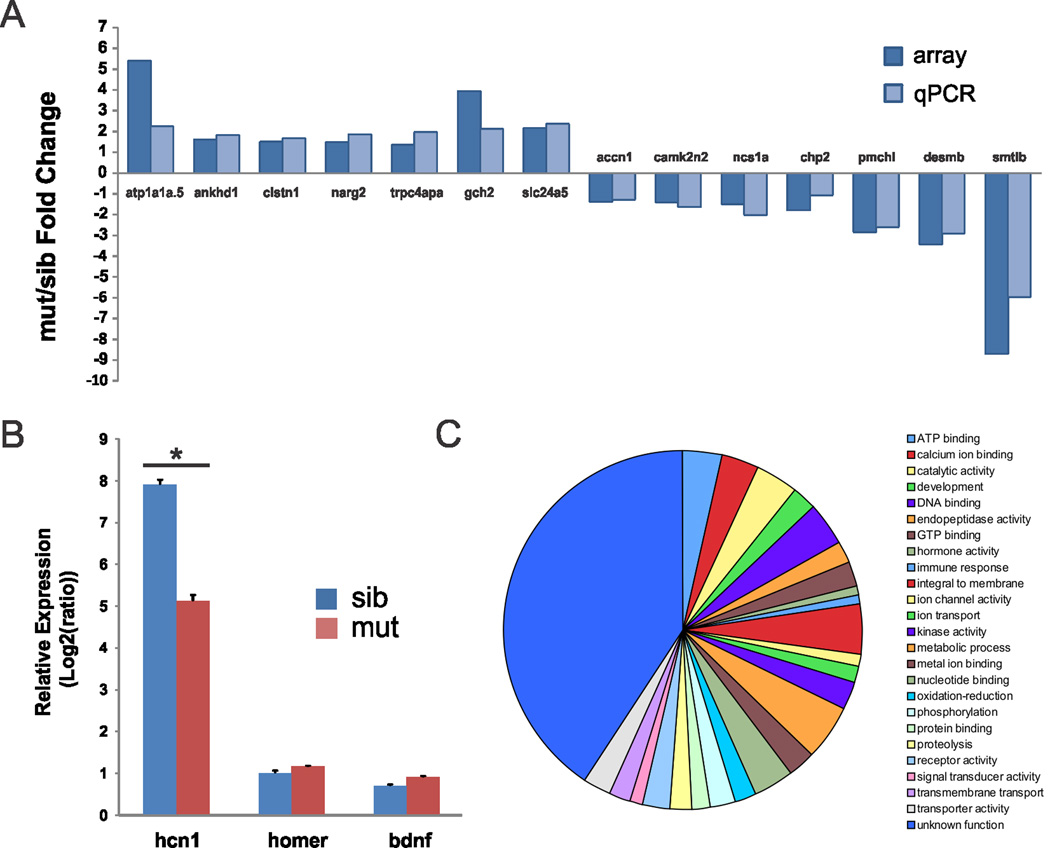
Although inherited disorders of voltage-gated ion channels are recognized as an etiology of epilepsy, investigation of transcriptional changes has not been reported for any epilepsy-related channelopathy. To detect differences in gene expression in an unbiased manner we used an Agilent Danio rerio chip covering ~44,000 probes (Figs. 2A, B). Hierarchical clustering analyses showed that ~2.5% (1099) of these probes (see Supplementary Data 1) were differentially expressed between mutants and sibling controls at 5 dpf (p ≤ 0.01, t test; 674 up-regulated and 425 down-regulated); 405 were assigned to an “unknown function” category. A list of 30 down- and up-regulated known genes showing the greatest differences in expression is shown in Fig. 2C. Surprisingly, these differences were modest as 90% (990/1099) of the identified genes exhibited fold-changes between 0.8 and 2.0. Similar to microarray analysis of Mecp2 single-gene mutant mice18, many of the genes identified had no obvious CNS-related function and/or expression. The two largest fold-changed genes, somatolactin β and a Na, K-ATPase, have expression primarily restricted to the pituitary (smtlb)19 or ear, intestinal bulb and pronephric duct (atp1a1a.5)20. Probes for several genes related to apoptosis (casp8, casp8b and casp3b) did not reveal any statistically significant changes in the microarray studies. Of the genes with altered expression in scn1Lab mutants, six were previously implicated in neurological disorders e.g., pcdh19 (infantile epileptic encephalopathy), cyfip1 and fxr2 (Fragile X syndrome), ocrl (Lowe syndrome), ubap2l (Parkinson’s disease) and oca2 (Angelman syndrome). Microarray-based gene expression measurements were verified for 14 randomly selected genes using qPCR (Fig. 3A). Biological functions were assigned to all genes using gene ontology (GO) annotations and the 482 genes showing at least a 1.5-fold change in expression and a p value < 0.01 were categorized further (Fig. 3C). Calcium ion binding genes include annexinA1c, A1b and 2a, spectrin α2, neurexin 2a, calsyntenin 1 and parvalbumin 3. Significant changes in a gap junction channel (cx43), a gene involved in clustering of voltage-gated sodium channels at the axon initial segment (spna2) and the ubiquitin domain of a GABA receptor (map1lc3b) were also noted. Three additional genes not found on the microarray were chosen for qPCR analysis (Fig. 3B): hcn1, a gene shown to be correlated with SCN1A using data mining (Starnet2; http://vanburenlab.medicine.tamhsc.edu/starnet2.shtml) and down-regulated in several seizure models21 was significantly reduced in scn1Lab mutants compared to sibling control (p < 0.05 2-tail Student’s t-test). However, homer and bdnf, e.g., genes involved in synaptogenesis related to the formation of recurrent excitatory synapses and epilepsy22,23 were unchanged.
Figure 2. Microarray analysis of scn1Lab zebrafish mutants.
(A) Heat maps depicting the expression of genes differentially expressed between scn1Lab mutant and sibling control larvae at 5 dpf. Rows represent individual genes. Columns represent different larvae. Genes that are highly expressed in scn1Lab mutants relative to controls are shown in red. (B) MA plot of normalized microarray data for all 44,000 genes. The log-ratio M and the mean fluorescence intensity A were calculated as the averages for all replicates. (C) A list of the top 30 genes showing the greatest differences (up-regulated genes are shown in red and down-regulated genes are shown in blue) in expression between scn1Lab mutants and sibling controls.
Figure 3. Quantitative RT-PCR analysis of scn1Lab zebrafish mutants.
(A) Comparison of the gene expression fold changes obtained by microarray analysis (array) and real-time qPCR analysis. The y-axis represents the average fold change in gene expression of each gene from zebrafish at 5 dpf. The x-axis represents different genes. (B) qPCR analysis of three genes involved in epileptogenesis. The relative gene expression is presented as log2 ratios to the least abundant transcript (log2ΔΔct). Data were normalized to internal reference gene β-actin. Values represent averages from five independent biological samples (1 sample = 10 pooled larvae). Bars indicate S.E.M; *p < 0.05 t-test. (C) Gene ontology classification of differentially expressed genes detected in scn1Lab mutants at 5 dpf (p < 0.05 ANOVA one-way and fold changes >1.5). Biological processes representing at least 5 gene annotations in at least one category are displayed
Spontaneous seizures in scn1Lab mutant zebrafish
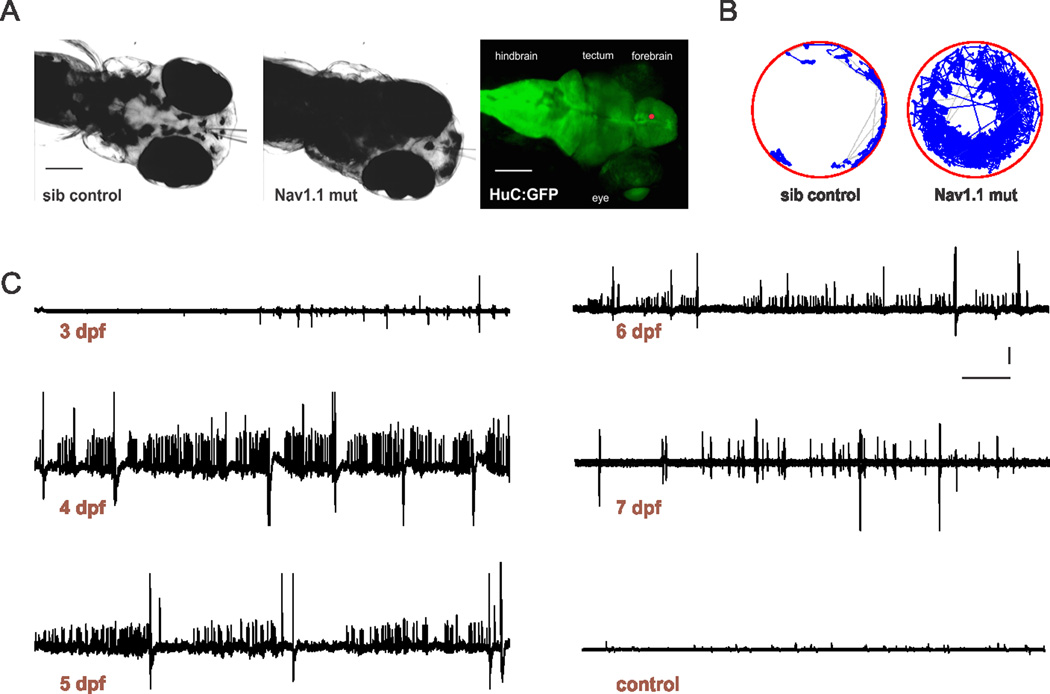
Next, we monitored scn1Lab mutants for evidence of spontaneous electrographic seizures starting at 3 dpf e.g., the first larval stage at which epileptiform discharge can be detected24–28. Mutant larvae were identified by their “black” appearance (Fig. 4A), which is indicative of a defect in pigment aggregation and die prematurely between 10 and 12 dpf, as reported previously15. Forebrain extracellular field recordings from paralyzed and agar-immobilized scn1Lab mutants were marked by frequent brief interictal-like bursts and large-amplitude long duration ictal-like events starting at 3 dpf (n = 4) and progressively becoming more prominent between 4 and 7 dpf (n = 132) (Fig. 2C). These events were confirmed in 100% of mutants at 3 dpf, 100% at 4 dpf, 97% at 5 dpf, 98% at 6 dpf and 100% at 7 dpf. Temporal expansions of seizure activity and comparisons with single action potential and twitch artifact recordings are shown in Supplemental Figure S1. Abnormal electrical events were never observed in age-matched sibling controls at any developmental stage (n = 36). Hyperthermia-induced seizures26 could be evoked in 5 dpf scn1Lab mutants and controls at apparently similar temperature thresholds (mutant: 26.9 ± 0.5 C°; n = 14; control: 25.9 ± 0.5 C°; n = 14; p = 0.164 t-test). However, these measurements were complicated, in mutants, by simultaneous occurrence of high frequency spontaneous epileptiform discharges. Mutants had elevated levels of swim activity and exhibited unprovoked seizure-like behavior consisting of whole-body convulsions and rapid undirected movement starting at 4 dpf (n = 36). A representative locomotion tracking plot of a scn1Lab mutant showing hyperactivity and convulsive behavior is shown in Fig. 4B. This behavior is similar to that classified as a Stage III seizure in larvae exposed to pentylenetetrazole27. Seizure behaviors were never observed in controls at any stage of development (n = 36). In pools of mutant and sibling control larvae, scn1Lab mutants stay close to the sides of the petri dish, which is considered a form of thigmotaxis in fish29. These results reveal a striking epilepsy phenotype in scn1Lab mutant zebrafish.
Figure 4. Spontaneous seizures in scn1Lab zebrafish mutants.
(A) Immobilized and agar-embedded zebrafish larvae are shown. Images were obtained using a 4× objective and 2× magnifier on an Olympus upright microscope during forebrain electrophysiological recordings in sibling control (A, left) and scn1Lab mutant (A, middle) larvae at 5 dpf. Note the dark pigmentation for mutants. Recording electrodes can be seen in panels A1-2 and the approximate site of the recording electrode tip in the forebrain (red circle) is shown using a representative HuC:GFP labeled larvae in A, right. Scale bar: 100 µm. (B) Sample locomotion tracking plot for sibling control (B, left) and scn1Lab mutant (B, right) larvae at 5 dpf. (C) Representative 10 min recording epochs obtained in the forebrain of paralyzed, immobilized and agar-embedded scn1Lab mutant larvae between 3 and 7 dpf. Note the presence of small and large amplitude spontaneous burst discharge; additional temporal expansions of seizure activity are shown in Supplemental Figure S1. A representative recording, under identical recording conditions, from a sibling control larvae at 5 dpf is also shown. Scale bar: 2 mV; 30 sec.
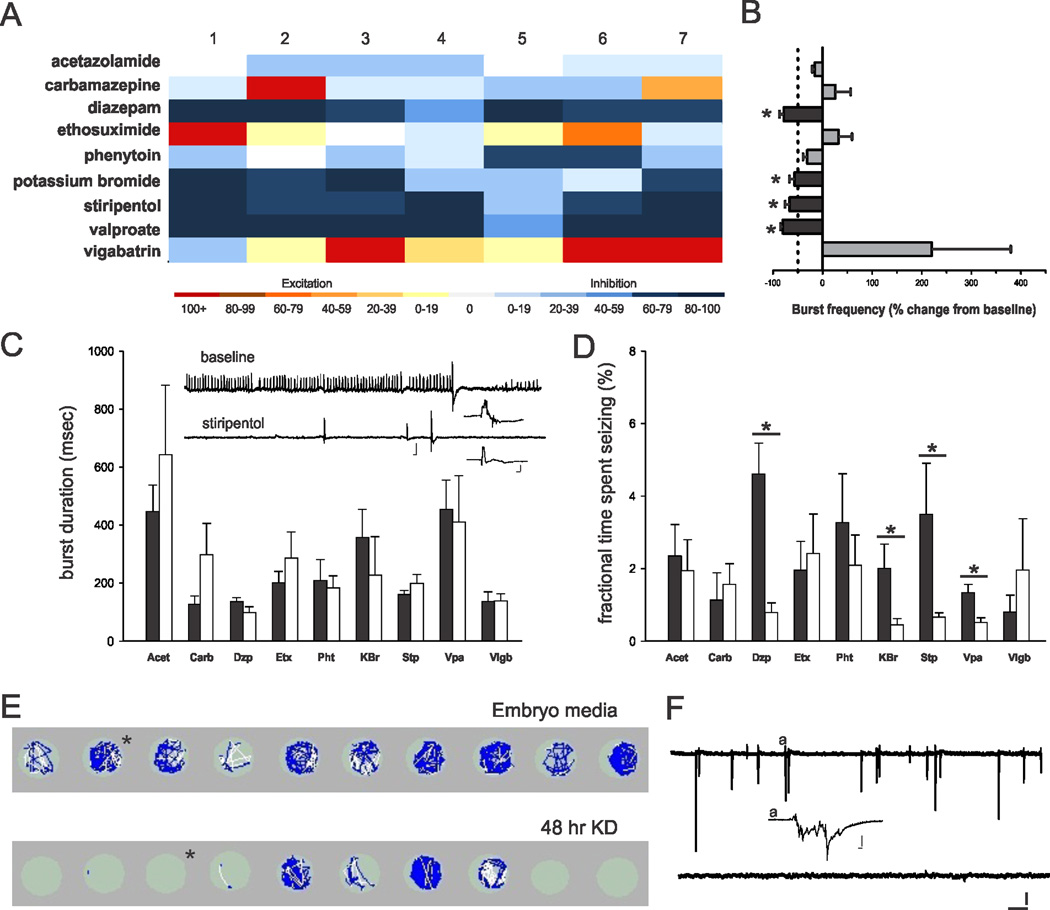
Pharmacological evaluation of scn1Lab mutant zebrafish
Seizures associated with SCN1A mutations are poorly responsive to most AEDs. To evaluate pharmaco-sensitivity we recorded spontaneous electrographic seizures in agar-embedded scn1Lab mutants (5–6 dpf) under baseline conditions, and again after application of a commercially available AED. All drugs were bath applied at a concentration of 1 mM; seven fish were tested for each drug. Epileptiform event frequency (including interictal- and ictal-like discharges) and the fractional time spent seizing in scn1Lab mutants were reduced by valproate, diazepam, potassium bromide and stiripentol (Figs. 5A, B, D). Burst durations were not significantly changed for any of these drug exposures (Fig. 5C). As expected, most AEDs had no effect and epileptiform activity became more frequent following exposure to carbamazepine (in 2 of 7 fish), ethosuximide (4 of 7 fish) or vigabatrin (6 of 7 fish). Because DS children often respond to the ketogenic diet (KD)30 we exposed a separate clutch of scn1Lab mutants, siblings and WT controls to a form of the diet31 for 48 hr starting at 4 dpf. Locomotion tracking data on KD-exposed larvae at 6 dpf confirm a reduction in seizure-like behavior to control levels in 7 of 10 mutants (Fig. E; mean velocity, treated mutants = 0.43 ± 0.09 mm/sec, n = 16; un-treated mutants = 0.81 ± 0.05 mm/sec, n = 28; p < 0.05 Kruskal-Wallis ANOVA on Ranks with a Dunn’s pairwise multiple comparison). No significant differences in swim behavior were noted in sibling controls treated with the KD (mean velocity = 0.63 ± 0.05 mm/sec, n = 20) compared to un-treated WT larvae at 6 dpf (mean velocity = 0.62 ± 0.07 mm/sec; n = 20). Acute exposure (20 min) to the diet had no effect on mutant seizure behavior in the locomotion assay (n = 14; change in mean velocity < 34%). Subsequent forebrain field recordings obtained from the same zebrafish used in the locomotion assay (Fig. 5F, top trace) confirmed the occurrence of spontaneous epileptiform discharge for embryo media exposed scn1Lab mutants and a suppression of burst activity in mutants exposed to the KD for 48 hr (Fig. 5F, bottom trace). These results demonstrate that the pharmacological profile for scn1Lab mutants resembles that seen in children with DS.
Figure 5. Pharmacological validation of scn1Lab zebrafish mutants.
(A) Heat map showing the response to nine different AEDs. Each column represents the percent change in burst frequency (baseline − drug/baseline × 100) for one individual zebrafish mutant. Drugs that inhibit seizure events are shown in dark blue. All drugs were tested at a concentration of 1 mM. Note in some trials carbamazepine and vigabatrin increased burst frequency over the initial baseline levels. (B) Plot of the mean change in burst frequency and standard error for the data shown in the heat map. Paired t-test or Wilcoxon signed rank sum test for data that failed the normality test showed significance as follows: diazepam (p = 0.002; n = 7), potassium bromide (p = 0.016; n = 7), stiripentol (p = 0.024; n = 7), and valproate (p = 0.004; n = 7). (C) Plot of the burst duration for all trials shown in A. Data is presented as the mean ± S.E.M. for electrographic seizure events at baseline (black bars) and after drug exposure (white bars). Inset shows a representative 2 min recording during the stiripentol trial; scale bars: large trace 1 mV, 1 sec; small trace, 1 mV, 100 msec. (D) Plot of the fractional time spent seizing for all trials shown in A. Data is presented as the mean ± S.E.M. for electrographic seizure events at baseline (black bars) and after drug exposure (white bars). Student’s t-test or Mann-Whitney-Rank sum test for data that failed the normality test showed significance as follows: diazepam (p = 0.001; n = 7); potassium bromide (p = 0.043; n = 7); stiripentol (p = 0.007; n = 7) and valproate (p = 0.007; n = 7 (E) Locomotion tracking plots for 10 individual mutant larvae raised in embryo media (top row) or the ketogenic diet for 48 hr. Plots show swim velocity and locomotion tracks with darker colors indicative of higher velocities; 10 min trials are shown. (F) Representative 10 min extracellular recording epochs from the same fish shown in E; representative examples are indicated by an * in the locomotion plots. Scale bar: 1 mV, 30 sec. Inset shows burst at higher temporal resolution (indicated by #); scale bar: 1 mV, 100 msec.
High-throughput drug screening in scn1Lab mutants
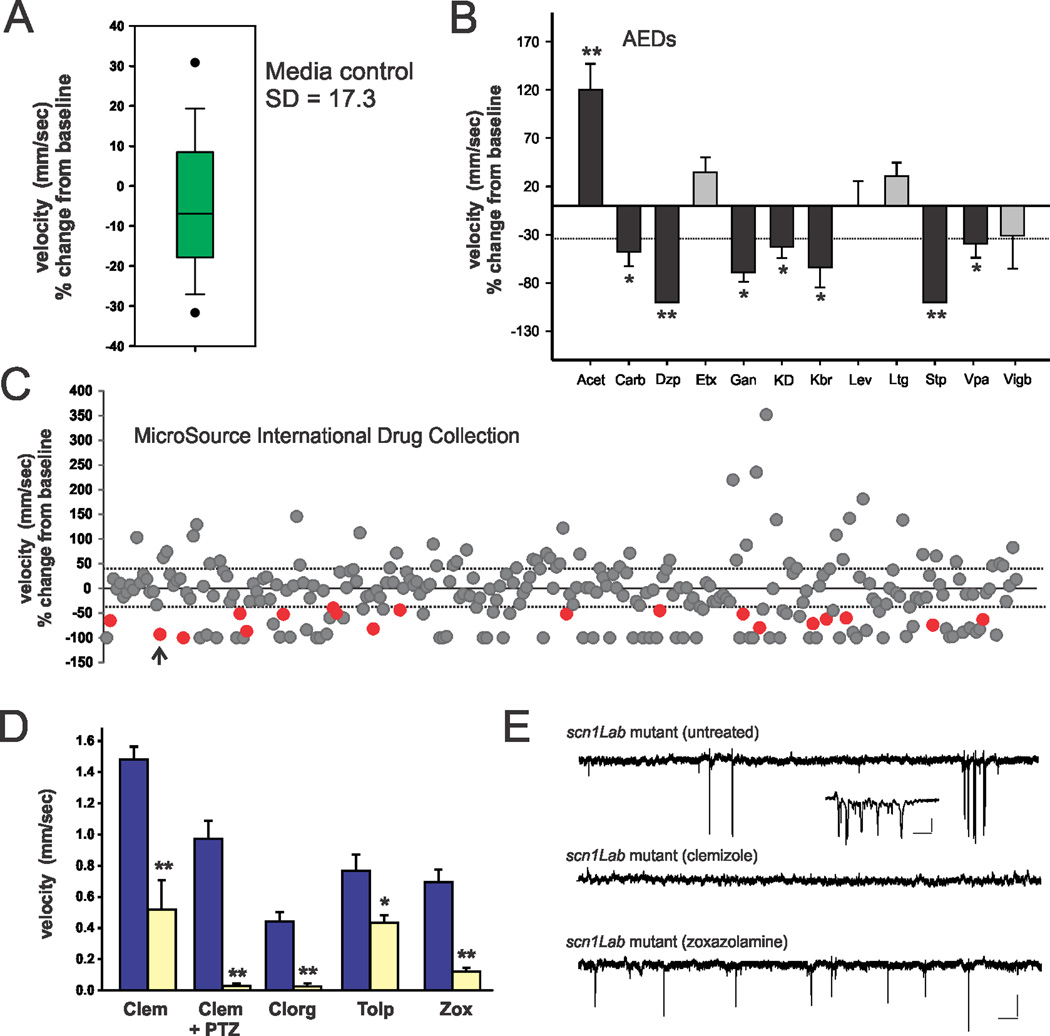
Because behavioral seizure activity is easily and rapidly monitored using a locomotion tracking format24,25,27,28,32–36 (Figs. 4B and 5B1), we designed a relatively high-throughput phenotype-based strategy to screen chemical libraries for compounds that reduce mutant behavior to Stage 0 (very little swim activity) or Stage I (increased, but non-convulsive, swim activity) e.g., behavior equivalent to that seen in normal WT mutants. Automated measurement of larval activity was achieved using EthoVision tracking software (Noldus Information Technology) and a high-speed camera. Previous studies confirmed that high velocity movement ≥ 20 mm/sec correspond to paroxysmal seizure-like convulsions (Stage III)35,36. Using a 96-well format, we automatically tracked mutant swim activity at baseline, and then again after addition of a test compound (100 µl); each compound was tested on 6 to 12 individual larvae at 5 dpf. The change in mutant swim activity between two consecutive recording epochs in embryo media was taken as baseline and is shown in Fig. 6A (n = 28). Based on a standard deviation of 17.3 for baseline recordings associated simply with a solution exchange, we screened for compounds that inhibited movement (measured as a change in mean velocity) by ≥ 34%. To validate this approach, we first screened eleven AEDs and the KD using this assay. As expected from electrophysiological assays (Fig. 5), diazepam, potassium bromide, stiripentol, valproate and a 48 hr exposure to KD effectively inhibited seizure behavior in the locomotion-based assay (Fig. 6B); ganaxolone, a neuroactive steroid related to allopregnalone, was also effective. Next, we screened test compounds at an initial concentration of 667 µM from a library that included US Food and Drug Administration (FDA) approved and toxicology tested drugs (The International Drug Collection; http://www.msdiscovery.com/spectrum.html). Among the 320 compounds screened in vivo, 18 were found to significantly inhibit spontaneous seizures in scn1Lab mutants to levels comparable to Stage 0 or Stage I behavior and/or reduce mean swim velocity (red circles in Fig. 6C). These 18 compounds were then re-tested on a separate clutch of scn1Lab mutants at concentrations of 667, 67 and 6.7 µM. In the initial screen, 81 compounds were identified as lethal i.e., no visible heartbeat or movement in response to touch after a 30 min exposure (see Supplementary Table S1) and were re-evaluated at a dilution of 1:100; none of these advanced further. The drug library included a number of additional compounds with putative anticonvulsant properties (beclamide, aminohydroxybutyric acid, and tiletamine) that were also ineffective in the 96-well locomotion assay at 667 µM. 14 of the re-tested compounds either failed to successfully inhibit seizure behavior in a second clutch of scn1Lab mutants or only suppressed behavior at the highest drug concentration. Next we selected 4 (out of 18) compounds that were effective in reducing seizure-induced swim activity and mean velocity at all three drug concentrations for further testing: zoxazolamine, clemizole HCl, clorgiline HCl and tolperisone HCl (Fig. 6D). Each of these compounds was evaluated a third time in the locomotion assay at a concentration of 100 µM, and subsequently monitored for forebrain electrographic activity. Clorgiline (a monoamine oxidase A inhibitor) and the muscle relaxants zoxazolamine37 and tolperisone38 were identified as “false positives” because they reduced swim activity at this concentration but when the same mutant was embedded in agar electrographic seizure events were still observed (see Fig. 6E). Only one compound, clemizole (antihistamine and NS4B RNA binding inhibitor)39,40, was effective in suppressing spontaneous seizure activity in both assays (Fig. 6D–E). A comparison of electrophysiological burst characteristics for un-treated (n = 3) and clemizole-treated (n = 7) scn1Lab mutants shown in Figure 6E zebrafish indicates a statistically significant suppression of activity (un-treated: burst frequency = 1.5 ± 0.3 bursts/min; burst duration = 926 ± 414 msec; fractional time spent seizing = 0.73 ± 0.17% vs. clemizole: burst frequency = 0.2 ± 0.01 bursts/min; burst duration = 154 ± 127 msec; fractional time spent seizing = 0.03 ± 0.02%; p = 0.001 for all comparisons, Kruskal-Wallis ANOVA with a Dunn’s pairwise multiple comparison test). Clemizole had no significant effect on seizure behavior in the locomotion assay at concentrations between 6.25 and 50 µM (n = 33). As an additional evaluation of the therapeutic potential for acute clemizole treatment, we also demonstrated that 100 µM clemizole was effective in reducing seizure behavior in WT zebrafish exposed to 15 mM pentylenetetrazole (Fig. 6D; n = 10) i.e., a model of acute seizures based on GABA receptor antagonism. These results suggest that scn1Lab mutants can be used in a high-throughput screen to identify potential lead compounds for Dravet syndrome.
Figure 6. Screen to identify drugs that rescue scn1Lab mutant epilepsy phenotype.
(A) Box plot of mean velocity (in mm/sec) for two consecutive recordings of mutant larvae in embryo media. The percent change in velocity from baseline (recording #1) vs. experimental (recording #2) is shown. The bottom and top of box represent 25th and 75th percentile, respectively. The horizontal line represents median value; vertical lines encompass the entire range of values. (B) Plot of the effect of AEDs on seizure behavior in scn1Lab mutants.. Bars represent percent change in mean velocity; 6–12 fish per experiment. Drugs tested at 1 mM; diazepam (Dzp; p < 0.001), carbamazepine (Carb; p = 0.024), ganaxolone (Gan; p = 0.003), stiripentol (Stp; p = 0.001), valproate (Vpa;, p = 0.026) and a 48 hr ketogenic diet (KD; p = 0.003) exposure reduced seizure activity, measured as a change in velocity, by more than the standard deviation in control recordings (34 %, dotted line). Acetazolamide (Acet; p < 0.001) and ethosuximide (Etx; p = 0.250) increased seizure behavior; levetiracetam (Lev; p = 0.243), and lamotrigine (Ltg; p = 0.058) had no effect. (C) Plot for all 320 compounds tested. Colored circles (red) represent positive hits; compounds that decreased activity by 100% were generally toxic; 6–12 fish per trial. Arrowhead; first clemizole trial. (D) Plot of drug re-trials on separate mutant clutches; 100 µM per drug; 10 fish per trial. Abbreviations: Clem, clemizole; Clem + PTZ, clemizole + 15 mM PTZ; Clorg, clorgiline; Tolp, tolperisone; Zox, zoxazolamine. Effect of acute clemizole on PTZ-induced seizures shown for wild-type larvae. Bars represent mean ± S.E.M. For panels B and D: Student’s paired t-test or Mann-Whitney Rank Sum test with significance set at p = 0.01 (*) or p < 0.001 (**). (E) Sample electrophysiology recordings from scn1Lab mutants exposed to clemizole in the locomotion assay (D) and then monitored using a forebrain recording (top trace; ictal-like burst shown in inset). Similar traces for an un-treated mutant (middle) and mutant treated with zoxazolamine (bottom). Scale bars: large traces 0.5 mV, 10 s; inset 0.5 mV, 100 msec.
Discussion
The scn1Lab zebrafish mutant described here is the first simple vertebrate model of a sodium channel mutation that recapitulates features of Dravet syndrome, a catastrophic form of drug-resistant epilepsy in children. We have shown that these mutants exhibit hyperactivity, including convulsive behavior, spontaneous electrographic seizures, shortened lifespan and a pharmacological profile similar to the human condition. Additional molecular analysis of scn1Lab mutants suggests the absence of gross changes in global gene expression and a lack of compensation, at the RNA level, by other voltage-gated Na+ channel subunits. A two-stage phenotype-based drug screening strategy to identify lead compounds with the potential to ameliorate epilepsy phenotypes associated with SCN1A mutation identified one FDA-approved drug (clemizole).
Electroencephalographic (EEG) activity is typically normal in the first year of life for DS patients with an evolution to abnormal paroxysmal and polyspike activity between 1 and 9 years of age. This age-dependent pattern was mimicked in developing zebrafish larvae at ages where scn1a expression was significant. Forebrain extracellular recordings in very young larvae (3 dpf) appeared largely normal with the occasional small burst of polyspike activity. Frequent brief interictal-like activity with large amplitude polyspike burst discharges became more prominent as larvae aged. The architecture of these electrical events resembled those previously described in wild-type larvae exposed to pentylenetetrazole27, 4-aminopyridine24, linopirdine28 or hyperthermia26. The appearance of electrographic seizure activity corresponds with hyperactivity, full-body convulsions with associated high-velocity swim activity and brief loss-of-posture in freely behaving mutants. These types of spontaneous behaviors are never observed in wild-type larvae and, again, resemble those previously observed only during exposure to convulsant drugs. These behaviors are an indirect indicator of seizure activity and could be used for rapid in vivo evaluation of drug treatments and lethality in a multi-well format using automated locomotion tracking software32,33,35. We also show that seizures in scn1Lab zebrafish mutants are responsive to the ketogenic diet and four AEDs (e.g., valproate, benzodiazepine, potassium bromide and stiripentol) prescribed clinically for patients with DS41.
Interestingly, electrographic seizure events in scn1Lab mutants remained unchanged (or perhaps worsened) in response to several commercially available AEDs. While it is possible that drug concentrations higher than 1 mM could be required to abolish electrical events, these would be considered high and potentially non-selective concentrations. In drug trials using an acute PTZ-induced seizure model in larval zebrafish24,32,33,42, AED concentrations of 1 mM and below were often sufficient for assessing antiepileptic activity. With a failure to respond to seven different AEDs this model fits the clinical definition of drug-resistant epilepsy43. For nearly 40 years, the discovery and identification of new AEDs has almost entirely been based upon preclinical animal models of acquired or acute seizures in rodents44. This approach successfully identified drugs that block generalized tonic-clonic seizures in humans45 but remains time-consuming, resource intensive, expensive and laborious. While testing against PTZ or other types of acquired seizures in zebrafish larvae may be more efficient than similar assays in rodents32,33,42, they ultimately should identify the same classes of compounds. In contrast, here we describe an alternative screening strategy using a 96-well format for rapid automated behavioral monitoring followed by a sensitive electrophysiological assay of spontaneous electrographic seizure activity in a mutant fish mimicking a known human genetic disorder. Our in vivo strategy simultaneously monitors lethality and is not limited to SCN1A, but could be applied to any monogenic epilepsy disorder. Indeed, this phenotype-based approach could form the basis of a genetically informed or “personalized” approach to drug discovery. While genetically modified mice mimicking known SCN1A mutations and exhibiting epilepsy have been developed, breeding can be complicated, background strain can modify seizure phenotypes and AEDs are rarely tested in these animals. For example, in Scn1aRX/+ mutant mice stiripentol and clobazam were only evaluated for effects on hyperthermia-induced seizure thresholds46. Treatment of Scn1a+/− mutant mice with clonazepam, an allosteric modulator of GABA-A receptors, rescued some of the autistic-like behaviors but was not evaluated as an antiepileptic43. Where drug-resistant rodent epilepsy models have been described, such as the subgroup of wild-type rats selected from kindling or post-status epilepticus models47, they remain only poorly characterized and are not suitable to initial high-throughput stages of drug screening. In contrast, using a zebrafish scn1Lab mutant with greater than 75% sequence identity for a human sodium channel mutation, we completed a large-scale transcriptomic profiling of over 44,000 probes, demonstrated a developmental progression of scn1Lab channel expression and epileptic phenotypes, analyzed the effects of available antiepileptic therapies, and screened a 320 compound chemical library against spontaneous unprovoked seizures. Although this first proof-of-principle screen was accomplished at one fish per well, 6 to 12 fish per trial and one trial per week, the ease with which zebrafish could be scaled upward (especially in a commercial setting) to study 100s to 1000s of larvae per week make this an attractive system for a rapid large-scale first-stage in vivo drug discovery program. Simultaneous in vivo evaluation of toxicity - one of the greatest sources of failure in moving lead compounds from the bench to the clinic - is a critical advantage of our approach over available organotypic hippocampal culture- or in silica-based screening strategies.
Although any animal model drug discovery data should be treated cautiously, clemizole, a compound with H1 antagonist and NS4B RNA inhibiting properties39,40, is an FDA-approved drug with a safe toxicology profile emerged from this screen and offers a exciting starting point for further research. For example, although it was recently recognized that antihistamines inhibit induced seizures in neonatal rats48, this may not be the mechanism of action here as four other H1 antihistamines (pimethixene maleate, chloropyramine HCl, mebhydrolin napthalenesulfonate and iproheptine) failed to suppress convulsive behavior in scn1Lab mutants. Furthermore, evidence suggests the potential for H1 antihistamines to adversely modify seizures in children49 indicating that more detailed analysis will be required to identify a mechanism of action. Given that clemizole was also effective in a zebrafish version of the Metrazol test it may be worthwhile to pursue additional pre-clinical testing in the NIH-sponsored Anticonvulsant Drug Development Program at the University of Utah. Most importantly, our studies suggest that in vivo drug screening and experimental analysis of scn1Lab mutant zebrafish could prove extremely valuable to understanding (and treatment) of Dravet syndrome.
Methods
Animals
Scn1Lab (didys552) zebrafish embryos were a kind gift from Herwig Baier. Adult HuC:GFP zebrafish were a kind gift from Stephen Ekker. Zebrafish were generated and maintained in accordance with the guidelines of the University of California, San Francisco Committee on the Use and Care of Animals. Zebrafish larvae were maintained in “embryo medium” consisting of 0.03% Instant Ocean (Aquarium Systems, Inc., Mentor, OH, U.S.A.) in deionized water containing 0.002% Methylene Blue as a fungicide. Larval zebrafish clutches were bred from scn1Lab heterozygous animals that had been backcrossed to TL wild-type or HuC:GFP zebrafish for at least 7 generations. Homozygous mutants (sorted based on pigmentation) and age-matched sibling larvae were used. Although the precise genetic defect responsible for the skin pigmentation issue is unknown, it is interesting that a 1.5-fold up-regulation of a gene encoding the melanocortin 5a receptor was noted in our microarray data.
Seizure monitoring
Procedures for locomotion tracking and electrophysiology were described24,27. In pilot experiments, HuC:GFP zebrafish were used in electrophysiology experiments to obtain an estimation of the location of recording electrodes. Locomotion plots were obtained for one fish per well at a recording epoch of 10 min using a DanioVision system running EthoVision XT software (Noldus Information Technology; Leesburg, VA). Seizure scoring was performed as described27. Locomotion plots were analyzed for distance traveled (in mm) and mean velocity (in mm/sec). Epileptiform events were analyzed in pClamp (Molecular Devices; Sunnyvale, CA) and defined as upward or downward membrane deflections greater than 2× baseline noise level and classified as interictal-like (100 to 300 msec duration) or ictal-like (1000 to 5000 msec duration). Burst frequency was determined by counting the number of epileptiform events per minute during a 10-min recording epoch. Burst duration was determined by measuring the onset-to-offset interval for all events during the same epoch.
Drugs
Drugs were obtained from Sigma-Aldrich and dissolved in embryo media. Stock solutions were prepared in embryo media at 1 mM and pH adjusted to ~7.5. Ganaxolone was a kind gift from BioCrea GmbH (Radebeul, Germany). Compounds for drug screening were purchased from MicroSource Discovery Systems, Inc. (International Drug Collection; Gaylordsville, CT) and were provided as 10 mM DMSO solutions. Test compounds were dissolved in embryo media and tested at concentrations between 6.7 and 667 µM; final DMSO concentration ~7%. An initial screen concentration of 667 µM was chosen for behavioral studies in freely swimming fish as this falls on the lower range of AED concentrations previously reported in to be effective against PTZ (10–20 mM) induced seizures in larval zebrafish (0.1 to 25 mM)27,32,42 and was the most efficient use of the small volume of stock solution (250 µL) provided by MicroSource Discovery Systems, Inc. A slightly higher concentration (1 mM) was chosen for the initial AED validation assays in Figures 5 and 6 to account for any potential complications associated with diffusion through the agar. DMSO was evaluated for toxicity at dilutions between 0.01 and 100% using wild-type larvae (n = 12 fish per concentration); DMSO at > 25% was lethal. In all drug screening studies compounds were coded and experiments were performed by investigators blind to the nature of the compound. Baseline recordings of seizure activity were obtained from mutants bathed in embryo media; a second plot was then obtained following a solution change to a test compound. Each test compound classified as a “positive hit” in the locomotion assay was visually confirmed as alive based on movement in response to touch and visible heartbeat. WT fish exhibit little to no spontaneous swim activity during these 10 min recording epochs (see Fig. 3B) and were not used in the drug discovery assay.
Molecular biology
Procedures for microarray, quantitative PCR and whole-mount in situ hybridization were described25. See Supplementary Tables S2 and S3 for primer sequence data.
Data analysis
Data are presented as mean and SEM, unless stated otherwise. Pairwise statistical significance was determined with Student’s two-tailed unpaired t-test, ANOVA or Mann-Whitney rank sum test, as appropriate, unless stated otherwise. Results were considered significant at P < 0.05, unless otherwise indicated.
Supplementary Material
Acknowledgments
The authors would like to thank Herwig Baier and Stephen Ekker for zebrafish lines used in this study; the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility and NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131 for the microarray support; Wyatt Potter, Robert Hunt and MacKenzie Howard for critical comments on the manuscript; and Rosanne Estrada for assistance with qPCR. This work was supported by a EUREKA grant from the NINDS (1 R01 NS079214), a Research Award from the Dravet Syndrome Foundation (http://www.dravetfoundation.org/) and a Challenge Award from Citizens United for Research in Epilepsy (http://www.cureepilepsy.org/home.asp) to S.C.B.
Footnotes
The authors have no conflicts of interest to report.
Author contributions
S.C. Baraban designed the experiments, performed all electrophysiology studies, developed and screened compounds using the locomotion assay and wrote the manuscript; M.T. Dinday maintained the zebrafish colonies and assisted in the drug screening assays; G.A. Hortopan performed all the molecular biology experiments and data analyses.
Competing financial interests: The authors declare no competing financial interests
References
- 1.Harkin LA, et al. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. doi: 10.1093/brain/awm002. [DOI] [PubMed] [Google Scholar]
- 2.Mulley JC, et al. SCN1A mutations and epilepsy. Hum. Mutat. 2005;25:535–542. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- 3.Ceulemans BP, Claes LR, Lagae LG. Clinical correlations of mutations in the SCN1A gene: from febrile seizures to severe myoclonic epilepsy in infancy. Pediatric Neurol. 2004;30:236–243. doi: 10.1016/j.pediatrneurol.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Bender AC, et al. SCN1A mutations in Dravet syndrome: Impact of interneuron dysfunction on neural networks and cognitive outcome. Epilepsy Beh. 2012;23:177–186. doi: 10.1016/j.yebeh.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catterall WA, Kalume F, Oakley JC. NaV1.1 channels and epilepsy. J. Physiol. 2010;588:1849–1859. doi: 10.1113/jphysiol.2010.187484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogiwara I, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J. Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DY, et al. Reduced sodium channel Na(v)1.1 levels in BACE1-null mice. J. Biol. Chem. 2011;286:8106–8116. doi: 10.1074/jbc.M110.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, et al. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J.Neurosci. 2004;24:4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu FH, et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat. Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- 10.Oakley JC, et al. Temperature- and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc. Natl. Acad. Sci. USA. 2009;106:3994–3999. doi: 10.1073/pnas.0813330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheah CS, et al. Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proc. Natl. Acad. Sci. USA. 2012;109:14646–14651. doi: 10.1073/pnas.1211591109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessman CA. The developing zebrafish (Danio rerio): a vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res. C. Embryo Today. 2011;93:268–280. doi: 10.1002/bdrc.20212. [DOI] [PubMed] [Google Scholar]
- 13.Delvecchio C, Tiefenbach J, Krause HM. The zebrafish: a powerful platform for in vivo, HTS drug discovery. Assay Drug Dev. Technol. 2011;9:354–361. doi: 10.1089/adt.2010.0346. [DOI] [PubMed] [Google Scholar]
- 14.Rinkwitz S, Mourrain P, Becker TS. Zebrafish: an integrative system for neurogenomics and neurosciences. Prog. Neurobiol. 2011;93:231–243. doi: 10.1016/j.pneurobio.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Novak AE, et al. Embryonic and larval expression of zebrafish voltage-gated sodium channel alpha-subunit genes. Dev. Dyn. 2006;235:1962–1973. doi: 10.1002/dvdy.20811. [DOI] [PubMed] [Google Scholar]
- 16.Schoonheim PJ, Arrenberg AB, Del Bene F, Baier H. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J. Neurosci. 2010;30:7111–7120. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MS, et al. The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum. Mol. Gen. 2007;16:2892–2899. doi: 10.1093/hmg/ddm248. [DOI] [PubMed] [Google Scholar]
- 18.Jordan C, Kwan HC, Francke U. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med. Genet. 2007;8:36. doi: 10.1186/1471-2350-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez M, et al. Expression of the somatolactin β gene during zebrafish embryonic development. Gene Expr. Patterns. 2006;6:156–161. doi: 10.1016/j.modgep.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Blasiole B, et al. Cloning, mapping, and developmental expression of a sixth zebrafish Na, K-ATPase alpha1 subunit gene (atp1a1a.5) Mech. Dev. 2002;119(Suppl 1):S211–S214. doi: 10.1016/s0925-4773(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 21.Noam Y, Bernard C, Baram TZ. Towards an integrated view of HCN channel role in epilepsy. Curr. Opin. Neurobiol. 2011;21:873–879. doi: 10.1016/j.conb.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avedissian M, et al. Hippocampal gene expression analysis using the ORESTES methodology shows that homer 1a mRNA is upregulated in the acute period of the pilocarpine epilepsy model. Hippocampus. 2007;17:130–136. doi: 10.1002/hipo.20248. [DOI] [PubMed] [Google Scholar]
- 23.Tongiorgi E, et al. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J. Neurosci. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraban SC, et al. A large-scale mutagenesis screen to identify seizure-resistant zebrafish. Epilepsia. 2007;48:1151–1157. doi: 10.1111/j.1528-1167.2007.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hortopan GA, Dinday MT, Baraban SC. Spontaneous seizures and altered gene expression in GABA signaling pathways in a mind bomb mutant zebrafish. J. Neurosci. 2010;30:13718–13728. doi: 10.1523/JNEUROSCI.1887-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt RF, Hortopan GA, Gillespie A, Baraban SC. A novel zebrafish model of hyperthermia-induced seizures reveals a role for TRPV4 channels and NMDA-type glutamate receptors. Exp. Neurol. 2012;237:199–206. doi: 10.1016/j.expneurol.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baraban SC, Taylor MR, Castro PA, Baier H. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 2005;131:759–768. doi: 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 28.Chege SW, Hortopan GA, Dinday MT, Baraban SC. Expression and function of KCNQ channels in larval zebrafish. Dev. Neurobiol. 2012;72:186–198. doi: 10.1002/dneu.20937. [DOI] [PubMed] [Google Scholar]
- 29.Ellis LD, Seibert J, Soanes KH. Distinct modes of induced hyperactivity in zebrafish larvae. Brain Res. 2012;1449:46–59. doi: 10.1016/j.brainres.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Dravet C, et al. Severe myoclonic epilepsy in infancy: Dravet syndrome. Adv. Neurol. 2005;95:71–102. [PubMed] [Google Scholar]
- 31.Taylor MR, Hurley JB, Van Epps HA, Brockerhoff SE. A zebrafish model for pyruvate dehydrogenase deficiency: rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc. Natl. Acad. Sci. USA. 2004;101:4584–4589. doi: 10.1073/pnas.0307074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berghmans S, Hunt J, Roach A, Goldsmith P. Zebrafish offer the potential for a primary screen to identify a wide variety of potential anticonvulsants. Epilepsy Res. 2007;75:18–28. doi: 10.1016/j.eplepsyres.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Baxendale S, et al. Identification of compounds with anti-convulsant properties in a zebrafish model of epileptic seizures. Dis. Model. Mech. 2012;5:773–774. doi: 10.1242/dmm.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cario CL, Farrell TC, Milanese C, Burton EA. Automated measurement of zebrafish larval movement. J. Physiol. 2011;589:3703–3708. doi: 10.1113/jphysiol.2011.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter MJ, et al. Validation of a larval zebrafish locomotor assay for assessing the seizure liability of early-stage development drugs. J. Pharm. Tox. Methods. 2008;5:176–187. doi: 10.1016/j.vascn.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Orellana-Paucar AM, et al. Anticonvulsant activity of bisabolene sesquiterpenoids of Curcuma longa in zebrafish and mouse seizure models. Epilepsy Beh. 2012;24:14–22. doi: 10.1016/j.yebeh.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Hadra R, Millichap JG. Quantitative assessment of motor function in cerebral palsy: evaluation of zoxazolamine (flexin), a new muscular relaxant agent. Neurology. 1956;6:843–852. doi: 10.1212/wnl.6.12.843. [DOI] [PubMed] [Google Scholar]
- 38.Sakitama K. The effects of centrally acting muscle relaxants on the intrathecal noradrenaline-induced facilitation of the flexor reflex mediated by group II afferent fibers in rats. Jpn. J. Pharmacol. 1993;63:369–736. doi: 10.1254/jjp.63.369. [DOI] [PubMed] [Google Scholar]
- 39.Finkelstein M, Kromer CM, Sweeney SA, Delahunt CS. Some aspects of the pharmacology of clemizole hydrochloride. J. Am. Pharm. Assoc. Am. Pharm. Assoc. 1960;49:18–22. [PubMed] [Google Scholar]
- 40.Einav S, Sobol HD, Gehrig E, Glenn JS. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat. Biotechnol. 2008;26:1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller IO, Sotero de Menezes M. SCN1A-Related Seizure Disorders. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. GeneReviews™ [Internet] Seattle (WA): University of Washington, Seattle; 2007. [Google Scholar]
- 42.Afrikanova T, et al. Validation of the zebrafish pentylenetetrazol seizure model: locomotor versus electrographic responses to antiepileptic drugs. PLoS One. 2013;8:e54166. doi: 10.1371/journal.pone.0054166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Toffol B, et al. ESPERA study: Applicability of the new ILAE criteria for antiepileptic drug resistance of focal epilepsies in current clinical practice. Epilepsy Beh. 2012;25:166–169. doi: 10.1016/j.yebeh.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Loscher W, Schmidt D. Modern antiepileptic drug development has failed to deliver: Ways out of the current dilemma. Epilepsia. 2011;52:657–658. doi: 10.1111/j.1528-1167.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 45.Bialer M, White HS. Key factors in the discovery and development of new antiepileptic drugs. Nat. Rev. Drug Discov. 2012;9:10–19. doi: 10.1038/nrd2997. [DOI] [PubMed] [Google Scholar]
- 46.Cao D, et al. Efficacy of stiripentol in hyperthermia-induced seizures in a mouse model of Dravet syndrome. Epilepsia. 2012;53:1140–1145. doi: 10.1111/j.1528-1167.2012.03497.x. [DOI] [PubMed] [Google Scholar]
- 47.Han S, et al. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada K, Takizawa F, Tamura T, Kanda T. The effect of antihistamines on seizures induced by increasing-current electroshocks: ketotifen, but not olopatadine, promotes the seizures in infant rats. Biol. Pharm. Bull. 2012;35:693–697. doi: 10.1248/bpb.35.693. [DOI] [PubMed] [Google Scholar]
- 49.Miyata I, Saegusa H, Sakurai M. Seizure-modifying potential of histamine H1 antagonists: a clinical observation. Pediatr. Int. 2011;53:706–708. doi: 10.1111/j.1442-200X.2011.03328.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.