Abstract
T lymphocytes from patients with systemic lupus erythematosus (SLE) display a complex array of cellular, molecular, and signaling anomalies, many of which have been attributed to increased expression of the transcriptional regulator cAMP responsive element modulator α (CREMα). Recent evidence indicates that CREMα, in addition to its regulatory functions on gene promoters in T lymphocytes, alters the epigenetic conformation of cytokine genes by interacting with enzymes that control histone methylation and acetylation as well as cytosine-phosphate-guanosine (CpG) DNA methylation. This review summarizes the most recent findings on CREM protein expression in various cell types, in particular its effects on T lymphocyte biology in the context of both health and SLE. We emphasize CREMα as a key molecule that drives autoimmunity.
Keywords: SLE, systemic lupus erythematosus, cytokine, transcription, epigenetics, CREM, lymphocyte
Genetic and epigenetic alterations in SLE immunopathogenesis
Numerous cellular and molecular abnormalities in immune cells, including T and B lymphocytes, monocytes, and dendritic cells (DCs), have been linked to the pathogenesis of SLE. SLE isa prototypicautoimmunedisease that can affect almost every organ of the human body. Factors that cause or contribute to disease range from endogenous genetic polymorphisms to environmental and infectious agents, all of which result in a severely dysregulated immune system [1–3]. The strong influence of genetic predispositions can be observed from the detection of mutations in single genes that are linked to the development of lupus-like symptoms in an array of disorders: mutations within 3′-repair DNA exonuclease 1 (TREX1), sterile α motif domain and HD domain-containing protein 1 (SAMDH1), and RNase H2 genes, among others. Rare gene deficiencies, including those of complement factors C1q, C2, and C4, result in lupus-like disorders, and aberrant splicing patterns [e.g., of the interferon-regulatory factor (IRF)-5 gene] have been linked to the development of SLE [4–7]. Of the genetic susceptibility regions identified, many relate to alleles that are well-established contributors to immune cell pathways, such as IRF-5 or tumor necrosis factor superfamily member 4 (TNFSF4) [8,9]. These discoveries have largely shaped our understanding of a genetic predisposition to SLE, although the clinical discordance between monozygotic twins strongly indicates that genetic factors are not sufficient to lead to clinically defined SLE.
More recently, it has become clear that epigenetic factors play a key role in the immune pathogenesis of SLE [10–12]. Methylation of CpG (meCpG) DNA motifs, post-transcriptional modifications to histone tails, and micro-RNA (miRNA)-mediated effects alter nucleosome conformations, eventually affecting gene expression and subsequent cellular processes. UV radiation, drug exposure, and viral infections have all been documented to influence meCpG patterns in T and B cells because they trigger important interactions between environment and host [13]. In general, SLE is associated with (global) CpG hypomethylation in B and T lymphocytes [14,15]. Histone modifications in immune cells from SLE patients are complex and display patterns that are tissue- and cell type-specific. Epigenetic alterations in SLE patients have been attributed to the abundance and/or activity of DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) [10]. Through their effects on chromatin conformation, DNMTs and HDACs modify DNA accessibility, controlling access by transcription factors and RNA polymerases, and thereby regulating gene expression [10]. However, the precise mechanisms by which these enzymes are specifically recruited to genes in immune cells from SLE patients remain poorly understood.
Recent evidence indicates that the nuclear protein CREMα has key functions as both an epigenetic and transcriptional regulator of cytokine expression in T lymphocytes from SLE patients. This review summarizes the most recent findings linking CREMα signaling with T lymphocyte effector functions in healthy individuals and those with SLE and other autoimmune disorders.
Molecular basis of CREM signaling
CREMα belongs to a superfamily of transcription factors that also includes other CREM homologs, such as inducible cAMP early repressor (ICER), cAMP responsive element binding proteins (CREB)-1 and -2, and the CREM/activating transcription factors (ATF)-1, -2, and -3 (Figure 1). All members share high sequence homology within their DNA binding domains (a basic leucine zipper domain) and bind to the common palindromic consensus element 5′-TGACGTCA-3′, the cAMP responsive element (CRE), or its 5′-half-site [16]. The name ‘CRE’ originates from the observation that CREB and CREM proteins are activated upon an increase of intracellular cAMP levels. The CREM/ CREB signaling cascade comprises various extracellular signals including growth factors or hormones that bind to transmembrane receptors which, in turn, drive adenylate cyclase to generate high levels of cAMP. Cyclic AMP can then promote the enzymatic properties of protein kinases, such as PKA, PKC, and casein kinases I and II that can phosphorylate and thereby activate CREB/CREM proteins [17]. Serine residue 117 of CREM is one target for activating protein kinases [18]. In the context of T cell biology, T cell receptor (TCR) activation and increased calcium influx, as induced by ionomycine, are alternative pathways that activate protein kinases that phosphorylate CREB/ CREM proteins, such as calcium/calmodulin-dependent kinases (CaMKs). Sera of SLE patients display increased CaMKIV activity, which activates CREMα and leads to transcriptional effects on the IL2 promoter [19].
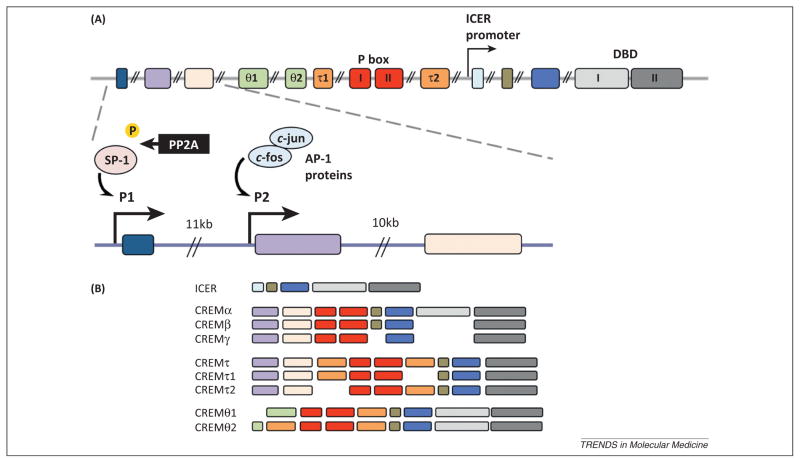
Figure 1.
(A) Schematic of the human CREM gene sequence and promoters. The upper graph displays the genomic structure of the human CREM gene comprising 14 exons. The fourth and fifth exons (light green θ1 and θ2) are preferentially expressed in testicular CREM isoforms, although the functions are not yet known. Exons six and nine (orange τ1 and τ2) are supposed to be trans-activating glutamine-rich domains. The red exons, seven and eight, encode the kinase-inducible P box domains I and II. The light and dark gray exons encode DNA binding domains (DBD) 1 and 2. Below is an amplified section of the three 5′ exons with CREM promoters P1 (in front of the first exon) and P2 (between the first and the second exon). P1 is mainly controlled by specificity protein (SP)-1 which is activated through de-phosphorylation exerted by protein phosphatase (PP)-2A, whereas P2 is trans-regulated by transcription factors of the AP-1 protein family (c-jun/c-fos homo- and heterodimers) [34,38]. (B) Schematic of selected CREM isoforms. A large number of CREM/ICER isoforms is generated by the usage of different promoters, transcription factor activities, and alternative splicing mechanisms [18,27,31].
When the crem gene was discovered in 1991 from a murine pituitary cDNA library, three different CREM isoforms were described that are generated by alternative splicing processes [20]. CREM is structurally related to CREB, which had been identified a few years earlier, and at the time of its discovery was thought to exert dominant-negative effects on CREB-mediated gene transcription [20,21]. Over time, additional CREM isoforms have been described (>20 variants in humans), with some activating and some repressing gene transcription.
Whereas CREB appears to be ubiquitously expressed, CREM expression is tightly regulated through cell-, tissue-and development-specific mechanisms. Thus, it is not surprising that CREM-mediated gene transcription has been extensively studied in cell systems that display selective expression of single CREM variants, such as male germ cells, cells of the adrenal and pituitary glands, and T lymphocytes. Table 1 summarizes CREM target genes in various cell systems; however, most of these findings were generated in vitro (see also Box 1).
Table 1.
Examples of CREM/ICER target genes
| Gene name | Regulationa | Isoform | Refs | |
|---|---|---|---|---|
| T lymphocytes | IL-2 | ⇓ | CREMα/ICER | [22,36,42,52,59] |
| IL-17A | ⇑ | CREMα | [52,44] | |
| IL-17F | ⇓ | CREMα | [51] | |
| spleen tyrosine kinase (syk) | ⇓ | CREMα | [54] | |
| CD3 ζ chain | ⇓ | CREMα | [53] | |
| fas ligand | ⇓ | ICER | [60] | |
| AP-1/c-fos | ⇓ | CREMα/ICER | [33,61] | |
| NFAT-c1 | ⇓ | ICER | [62] | |
| macrophage inflammatory protein (MIP)-1β | ⇓ | ICER | [63] | |
| Testis | proacrosin, protamine-1, transition protein-1, mitochondrial capsule selenoprotein (MCS) | ⇑ | CREMτ | [58] |
| phospholipid hydroperoxide glutathione peroxidase | ⇑ | CREMτ | [64] | |
| testis-specific angiotensin I converting enzyme | ⇑ | CREMτ | [64] | |
| transcript induced in spermiogenesis (Tisp)-40 | ⇑ | CREMτ | [65] | |
| Adrenal gland | Cyp17 | ⇑ | CREM (all)b | [66] |
| tyrosine hydroxylase (TH) | ⇓ | ICER | [67] | |
| Pineal gland | serotonin N-acetyltransferase | ⇓ | ICER | [68] |
| Liver | Cyp51 | ⇑ | ICERb | [69] |
| Heart | transcription factors dHAND and RhoB | ⇓ | CREM (all) | [70] |
⇓, repression of the indicated target gene(s); ⇑, induction/activation of the denoted target gene(s).
Results from CREM knockout (KO) mice where all CREM/ICER isoforms are not functional.
Box 1. CREM-directed gene regulation in spermatogenesis and the endocrine system.
Apart from T lymphocytes, members of the CREM protein family have also been described to play critical roles in other cell systems. The inducible and stage-dependent expression of CREM/ICER isoforms during spermatogenesis and in adrenal/pituitary cell physiology unravels mechanisms that may also be applied during T lymphocyte differentiation; however, this remains to be elucidated in future studies.
During spermatogenesis, CREM expression switches from a foremost ‘repressor isoform pattern’ in pre-meiotic germ cells to the CREMτ activator isoform in post-meiotic round spermatids. Male mice with a homozygous disruption of the crem gene are infertile due to the increased apoptosis of round spermatids emphasizing its central role in male germ cell differentiation [58,71]. CREM target genes in spermatids comprise proacrosin, various selenoproteins and testis-specific angiotensin I converting enzyme [71,72] (Table 1). A systematic ChiP-seq analysis performed in these cells demonstrated that more than 6700 gene loci are occupied by CREMτ with 80% of them being located within proximal promoter regions of annotated genes, but also in intergenic regions some of which encode for miRNAs. Further analyses revealed that most of these loci contained CRE half-sites. However, only and approximately 1% of the occupied promoters were trans-regulated through CREM proteins [69].
Circadially regulated processes, such as the synthesis of cholesterol and adrenal hormones, are under the control of CREM/ICER proteins. ICER is expressed rhythmically in the pineal gland peaking at night. It controls the expression of the serotonin N-acetyltransferase and thereby modulating the oscillatory synthesis of the hormone melatonin [66]. CREM/ICER proteins trans-activate cholesterogenic enzymes including Cyp51 and Cyp17 as we learned from studies in CREM-deficient mice [68,73]. One might speculate that in these mice particularly the absence of CREMα accounts for an epigenetic de-methylation of the Cyp17 promoter. Furthermore, ICER expression peaks rapidly upon stimulation with corticotrophin-releasing hormone in pituitary gland cells [74].
Human T lymphocytes predominantly express the CREMα isoform, and these levels are increased in T cells from SLE patients [22–24]. Until recently, CREMα was considered to act as a transcriptional repressor, but recent evidence indicates that CREMα can activate the transcription of certain cytokine genes in CD4+ T cells. Although it has been proposed that the presence of the τ domains within CREM isoforms determines their capacity as transcriptional activators [25], experiments measuring the differential effects of CREM variants with and without τ domains on the same gene promoter have yet to be performed.
Regulating CREM transcription
The human CREM gene spans 14 exons and is highly conserved throughout evolution (Figure 1A); the human and mouse CREM genes are approximately 90% homologous at the level of the coding nucleotides [26]. The multitude of CREM isoforms (>20 in humans) is achieved with differential splicing processes and the use of alternative promoters, transcription factor repertoires, and initiation codons (Figure 1B). Several stimuli, including adrenergic signals or an increase in the levels of intracellular cAMP or glucose induce the expression of the ICER isoform, which was initially thought to be the only inducible CREM isoform [27]. Transcription of ICER depends on an intronic promoter within the 3′-region of the CREM gene (Figure 1A) [28]. Transcription of the longer CREM variants, including CREMα, is orchestrated by two promoters at the 5′-end of the gene, one directly upstream of exon I (denoted P1) and a second upstream of exon II (denoted P2). It has been documented that CREMα expression is increased in T cells from SLE patients at the transcriptional and protein levels [22–24], mediated by the promoter P1 whose activity level mirrors disease severity.
SLE patients also display increased expression and enzymatic activity of protein phosphatase (PP)-2A, which specifically de-phosphorylates specificity protein (SP)-1 at serine residue 59. De-phosphorylated SP-1 binds to and trans-activates CREM P1, contributing to the enhanced basal CREMα expression in T cells from SLE patients [24,29,30]. Given that SP-1 expression is significantly enhanced in response to estrogen receptor engagement, this mechanism may contribute to the female predominance in SLE [31]. In contrast to P1, the intronic CREM promoter P2 is under the tight transcriptional control of activating protein (AP)-1 in response to T cell activation mediated through stimulation with anti-CD3/anti-CD28 or phorbol myristate acetate (PMA)/ionomycine [32]. In T cells from healthy individuals, T cell activation increases AP-1 recruitment to P2 with subsequent promoter activation and enforced CREMα transcription. Because AP-1/c-fos expression is decreased in SLE T cells (largely due to transcriptional repression by CREMα itself [33]), these cells fail to upregulate CREMα expression following T cell activation. Thus, the AP-1:CREMα axis is an important autoregulatory loop that eventually limits a further increase of CREMα expression in SLE T cells. This may be beneficial in light of the multiple deleterious effects CREMα exerts on cytokine expression in SLE pathogenesis.
In addition to the aforementioned trans-activating events, CREM promoter P1 is subject to epigenetic modifications, which regulate gene expression. CpG methylation influences the tissue-specific regulation of CREM in human T lymphocytes. Specifically, effector memory CD4+ T lymphocytes and T cells from SLE patients exhibit significantly reduced meCpG levels at CREM P1, contributing to enhanced CREMα mRNA expression [34].
Taken together, both transcriptional and epigenetic regulation of human CREM exemplifies differential gene expression between effector and naïve CD4+ T lymphocyte subsets, but also between T lymphocytes from SLE patients and controls, respectively (Figure 1A). In this context, the PP2A:SP-1 pathway and variable meCpG levels of the CREM promoter P1 appear to control basal CREM transcription, whereas promoter P2 is regulated by AP-1 proteins which account for inducible CREM expression following T cell activation.
CREM effects on cytokine expression
Until recently, CREM signaling was considered inhibitory to T cells because many T cell-relevant target genes are transcriptionally repressed by CREM or ICER (Table 1) [35]. In human T cells, the expression of both CREMα and ICER is inducible following TCR activation or calcium mobilization [32,36–38]. A growing body of literature suggests that CREMα is involved in the antithetic regulation of the cytokine genes IL17A and IL2 in both health and disease (Figure 2 and Box 2). meCpG mediates tissue-specific silencing of CREMα, which may affect T lymphocyte differentiation [34]. Naïve CD45RA+CCR7+CD4+ and CD45R0+CCR7+CD4+ ‘central memory’ T lymphocytes share subset-specific homing to secondary lymphatic tissues and have gene expression patterns that are partially similar. Naïve and central memory CD4+ T cells express high levels of IL-2, but relatively low amounts of IL-17A in response to TCR stimulation. By contrast, CD45R0+CCR7−CD4+ ‘effector memory’ T lymphocytes are primarily detected in the peripheral blood, express perforin and effector cytokines (including IFN-γ, IL-4, and IL-17A) in response to TCR stimulation, but fail to produce IL-2 [34,39]. Recently, tissue-specific CREMα expression patterns have been documented in naïve and memory CD4+ T lymphocytes. CREMα is increased in cells that express IL-17A and fail to express IL-2 (‘effector memory’ CD4+ T cells), leading us to conclude that CREMα contributes to subset-specific CpG methylation of the IL2 and IL17A promoters, determining the amount of gene expression, and thus contributing to CD4+ T cell subset distribution [34].
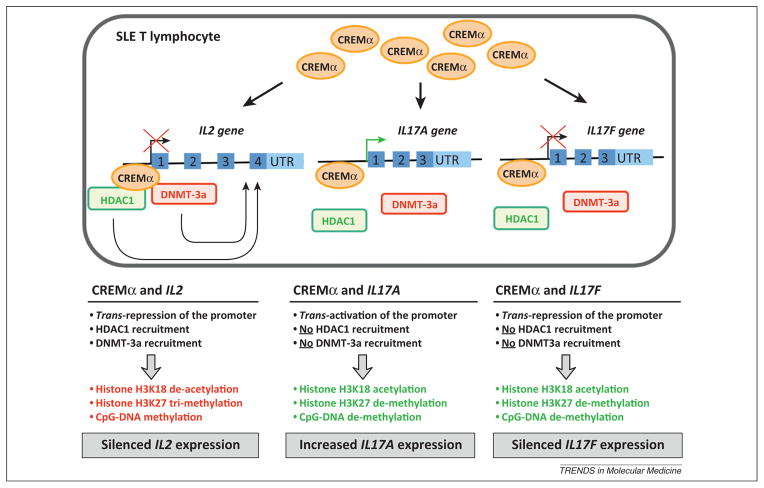
Figure 2.
The effects of CREMα on IL2 and IL17 gene transcription. Abbreviations: HDAC1, histone deacetylase 1; DNMT-3a, DNA methyl transferase-3a; CpG, cytosine-phosphate-guanosine.
Box 2. CREM regulates cytokine production.
The CREM/ICER family of transcription factors strongly impact transcriptional and epigenetic regulation of multiple target genes.
Under physiological conditions, the production of CREM and ICER are tightly regulated and involve the differential use of alternate promoters and splicing processes, resulting in cell- and tissue-specific expression patterns.
CREMα mRNA and protein expression is increased in T cells from SLE patients and significantly alters the expression of various T lymphocyte-specific target genes, including IL-2 and IL-17 family cytokines.
CREM constitutes a key factor in the pathogenesis of SLE.
CREMα and its interacting CpG- and histone-modifying enzymes may be promising targets in the search for disease biomarkers and novel treatment options.
Dysregulation of cytokine expression, especially of IL-2 and IL-17, is a hallmark of misguided immunity in SLE patients. T cells from SLE patients fail to express IL-2, an effect caused by multiple mechanisms. The first mechanism reported to affect IL-2 expression in SLE T lymphocytes is trans-repression at the IL2 gene promoter. CREMα and the activating transcriptional regulatory factor CREB share a cis-regulatory element 180 base pairs upstream of the transcriptional initiation site of IL2 (a site denoted −180CRE). Under physiological conditions, the −180CRE site is constitutively occupied by CREB. Upon T cell activation CREB undergoes phosphorylation, which further increases CREB binding to this site. In T lymphocytes from SLE patients, CREMα replaces CREB, and trans-represses IL2 gene transcription [22,40]. In addition to this trans-repression, CREMα recruits HDAC1 and DNMT-3a to the IL2 promoter, mediating epigenetic remodeling through histone deacetylation and de novo CpG methylation, respectively [41,42]. Similar to CREB, CREMα can bind histone acetyltransferase p300, mediating its recruitment to the IL2 promoter. However, unlike CREB, CREMα lacks a trans-activating domain and fails to activate p300 [41,43]. This contributes to reduced histone acetylation and condensation of the IL2 promoter.
In addition to the IL2 gene itself, CREMα influences gene expression of factors that regulate IL-2 expression. CREMα binds to and represses the promoter of the proto-oncogene c-fos in T cells from SLE patients, resulting in reduced activity of the transcription factor AP-1, which, under physiological conditions, activates IL2 gene transcription [33].
Furthermore, CREMα affects epigenetic modifications and transcriptional control of cytokines from the IL-17 family, namely IL17A and IL17F [32,44]. Both gene products, IL-17A and IL-17F, are strongly proinflammatory and are produced by T lymphocytes, natural killer cells, mast cells, and neutrophils [45]. Excessive IL-17 expression has been linked to several autoimmune diseases, including rheumatoid arthritis, multiple sclerosis, inflammatory bowel disease, and SLE [46–51]. The human IL17A and IL17F genes are located in close proximity to each other on chromosome 6 (within approximately 85 kb). Although the biological properties and regulation of IL-17F are less well studied than IL-17A, some information is available. It was recently demonstrated that increased CREMα expression in SLE T cells contributes to epigenetic remodeling of the IL17A/F gene locus. In SLE T cells recruitment of CREMα to the IL17A and the IL17F promoters is increased, as compared with T cells from healthy individuals. In contrast to the effects observed at the IL2 gene locus, CREMα recruits less HDAC1 and DNMT3a to the IL17A promoter, contributing to an overall increase in histone H3 acetylation and reduced histone H3 methylation, as well as CpG hypomethylation of the IL17 locus. Indeed, CREMα overexpression in human T cells increases histone H3 acetylation while it decreases histone H3 methylation and meCpG levels over the entire IL17 gene locus in a yet-to-be determined manner (T. Rauen and C.M. Hedrich, unpublished and [34]). This is in agreement with the finding that in addition to its transcriptional effects, the activating isoform CREMτ mediates increases in histone H3 acetylation in male germ cells [50]. Furthermore, forced CREMα expression in human T lymphocytes results in CpG de-methylation of the IL17A promoter, mimicking the situation in T lymphocytes from SLE patients, which display almost complete de-methylation of the IL17A proximal promoter [44,51]. Thus, CREMα regulates the remodeling of the IL17 gene into an open, accessible state, which allows additional transcription factors to bind. The exact mechanisms by which CREMα mediate histone acetylation in this region while mediating diametric histone modification around the IL2 gene remains to be elucidated. However, the ‘transcription factor environment’ of the two promoters may affect this process. At the transcriptional level, CREMα mediates differential effects on the IL17A (trans-activation) and the IL17F (trans-repression) promoters, which ultimately results in increased IL17A and decreased IL17F mRNA expression and protein levels in SLE patients. Although CREMα confers an activating, open pattern to the IL17F gene in SLE T cells, its repressive functions on the IL17F promoter prevail and dictate the ultimately decreased IL-17F production in these cells. The imbalanced IL-17A/IL-17F ratio in activated T cells, which may largely be attributed to the CREMα effects described above, appears to be specific for SLE T cells because T cells from healthy individuals and patients with other autoimmune diseases do not necessarily upregulate IL-17A and downregulate IL-17F production upon activation [46]. Notably, the observed effects of CREMα on IL-2 and IL-17A cytokine production in humans are also observed in transgenic mice with T cell-specific CREMα overexpression (under control of the cd2 promoter). These mice have decreased IL-2 and increased IL-17A levels and are more prone to develop signs of autoimmunity (including lymphadenopathy, elevated IL-17 production, and higher autoantibody titers against double-stranded DNA) when an additional genetic deletion of the cd95 gene is present [52]. These data support the idea of CREMα as an independent promoter of the ‘autoimmune phenotype’.
It has also been demonstrated that the TCR–CD3 complex that recognizes (auto-)antigens and mediates T cell activation is ‘rewired’ in SLE T lymphocytes, contributing to an amplified T cell activation in SLE [53]. Evidence suggests the involvement of CREMα in the ‘skewed’ TCR–CD3 composition in SLE T cells, which sees replacement of the normal TCR ζ chain by the ‘common’ FcRγ chain. CREMα trans-represses the TCR ζ chain promoter and mediates chromatin remodeling through histone deacetylation, contributing to defective ζ chain expression and its replacement by the common FcRγ chain in the TCR–CD3 complex. This has tremendous significance for the subsequent signaling events because FcRγ-mediated downstream signaling is far more potent than that mediated through the ζ chain.
It is well documented that T cells from SLE patients express increased levels of tyrosine kinase (Syk) which contribute to aberrant T cell signaling in these individuals. In vitro findings suggest CREMα as a strong repressor of Syk gene transcription that is caused by the recruitment to a CRE within the Syk promoter. At first, this might appear contradictory in the context of the classic ‘SLE T cell phenotype’. Yet, the Syk promoter exhibits reduced histone H3 acetylation in SLE T cells, translating into reduced accessibility for the transcriptional repressor CREMα [54].
Characteristics of CREM signaling in antigen presenting cells (APCs)
Recently, the involvement of CREB/ATF superfamily members in regulating APC functions has been demonstrated. Gilchrist et al. linked Toll-like receptor (TLR)-4 induced ATF-3 expression with transcriptional repression of Il6 and Il12b and concluded that ATF-3 is part of a physiological feedback loop [55]. A disruption of the tight balance between activating and repressing CREB/ATF family members may further contribute to autoimmune pathology. Indeed, CREMα negatively regulates mRNA and protein expression of the co-stimulatory molecule CD86 on bone marrow-derived DCs [56]. CD86 is expressed on the surface of APCs and provides activation and survival signals to T lymphocytes through engagement with CD28 and CTLA-4. CREMα binds to a CRE within the CD86 promoter, resulting in trans-repression and reduced histone acetylation. CREMα thus may mediate epigenetic remodeling of CD86 through the same mechanisms (HDAC1 recruitment and reduced p300 activation) as the aforementioned IL2 promoter. In turn, CREMα deficiency results in increased CD86 expression on DCs, which subsequently enforces co-stimulation to T lymphocytes. This probably accounts for the enhanced T lymphocyte proliferation in vitro and aggravated contact dermatitis in mice transferred with DCs from CREMα-deficient mice.
These findings extend CREMα effects to yet another immune cell subset, myeloid DCs, which express reduced levels of CD86 that is caused by trans-repression through CREMα. This in turn decreases the ability of these cells to transduce co-stimulatory signals towards T lymphocytes. In line with the trans-repressive effects of CREMα on IL-2 and TCR ζ chain production, this constitutes another (indirect) mechanism by which CREMα severely impairs T cell survival and function. It remains to be elucidated whether this mechanism plays a role in the pathogenesis of SLE, because lupus patients display increased CD86 levels on myeloid DCs [57]. Furthermore, it is yet unknown whether the expression of CREM itself is deregulated in myeloid DCs from lupus patients.
Concluding remarks and future perspectives
CREMα has been established as a central contributor to tissue-specific gene regulation in immune cells. Secondary to increased expression of CREMα in T cells from SLE patients, CREMα harbors the potential of being utilized as a disease biomarker and/or therapeutic target. However, several central questions remain to be answered in future studies, many of which are outlined below.
Modifying CREM expression in wild type or lupus-prone mice (e.g., B6.lpr and MRL.lpr mice) will provide a more global understanding of the involvement of CREMα and its isoforms in vivo. CREM-deficient mice have already unraveled the involvement of CREM during spermatogenesis [58]. CREM-deficient mice and transgenic approaches, such as overexpressing CREMα in T lymphocytes, will help to decipher whether CREMα plays a role during the priming and differentiation of immune cells in health and autoimmunity. Targeting CREMα with small interfering RNAs (siRNAs) in vivo will also help to assess its therapeutic potential in SLE.
It has become clear that CREMα orchestrates tissue-specific expression of the IL2 and IL17A genes, CREMα contributes to the silencing of IL2 through trans-repression and tissue- and region-specific recruitment of specific DNA and histone methyltransferases or HDACs. Conversely, CREMα trans-activates IL17A and mediates epigenetic remodeling with reduced meCpG and increased histone H3 acetylation in a yet-to-be determined manner [34,42,44]. At present, the molecular mechanisms directing these functional differences remain to be determined. Tissue- and region-specific differences could be mediated by blocking DNA methylation of the daughter-strand during cell division and subsequent translation into the histone code or through the tissue-specific interaction with DNA de-methylases and histone acetyltransferases. Those functional differences could be directed by the proximity to further transcription factors allowing or prohibiting the recruitment of epigenetic activators or repressors.
Furthermore, CREMα has been demonstrated to contribute to antithetic expression patterns in naïve and effector T lymphocytes through epigenetic remodeling processes [34]. However, it remains to be determined whether CREMα plays a role during the differentiation towards Th17 subset or whether CREMα rather plays a role in the tissue-specific expression of IL-17 and the silencing of IL-2 in non-naïve T lymphocyte subsets. This question will need to be addressed in genetically modified systems.
We previously demonstrated that the expression of CREMα is regulated by several mechanisms, including differential trans-activation of the CREM promoters and/or epigenetic remodeling of the promoter P1 [24,32,34]. However, the molecular mechanisms that mediate CpG-DNA de-methylation in effector T cells and T lymphocytes from SLE patients remain to be determined. Furthermore, the alternative splicing mechanisms that favor the production of CREMα over other CREM isoforms will be the focus of future research.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Tan W, et al. Association of PPP2CA polymorphisms with systemic lupus erythematosus susceptibility in multiple ethnic groups. Arthritis Rheum. 2011;63:2755–2763. doi: 10.1002/art.30452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grammatikos AP, Tsokos GC. Immunodeficiency and autoimmunity: lessons from systemic lupus erythematosus. Trends Mol Med. 2012;18:101–108. doi: 10.1016/j.molmed.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramantani G, et al. Expanding the phenotypic spectrum of lupus erythematosus in Aicardi–Goutieres syndrome. Arthritis Rheum. 2010;62:1469–1477. doi: 10.1002/art.27367. [DOI] [PubMed] [Google Scholar]
- 5.Moser KL, et al. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kavanagh D, et al. New roles for the major human 3′-5′ exonuclease TREX1 in human disease. Cell Cycle. 2008;7:1718–1725. doi: 10.4161/cc.7.12.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truedsson L, et al. Complement deficiencies and systemic lupus erythematosus. Autoimmunity. 2007;40:560–566. doi: 10.1080/08916930701510673. [DOI] [PubMed] [Google Scholar]
- 8.Ramos PS, et al. Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 2011;63:2049–2057. doi: 10.1002/art.30356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos PS, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011;7:e1002406. doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao M, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun. 2010;35:58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, et al. Histone modifications and methyl-CpG binding domain protein levels at the TNFSF7 (CD70) promoter in SLE CD4+ T cells. Lupus. 2011;20:1365–1371. doi: 10.1177/0961203311413412. [DOI] [PubMed] [Google Scholar]
- 13.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. epigenetics in autoimmunity – DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41:278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J Med. 2011;60:10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 15.Sunahori K, et al. Promoter hypomethylation results in increased expression of protein phosphatase 2A in T cells from patients with systemic lupus erythematosus. J Immunol. 2011;186:4508–4517. doi: 10.4049/jimmunol.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barcellos LF, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5:e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monaco L, et al. Specialized rules of gene transcription in male germ cells: the CREM paradigm. Int J Androl. 2004;27:322–327. doi: 10.1111/j.1365-2605.2004.00494.x. [DOI] [PubMed] [Google Scholar]
- 18.de Groot RP, et al. Multiple and cooperative phosphorylation events regulate the CREM activator function. EMBO J. 1993;12:3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juang YT, et al. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J Clin Invest. 2005;115:996–1005. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foulkes NS, et al. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 21.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 22.Solomou EE, et al. Molecular basis of deficient IL-2 production in T cells from patients with systemic lupus erythematosus. J Immunol. 2001;166:4216–4222. doi: 10.4049/jimmunol.166.6.4216. [DOI] [PubMed] [Google Scholar]
- 23.Kyttaris VC, et al. CAMP response element modulator α expression in patients with systemic lupus erythematosus. Lupus. 2006;15:840–844. doi: 10.1177/0961203306069985. [DOI] [PubMed] [Google Scholar]
- 24.Juang YT, et al. Transcriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. J Biol Chem. 2011;286:1795–1801. doi: 10.1074/jbc.M110.166785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulkes NS, et al. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 26.Masquilier D, et al. Human CREM gene: evolutionary conservation, chromosomal localization, and inducibility of the transcript. Cell Growth Differ. 1993;4:931–937. [PubMed] [Google Scholar]
- 27.Higai K, et al. Prolonged high glucose suppresses phorbol 12-myristate 13-acetate and ionomycin-induced interleukin-2 mRNA expression in Jurkat cells. Biochim Biophys Acta. 2009;1790:8–15. doi: 10.1016/j.bbagen.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Molina CA, et al. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 29.Vicart A, et al. Increased chromatin association of Sp1 in interphase cells by PP2A-mediated dephosphorylations. J Mol Biol. 2006;364:897–908. doi: 10.1016/j.jmb.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 30.Katsiari CG, et al. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest. 2005;115:3193–3204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulton VR, et al. Estrogen upregulates cyclic AMP response element modulator α expression and downregulates interleukin-2 production by human T lymphocytes. Mol Med. 2012;18:370–378. doi: 10.2119/molmed.2011.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauen T, et al. A novel intronic cAMP response element modulator (CREM) promoter is regulated by activator protein-1 (AP-1) and accounts for altered activation-induced CREM expression in T cells from patients with systemic lupus erythematosus. J Biol Chem. 2011;286:32366–32372. doi: 10.1074/jbc.M111.245811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyttaris VC, et al. Cyclic adenosine 5′-monophosphate response element modulator is responsible for the decreased expression of c-fos and activator protein-1 binding in T cells from patients with systemic lupus erythematosus. J Immunol. 2004;173:3557–3563. doi: 10.4049/jimmunol.173.5.3557. [DOI] [PubMed] [Google Scholar]
- 34.Hedrich CM, et al. cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc Natl Acad Sci USA. 2012;109:16606–16611. doi: 10.1073/pnas.1210129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broecker S, et al. Combined use of liquid chromatography-hybrid quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) and high performance liquid chromatography with photodiode array detector (HPLC-DAD) in systematic toxicological analysis. Forensic Sci Int. 2011;212:215–226. doi: 10.1016/j.forsciint.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273:9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 37.Bodor J, et al. Suppression of T cell function: a potential role for transcriptional repressor ICER. J Leukoc Biol. 2000;67:774–779. doi: 10.1002/jlb.67.6.774. [DOI] [PubMed] [Google Scholar]
- 38.Korf HW, et al. Signal transduction molecules in the rat pineal organ: Ca2+, pCREB, and ICER. Naturwissenschaften. 1996;83:535–543. doi: 10.1007/BF01141978. [DOI] [PubMed] [Google Scholar]
- 39.Sallusto F, et al. Switch in chemokine receptor expression upon TCR stimulation reveals novel homing potential for recently activated T cells. Eur J Immunol. 1999;29:2037–2045. doi: 10.1002/(SICI)1521-4141(199906)29:06<2037::AID-IMMU2037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 40.Tenbrock K, et al. Antisense cyclic adenosine 5′-monophosphate response element modulator up-regulates IL-2 in T cells from patients with systemic lupus erythematosus. J Immunol. 2002;169:4147–4152. doi: 10.4049/jimmunol.169.8.4147. [DOI] [PubMed] [Google Scholar]
- 41.Tenbrock K, et al. The cyclic adenosine 5′-monophosphate response element modulator suppresses IL-2 production in stimulated T cells by a chromatin-dependent mechanism. J Immunol. 2003;170:2971–2976. doi: 10.4049/jimmunol.170.6.2971. [DOI] [PubMed] [Google Scholar]
- 42.Hedrich CM, et al. cAMP-responsive element modulator (CREM)α protein signaling mediates epigenetic remodeling of the human interleukin-2 gene: implications in systemic lupus erythematosus. J Biol Chem. 2011;286:43429–43436. doi: 10.1074/jbc.M111.299339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asahara H, et al. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol Cell Biol. 2001;21:7892–7900. doi: 10.1128/MCB.21.23.7892-7900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rauen T, et al. cAMP-responsive element modulator (CREM)α protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J Biol Chem. 2011;286:43437–43446. doi: 10.1074/jbc.M111.299313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaffen SL. Recent advances in the IL-17 cytokine family. Curr Opin Immunol. 2011;23:613–619. doi: 10.1016/j.coi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nistala K, Wedderburn LR. Th17 and regulatory T cells: rebalancing pro- and anti-inflammatory forces in autoimmune arthritis. Rheumatology. 2009;48:602–606. doi: 10.1093/rheumatology/kep028. [DOI] [PubMed] [Google Scholar]
- 47.Zepp J, et al. IL-17 receptor signaling and T helper 17-mediated autoimmune demyelinating disease. Trends Immunol. 2011;32:232–239. doi: 10.1016/j.it.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hundorfean G, et al. Functional relevance of T helper 17 (Th17) cells and the IL-17 cytokine family in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:180–186. doi: 10.1002/ibd.21677. [DOI] [PubMed] [Google Scholar]
- 49.Crispin JC, Tsokos GC. IL-17 in systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:943254. doi: 10.1155/2010/943254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajkovic M, et al. Functional cooperation between CREM and GCNF directs gene expression in haploid male germ cells. Nucleic Acids Res. 2010;38:2268–2278. doi: 10.1093/nar/gkp1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hedrich CM, et al. cAMP-responsive element modulator α (CREMα) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE) J Biol Chem. 2012;287:4715–4725. doi: 10.1074/jbc.M111.323261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lippe R, et al. CREMα overexpression decreases IL-2 production, induces a TH17 phenotype and accelerates autoimmunity. J Mol Cell Biol. 2012;4:121–123. doi: 10.1093/jmcb/mjs004. [DOI] [PubMed] [Google Scholar]
- 53.Tenbrock K, et al. The cyclic AMP response element modulator regulates transcription of the TCR ζ-chain. J Immunol. 2005;175:5975–5980. doi: 10.4049/jimmunol.175.9.5975. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh D, et al. CREMα suppresses spleen tyrosine kinase expression in normal but not systemic lupus erythematosus T cells. Arthritis Rheum. 2012;64:799–807. doi: 10.1002/art.33375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 56.Ahlmann M, et al. The cyclic AMP response element modulator α suppresses CD86 expression and APC function. J Immunol. 2009;182:4167–4174. doi: 10.4049/jimmunol.0802976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerl V, et al. Blood dendritic cells in systemic lupus erythematosus exhibit altered activation state and chemokine receptor function. Ann Rheum Dis. 2010;69:1370–1377. doi: 10.1136/ard.2009.111021. [DOI] [PubMed] [Google Scholar]
- 58.Nantel F, et al. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 59.Tenbrock K, Tsokos GC. Transcriptional regulation of interleukin 2 in SLE T cells. Int Rev Immunol. 2004;23:333–345. doi: 10.1080/08830180490452558. [DOI] [PubMed] [Google Scholar]
- 60.Bodor J, et al. Differential inducibility of the transcriptional repressor ICER and its role in modulation of Fas ligand expression in T and NK lymphocytes. Eur J Immunol. 2002;32:203–212. doi: 10.1002/1521-4141(200201)32:1<203::AID-IMMU203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 61.Misund K, et al. Inducible cAMP early repressor splice variants ICER I and IIγ both repress transcription of c-fos and chromogranin A. J Cell Biochem. 2007;101:1532–1544. doi: 10.1002/jcb.21267. [DOI] [PubMed] [Google Scholar]
- 62.Vaeth M, et al. Regulatory T cells facilitate the nuclear accumulation of inducible cAMP early repressor (ICER) and suppress nuclear factor of activated T cell c1 (NFATc1) Proc Natl Acad Sci USA. 2011;108:2480–2485. doi: 10.1073/pnas.1009463108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barabitskaja O, et al. Suppression of MIP-1β transcription in human T cells is regulated by inducible cAMP early repressor (ICER) J Leukoc Biol. 2006;79:378–387. doi: 10.1189/jlb.0505255. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, et al. cAMP-response element modulator τ is a positive regulator of testis angiotensin converting enzyme transcription. Proc Natl Acad Sci USA. 1996;93:12262–12266. doi: 10.1073/pnas.93.22.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagamori I, et al. Transcription factors, cAMP-responsive element modulator (CREM) and Tisp40, act in concert in postmeiotic transcriptional regulation. J Biol Chem. 2006;281:15073–15081. doi: 10.1074/jbc.M602051200. [DOI] [PubMed] [Google Scholar]
- 66.Foulkes NS, et al. Rhythmic transcription: the molecular basis of circadian melatonin synthesis. Biol Cell. 1997;89:487–494. doi: 10.1016/s0248-4900(98)80004-x. [DOI] [PubMed] [Google Scholar]
- 67.Tinti C, et al. Inducible cAMP early repressor can modulate tyrosine hydroxylase gene expression after stimulation of cAMP synthesis. J Biol Chem. 1996;271:25375–25381. doi: 10.1074/jbc.271.41.25375. [DOI] [PubMed] [Google Scholar]
- 68.Acimovic J, et al. CREM modulates the circadian expression of CYP51, HMGCR and cholesterogenesis in the liver. Biochem Biophys Res Commun. 2008;376:206–210. doi: 10.1016/j.bbrc.2008.08.126. [DOI] [PubMed] [Google Scholar]
- 69.Martianov I, et al. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics. 2010;11:530. doi: 10.1186/1471-2164-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller FU, et al. Heart-directed expression of a human cardiac isoform of cAMP-response element modulator in transgenic mice. J Biol Chem. 2005;280:6906–6914. doi: 10.1074/jbc.M407864200. [DOI] [PubMed] [Google Scholar]
- 71.Blendy JA, et al. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 72.Tramer F, et al. cAMP-response element modulator-τ activates a distinct promoter element for the expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Biochem J. 2004;383:179–185. doi: 10.1042/BJ20040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kosir R, et al. Circadian expression of steroidogenic cytochromes P450 in the mouse adrenal gland – involvement of cAMP-responsive element modulator in epigenetic regulation of Cyp17a1. FEBS J. 2012;279:1584–1593. doi: 10.1111/j.1742-4658.2011.08317.x. [DOI] [PubMed] [Google Scholar]
- 74.Della Fazia MA, et al. Stress-induced expression of transcriptional repressor ICER in the adrenal gland. FEBS Lett. 1998;434:33–36. doi: 10.1016/s0014-5793(98)00944-2. [DOI] [PubMed] [Google Scholar]